Abstract
Montane environments around the globe are biodiversity ‘hotspots’ and important reservoirs of genetic diversity. Montane species are also typically more vulnerable to environmental change than their low-elevation counterparts due to restricted ranges and dispersal limitations. Here we focus on two abundant congeneric mayflies (Baetis bicaudatus and B. tricaudatus) from montane streams over an elevation gradient spanning 1400 m. Using single-nucleotide polymorphism genotypes, we measured population diversity and vulnerability in these two species by: (i) describing genetic diversity and population structure across elevation gradients to identify mechanisms underlying diversification; (ii) performing spatially explicit landscape analyses to identify environmental drivers of differentiation; and (iii) identifying outlier loci hypothesized to underlie adaptive divergence. Differences in the extent of population structure in these species were evident depending upon their position along the elevation gradient. Heterozygosity, effective population sizes and gene flow all declined with increasing elevation, resulting in substantial population structure in the higher elevation species (B. bicaudatus). At lower elevations, populations of both species are more genetically similar, indicating ongoing gene flow. Isolation by distance was detected at lower elevations only, whereas landscape barriers better predicted genetic distance at higher elevations. At higher elevations, dispersal was restricted due to landscape effects, resulting in greater population isolation. Our results demonstrate differentiation over small spatial scales along an elevation gradient, and highlight the importance of preserving genetic diversity in more isolated high-elevation populations.
Introduction
Dispersal is a vital process driving patterns of demographic and genetic connectivity across landscapes (Baguette et al., 2013). The scale and extent of connectivity among populations are key factors determining a species’ vulnerability to environmental change (Thomas et al., 2004; Bálint et al., 2011), and they are determined primarily by the dispersal ability of the organism and the distribution of suitable habitat across the landscape. Dispersal promotes resilience, as organisms are free to move away from unsuitable sites in search of favorable conditions, and exchange of alleles maintains genetic diversity across the landscape.
For species inhabiting montane regions, elevation gradients have an important role in shaping dispersal and population substructure due to isolation of high-elevation sites, local adaptation to extreme environments, or both (Hughes et al., 2009). Knowing how genetic diversity is distributed across elevation gradients is important for our understanding of population dynamics in alpine systems, and it has implications for conservation of montane diversity (Clarke et al., 2008; Múrria et al., 2013). Recent studies of intra- and inter-specific diversity in montane streams highlight both the importance of high-elevation sites as reservoirs of diversity and their vulnerability (Hughes, 2007; Finn et al., 2011; Vuataz et al., 2016). Loss of isolated populations at high elevations could result in disproportionate loss of genetic diversity and perhaps more importantly, loss of locally adapted genotypes (Schiffers et al., 2013). In fact, a growing body of theory predicts that diversification and adaptation may be accelerated at the edges of a species’ elevational range (Halbritter et al., 2015), and numerous studies provide examples of both isolation and differentiation with increasing elevation (Funk et al., 2005; Hodkinson, 2005).
The distribution of habitats in a landscape can constrain dispersal to generate population structure in three primary ways—isolation by distance (IBD), isolation by resistance (IBR) or isolation by environment (IBE). When habitat is available over broad geographic scales and distance is the primary constraint on dispersal, a pattern of IBD arises, which is characterized by increasing genetic distance at larger geographic scales. An alternative to this model is IBR, which predicts that intervening barriers to dispersal, such as steep mountains or human development, restrict connectivity. These landscape features divide populations, and genetic drift in populations on opposite sides of the barrier gradually leads to differentiation (Cushman et al., 2006). The concept of IBR has been developed to account for complex landscapes where portions of a heterogeneous habitat matrix may inhibit but not completely restrict gene flow (Shah and McRae, 2008). Finally, IBE applies when habitats are distributed over environmental gradients (for example, elevation change along a mountainside), such that populations can experience variable selective pressures and adaptation can drive differentiation across the gradient, leading to patterns in which genetic and environmental distances are positively correlated, independent of geographic distance or intervening landscape resistance (Wang and Bradburd, 2014). Although the model of IBE is not mutually exclusive from the other models, it is unique because natural selection, rather than restricted gene flow and genetic drift, is the evolutionary force driving population divergence.
Aquatic insects such as Baetis bicaudatus and B. tricaudatus are primary consumers in stream ecosystems and have a vital role in stream communities. Mayflies in general are considered indicators of habitat quality because of their sensitivity to environmental change (Bauernfeind and Moog, 2000). Dispersal varies significantly among mayfly species but typically is accomplished by both larval drift (downstream movement) and adult flight (Bilton et al., 2001; Gattolliat, 2004; Rutschmann et al., 2014). Species-specific traits, such as adult longevity, wing structure and breeding habitat are important predictors of variation in dispersal capacity, thus influencing genetic structure (Brittain, 1982; Hughes et al., 2009; Alp et al., 2012; Paz‐Vinas et al., 2015). In species with flight, connectivity can be maintained over broad geographic scales, and is not constrained by the structure of stream networks (Bunn and Hughes, 1997; Hughes et al., 2003). However, in mayflies, dispersal seems to be more frequent in species that spend larval stages in standing water, whereas montane stream species typically show higher site fidelity (Monaghan et al., 2005).
We used genome-wide single-nucleotide polymorphism (SNP) markers to test the overarching hypothesis that elevation gradients in the Colorado Rockies affect levels of gene flow and population structure in these two montane mayfly species. We focused on two species that have different elevational ranges, to compare population structure and gene flow along an entire elevation gradient. Our objectives were threefold. First, we assessed genetic diversity and divergence among populations along the elevation gradient, to identify the mechanisms underlying population diversification. Second, we applied novel landscape genomic approaches to select among models of isolation (IBD, IBR and IBE) and identify landscape features contributing to genetic differentiation. Third, we tested for the potential role of local adaptation at high elevations by identifying outlier loci in our genome-wide SNP data and their correlation with specific environmental variables. Our results provide insights into patterns of dispersal and connectivity in montane stream biota distributed over elevation gradients, identify landscape features and investigate the potential for local adaptation as a promoter of population divergence. More broadly, integrative landscape genomic studies contribute to our understanding of how montane regions maintain genetic diversity within species, promote diversification and, in turn, increase species vulnerability to extinction.
Methods
Target species
The species B. tricaudatus and B. bicaudatus are both found in swiftly moving high-elevation streams of the Colorado Rockies (Dodds, 1923; Ward and Kondratieff, 1992). Baetis species spend the majority of their lifecycle in an aquatic larval form, and thus fitness is sensitive to water temperature and hydrological disturbances. B. tricaudatus is found throughout North America, whereas the range of B. bicaudatus is restricted to the western United States and Canada (Morihara and McCafferty, 1979; McCafferty et al., 2012). Although morphologically distinguishable, the two species hybridize in areas where they overlap (Dodds, 1923; Supplementary File 1), and at least B. tricaudatus may currently include cryptic species (Gill et al., 2014). In Colorado, we collected B. tricaudatus from lower-elevation foothills at ~1500–2900 m and B. bicaudatus from 2300 to 3400 m. The elevational ranges of both species overlapped from ~2300 to 2900 m.
Empirical data on the flight abilities of our focal species are not available. We expect that dispersal potential should be comparable due to their similarities in terms of life history and morphological traits (Edmunds et al., 1976; Morihara and McCafferty, 1979); however, variation in dispersal ability can be driven by many factors including timing, habitat conditions and biotic interactions (Bilton et al., 2001). Thus, the use of genetic methods as applied here provides a useful means to assess realized dispersal in natural populations.
Sample collection
Insect collections were performed as described by Gill et al. (2014). Samples were collected from small (2nd–3rd order), minimally disturbed tributaries to main-stem rivers from three adjacent drainages along the eastern slope of the Colorado Rockies (Figure 1)—the Cache la Poudre, Big Thompson and St. Vrain. Sampled elevations ranged from 1500 to 3400 m, with ~200 m intervals between adjacent tributaries. Collection of samples from similarly sized low-order tributaries rather than main-stem rivers at all elevations ensures that sample elevation is not confounded with stream size. At all sites, aquatic larvae were collected along a 100 m reach using Surber samplers and kicknets with 500 μm mesh and by searching under-submerged stones. A target sample size of n⩾9 was required to include a site in all of our analyses. This target was reached in all 17 sites for B. bicaudatus, but was not reached in 4 out of 12 sites for B. tricaudatus (Figure 2). All specimens were preserved in 100% ethanol, which was replaced after 24 h to ensure long-term preservation. Individual insects were morphologically sorted in the laboratory to species level as either B. bicaudatus or B. tricaudatus as in Gill et al. (2014), and reference images were collected using the cellSens digital imaging software (Olympus, Center Valley, PA, USA).
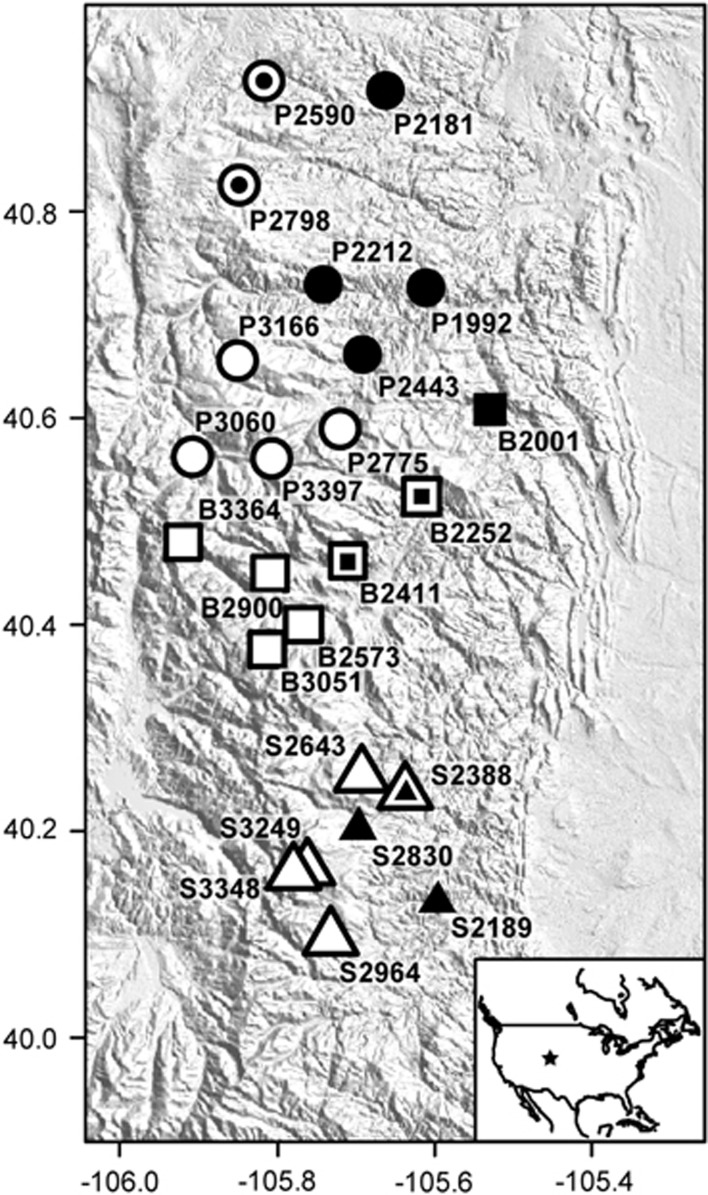
Figure 1.
Sampling locations for B. tricaudatus (black), B. bicaudatus (white) or both species (black center and white outline). Sampling spanned three drainages in the Colorado Rockies (represented by different shapes, circles: Poudre [P], squares: Big Thompson [B], triangles: St. Vrain [S]). Site IDs indicate drainage (P, B or S) followed by elevation in meters. The star in the inset figure shows the sampling location in North America. Axes represent longitude and latitude, and the map is oriented with north at the top.
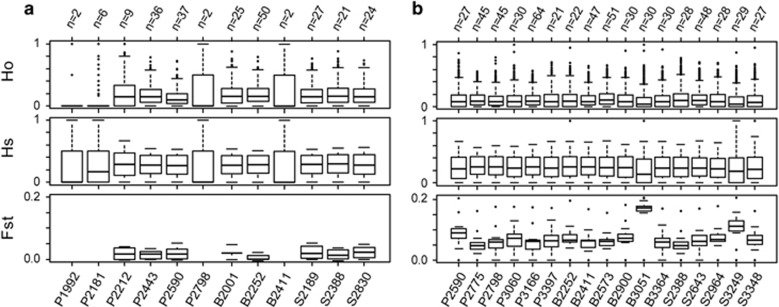
Figure 2.
Sample sizes and genetic diversity estimates by site for (a) B. tricaudatus and (b) B. bicaudatus. Site IDs indicate drainage (P, B or S) followed by elevation in meters. Plots of observed heterozygosity (Ho) and gene diversity (Hs) represent the distribution of heterozygosity for each site. Plots of pairwise FST represent the distribution of pairwise relationships for a given site with all other sites. Pairwise FST values were not computed for sites with low sample numbers (n⩽8). In general, B. bicaudatus populations show reduced genetic diversity and elevated pairwise FST, reflecting the combined effects of isolation, drift and local adaptation.
DNA Extraction and ddRAD Library Sequencing
We isolated total genomic DNA from each insect using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA). Double-digest restriction site-associated DNA (ddRAD) sequence libraries were generated following methods in Peterson et al. (2012). Each library included 288 uniquely barcoded/indexed individuals, an appropriate level of multiplexing given the small genome size of mayflies (~700 Mb; Gregory, 2002). Digestion and adapter ligation steps were combined, and comprised of 100 ng genomic DNA (10 μl at 10 ng μl−1), 15 units of both SbfI-HF and MspI (NEB, Ipswich, MA; 0.75 μl each), 300 units of T4 DNA ligase (NEB, 0.75 μl), 3 μl of 10 × CutSmart ligase buffer (NEB), 1.0 μl each of a uniquely barcoded SbfI-P1 adapter (one of 24) and MspI-P2 adapter (Integrated DNA Technologies, Coralville, IA, USA; 0.25 μM each), 3 μl of 10 mM ATP (NEB) and 9.75 μl of ddH2O for a total reaction volume of 30 μl. The restriction-ligation mixture was incubated at 37 °C for 30 min, and 20 °C for 1 h, before pooling 20 μl from each of the 24 barcoded samples. Each pool was PCR-amplified using 20 ng P1/P2 adapted DNA template (10 μl at 2 ng μl−1), 12.5 μl 2X Phusion PCR Master Mix (2X, NEB) and 1.25 μl each of forward index primer and a uniquely indexed reverse primer (one of 12; 5 μM each, Integrated DNA Technologies). PCRs were pooled across index groups and size-selected to retain fragments from 300 to 1000 bp. Each library was single-end sequenced to 100 bp on one lane of the Illumina HI-SEQ 2500 at the Cornell University Genomics Facility.
Data processing and SNP calling
Sequence processing, alignment and SNP calling were performed with Stacks v 1.19 (Catchen et al., 2013). Reads for each index group were divided by barcode, trimmed to a standard length of 94 bp and sequences flagged by the Illumina quality filter were removed using the process_radtags script. After renaming the samples based on their unique index/barcode combinations, subsets of replicate samples were used to estimate error rates and optimize assembly parameters (Supplementary File 2). Three separate assemblies were performed. The first included all (n=1077) Baetis samples. Although morphological traits (that is, length of the middle caudal filament) can reliably distinguish B. bicaudatus from B. tricaudatus in most cases, a subset of specimens from elevations where the two species’ ranges overlap showed intermediate morphologies. Results from this first assembly yielded a hybrid index (h) for each individual. The hybrid index represents the proportion of alleles in an individual that are derived from the B. tricaudatus population. Therefore, h=0 indicates a B. bicaudatus genotype and h=1 indicates a B. tricaudatus genotype. The values of h were used to confirm morphological identification of species, identify species with hybrid ancestry and confirm identification of specimens with intermediate morphologies, small size or missing parts. This was followed by separate assemblies for samples classified with high confidence (based on morphology and hybrid indices, Supplementary File 1) as B. tricaudatus (n=241) or B. bicaudatus (n=602). For each assembly, the denovo_map script was used with the following parameters: a minimum of three reads required to create a stack (-m), two mismatches allowed between loci when processing an individual (-M), four mismatches allowed between loci when building the catalog (-n), and highly repetitive RAD tags removed or broken-up in ustacks.
Data filtering was performed using custom scripts in R (available upon request). The distribution of SNP number per position along the 94 bp sequences was quantified over all tags to identify positions with elevated numbers of SNPs at the beginning and end of the RAD tags that could be due to low-quality base calls. Positions with a number of SNPs beyond 1.5 × the interquartile range of the distribution were removed to avoid false positives due to sequencing error. Next, we removed SNPs from tags with >15 variable sites, which may have been the result of misassembled sequences, along with loci that had >2 alleles or minor allele frequencies <0.05. For the initial analysis of all samples, only loci that were typed in >70% of individuals were used to maximize the number of individuals retained in the analysis at the expense of the number of loci retained. A less stringent cutoff of 50% was used for the subsequent single species analyses. The data were further filtered to include only one SNP locus per tag and remove any loci where the genotyping success rate showed evidence of library bias (that is, a difference in the proportion of individuals typed in two different libraries >1.5 × the interquartile range of the distribution of differences in genotyping success for all loci). All filtering and analyses after this step were performed using the single representative SNP from each RAD locus (randomly selected in cases where multiple SNPs were present). RAD-tag sequences with homology with mitochondrial DNA or potential contaminants (plants, fungi, human and prokaryotes) were removed from further analysis (n⩽20 in both species). Tests for linkage disequilibrium and deviation from Hardy–Weinberg proportions were performed within each sampling site. Loci showing evidence of either in >50% of sites after false discovery rate correction were also removed. Finally, individuals with >50% missing data were removed from the analysis.
Genetic diversity and population structure along elevation gradients
We assessed genetic diversity and population structure across the elevational gradient to identify potential microevolutionary forces contributing to diversification in this system. Summary diversity statistics were computed using the R package hierfstat (Goudet, 2005). Diversity estimates for B. tricaudatus from sites with small sample sizes (n⩽8) were excluded from further landscape genetic analyses. Estimates of effective population size (Ne) provide another measure for comparison. It reflects the size of an idealized population that experiences genetic drift at the rate observed in the sampled population (Wright, 1969). We calculated Ne for each site using the molecular co-ancestry method, which provides an unbiased estimate from a single genetic cohort, in NeEstimator v2.01 (Nomura, 2008; Do et al., 2014).
We performed a principal components analysis along with discriminant analysis of principal components in the R package Adegenet (Jombart, 2008) to obtain an independent estimate of the number of population clusters (K) present in the sample (Supplementary File 3). We also carried out an analysis of molecular variance in the R package Poppr (Kamvar et al., 2014), partitioning genetic diversity among all sites, drainages (Big Thompson, St. Vrain and Cache la Poudre; Figure 1) and among sites within drainages. Pairwise FST values were obtained with Arlequin v3.5 (Excoffier and Lischer, 2010) using 1000 permutations to assess significance. We used the program Structure v 2.3.4 (Hubisz et al., 2009) to identify population clusters and estimate the amount of admixture within individuals. For each data set, we performed 3 replicate runs of 500 000 iterations and a burn-in of 100 000 for values of K (number of subpopulation clusters) from 1 to 10 using the admixture model with correlated allele frequencies. Convergence of alpha and LnLikelihood values was confirmed before submitting to the CLUMPAK server (Kopelman et al., 2015; http://clumpak.tau.ac.il/) to estimate the number of clusters, merge the replicate runs and visualize the results.
Landscape analysis: testing for isolation by distance and resistance
GIS raster layers, representing landscapes in terms of their potential to affect the movement of dispersing mayflies, were used as resistance surfaces for variables related to topography (elevation, slope and topology) and habitat (stream channels; Supplementary Table S1). Although we were also interested in the potential effects of precipitation and temperature, the data layers available for these variables were highly correlated with the elevation data for this region of Colorado. Thus the elevation layer was used as a proxy for temperature and precipitation.
Resistance layers representing stream channels, elevation, slope and topography (Supplementary Table S1,Supplementary Figure S1) were used to generate circuit resistances using Circuitscape v4.0 (Shah and McRae, 2008). These circuits represent the resistance encountered by a dispersing organism at each point on the map. Circuit resistances provide models of connectivity or resistance to predict patterns of dispersal among sites in a heterogeneous landscape. This approach considers multiple dispersal pathways at once, thus integrating over the entire landscape, rather than considering single straight line distances or least-cost paths among sites. We used a cell connection scheme linking each node to four neighbor cells to produce a resistance distance matrix based on the circuit resistances for each raster layer. These matrices were then read into R for downstream analysis.
We quantified the importance of each landscape variable in two ways. First, multiple regression on distance matrices (MRDM) was used to test for relationships between genetic distance and landscape parameters. This included a test for the null hypothesis of IBD using only the Euclidean distances, followed by tests for IBR that considered the circuit resistances for each of the predictor variables. MDRM performs regression of a response matrix (genetic distance) on multiple predictor matrices (landscape resistance distances) and assesses statistical significance via permutation. The MRM function in the R package ecodist (Goslee and Urban, 2007) was run with 1,000 permutations using the genetic distance matrix (FST) as the response variable and each of the landscape resistance matrices along with the Euclidean distance matrix as the predictors for each species. We followed these analyses by running MRDM including all predictor variables in a single model for each species. We assessed multicollinearity in the full models using variance inflation factors (VIF) to obtain a best reduced model for each species, removing variables with high VIF scores in turn until none of the remaining variables showed VIF values >4.
To confirm the results obtained from MRDM, the circuit resistance matrices were re-analyzed using a modified version of the mgLandscape function from the R package MEMGENE (Galpern et al., 2014; Supplementary File 4). This method uses Moran’s Eigenvector Maps to represent spatial patterns in the data. Redundancy analysis is then performed using the MEM eigenvectors as predictor variables and the genetic distances as the response. To identify significant eigenvectors derived from each of the resistance distance matrices, a stepwise procedure is used to add variables to the model based on their contribution as predictors until further additions no longer yield significant improvement of fit. This method has the ability to detect complex and relatively weak spatial patterns, and it has been recommended as a powerful alternative to other statistics commonly used in landscape genetic analyses (Legendre and Fortin, 2010).
FST outliers: testing for isolation by environment
Testing for evidence of IBE was accomplished by identifying loci that showed evidence of selective divergence that was correlated with environmental variation. First, overall FST outliers were detected; then tests to link outlier loci with environmental variables were performed. Three replicate runs of 50 000 simulations were performed using default settings and an FDR of 0.05 to detect SNPs showing evidence of positive selection using fdist (Beaumont and Nichols, 1996). These results were then compared to those obtained using BayeScan v2.1 (Foll and Gaggiotti, 2008) to avoid false positives. BayeScan was run for three replicates of 5000 iterations using default parameters, except for an increased prior odds of neutrality of 1000 to decrease false-positive rates. Loci with q-values <0.05 were considered statistically significant. Loci were only considered as outliers if both methods identified them as such.
Tests to associate outlier loci with environmental variation were carried out using BayeScEnv (Villemereuil and Gaggiotti, 2015). This method extends the capabilities of the BayeScan algorithm by including a model that incorporates environmental data from each collection site represented as environmental differentiation (that is, differences from the average environment divided by the s.d.). The model considering the environmental parameter (g) is then compared to the null F-model, and standard α model to determine which FST outlier loci show variation associated with environmental parameters. We considered six environmental parameters (see Supplementary Table S2 for additional details), consisting of two climatic variables (temperature seasonality and mean temperature of the driest quarter from 1950 to 2000), and three variables measured on site at the time of collection (elevation, stream gradient and canopy cover). Canopy cover was estimated from the mean of spherical densiometer measurements taken at 20, 60 and 100 m upstream, downstream and facing the left and right bank from the sampling site. The BayeScEnv model was run for each of the standardized environmental variables with the parameter settings as follows: g(upper bound)=10, α(mean prior)=−1.0, P=0.50, and π=0.10. After 20 pilot runs of 2000 iterations each and a burn-in of 50 000 iterations, 5000 Markov Chain Monte Carlo (MCMC) samples were taken with 10 steps between each sample. Diagnostics of the log likelihoods and FST values for the 5000 sampled iterations were checked using the R package coda (Plummer et al., 2006) to confirm convergence and sample sizes of at least 2500. Only loci that were confirmed FST outliers in the previous analysis were considered true outliers in this test of local adaptation.
Results
SNP genotyping and data filtering
Illumina sequencing yielded an average of 491 426 (SD 379 010) reads per individual for our sample of 1077 baetids after splitting by unique index/barcode combinations and filtering flagged sequences. The average number of RAD-tags per individual was 6265 (SD 2454) with a mean merged depth of coverage of 65 (SD 30) reads. The combined assembly that included all 1077 Baetis samples yielded a set of 85 loci that were present in >70% of all samples, unique to a single tag and free from library bias. The individual species assembly for B. tricaudatus yielded 2312 loci after initial filtering. Trimming to a single locus per tag and removing loci with evidence of LD resulted in 665 loci. Of the remaining loci, only one showed deviation from Hardy–Weinberg expectations in over half of the sample locations and was excluded from analyses that assume an equilibrium model. For B. bicaudatus, we obtained 3564 loci after initial filtering, and 1433 loci remained after trimming to a single locus per tag and removing loci with evidence of LD. Due to deviations from Hardy–Weinberg expectations, 420 loci were excluded from the Structure analysis. Data for the final sets of loci were then filtered by individual for >50% genotyping success over all loci. This resulted in final sample sizes of 961 individuals in the two species analysis, 241 individuals in the B. tricaudatus analysis and 602 individuals in the B. bicaudatus analysis.
Genetic diversity is reduced at high elevations
Summary statistics of genetic diversity were generally similar across sites and drainages for both species (Figure 2). B. tricaudatus had significantly higher levels of overall observed heterozygosity and gene diversity (Ho: 0.18±0.02; Hs: 0.28±0.01; n=12) relative to B. bicaudatus (Ho: 0.12±0.01; Hs: 0.25±0.01; n=17) across all sites and loci (t=−0.5, df=7717, P<0.001; Figure 3). Both Ho and Hs declined with increasing elevation in B. bicaudatus, and this relationship was significant for Ho (Ho: r2=0.34, P=0.01; Hs: r2=0.13, P=0.15). In B. tricaudatus no such relationship was observed (Ho: r2=0.01, P=0.73; Hs: r2=0.03, P=0.59). Estimates of Ne were also significantly higher (t=2.8, df=13.7, P=0.02) in B. tricaudatus (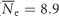 ) relative to B. bicaudatus (
) relative to B. bicaudatus (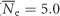 ). Within-species estimates of Ne displayed a declining trend with increasing elevation in B. tricaudatus, but were relatively constant in B. bicaudatus (Figure 3).
). Within-species estimates of Ne displayed a declining trend with increasing elevation in B. tricaudatus, but were relatively constant in B. bicaudatus (Figure 3).
Figure 3.
Relationship between (a) observed heterozygosity (Ho) and elevation, and (b) effective population size (Ne) and elevation in meters for each site and species. Solid circles and dashed lines represent B. tricaudatus; and open circles and dotted lines represent B. bicaudatus. B. tricaudatus samples with n<5 were excluded from the estimation of Ne. B. bicaudatus had significantly lower levels of both Ho and Ne, and a decline in Ho with increasing elevation. In B. tricaudatus, Ne displayed a declining trend with increasing elevation.
Overall population genetic structure was lower in B. tricaudatus with a global FST of 0.041 (Φst=0.03, P=0.003) relative to an FST of 0.093 in B. bicaudatus (Φst=0.086, P=0.001). Analysis of molecular variance found significant substructure in both species (Table 1), but the scale of the observed structure differed. Differentiation by drainage was roughly equivalent between the species (but note the difference in P-values; B. tricaudatus: Φct=0.01, P=0.06; B. bicaudatus: Φct=0.01, P=0.50), but diversity at smaller spatial scales (that is, among sites within drainages) was lower in B. tricaudatus (Φsc=0.02, P=0.003) relative to B. bicaudatus (Φsc=0.07, P=0.001). All values of pairwise FST were statistically significant (P<0.01) and were generally higher among the B. bicaudatus samples (Figure 2, Supplementary Tables S3 and S4).
Table 1. Analysis of molecular variance indicated significant population substructure in both B. bicaudatus and B. tricaudatus.
| (Φct) Among drainages | (Φsc) Among sites within drainages | (Φst) Among all sites | Total | ||
|---|---|---|---|---|---|
| B. bicaudatus | df | 2 | 14 | 585 | 601 |
| SumSq | 2604.2 | 9560.7 | 107740.4 | 119905.4 | |
| MeanSq | 1302.1 | 682.9 | 184.2 | 199.5 | |
| % | 1.4% | 7.1% | 91.4% | 100% | |
| Phi | 0.014 | 0.072 | 0.086 | – | |
| P-value | 0.5 | 0.001 | 0.001 | – | |
| B. tricaudatus | df | 2 | 5 | 221 | 228 |
| SumSq | 561.6 | 976.9 | 23699.2 | 25137.7 | |
| MeanSq | 280.8 | 175.3 | 107.2 | 110.2 | |
| % | 1.0% | 2.4% | 96.6% | 100% | |
| Phi | 0.010 | 0.023 | 0.034 | – | |
| P-value | 0.057 | 0.003 | 0.003 | – |
In B. bicaudatus, structure was significant within and among sites, but was not significant at the largest spatial scale (that is, drainages were not a meaningful grouping of genetic variation). In B. tricaudatus, population structure was also significant within and among sites, and was approaching significance even at the largest spatial scale. Only sites with n⩾9 were included in the analysis of molecular variance analysis. P-values were corrected for multiple tests.
Population structure is more extensive in the high-elevation species
For B. tricaudatus, the Structure results for K=1 to K=10 indicated an optimum at K=2 according to Evanno’s method (Evanno et al., 2005) implemented in CLUMPAK. Population subdivision according to Structure was largely consistent with the discriminant analysis of principal component results (Supplementary File 3), but with evidence of more extensive gene flow across the landscape. At K=2, a north–south split near the ridge separating the Poudre and Big Thompson drainages was apparent (Figure 4 and Supplementary Figure S3).
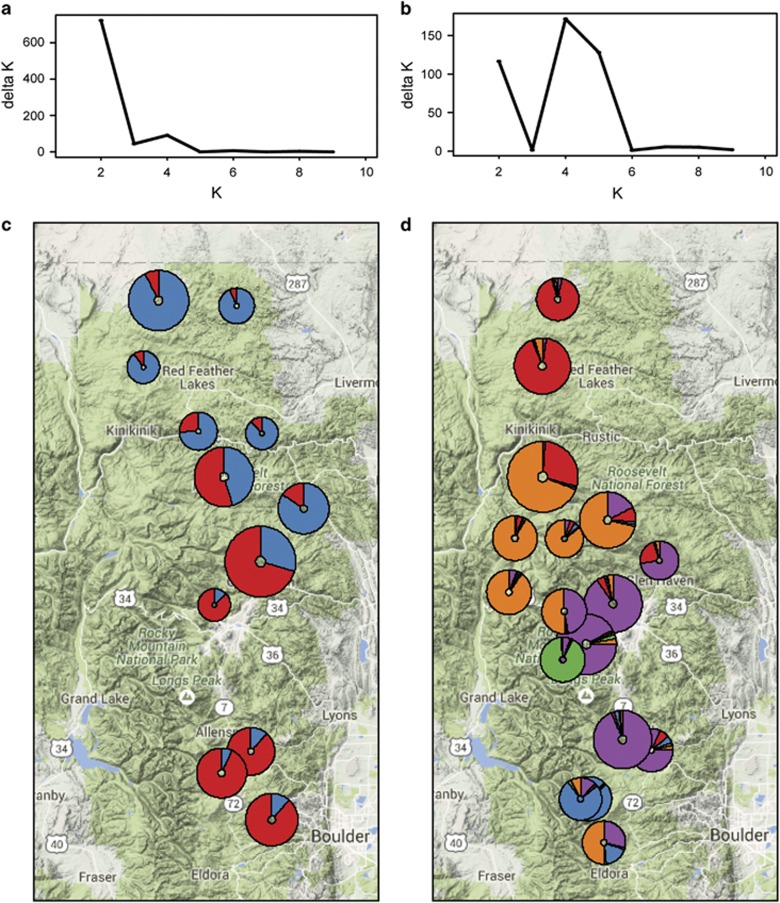
Figure 4.
Structure clustering results (upper panels) for (a) B. tricaudatus and (b) B. bicaudatus. Genetic clustering showed an optimum number of two clusters (K) for the B. tricaudatus data set and similar levels of support for four or five clusters in B. bicaudatus (K=5 shown in lower panel). The distribution of genetic clusters (represented by different colors, lower panels) over the landscape shows fewer population clusters in (c) B. tricaudatus distributed over wider geographic areas than those in (d) B. bicaudatus. Circle sizes are proportional to sample numbers for each species. Place names and road numbers are included on map for reference.
Structure results for B. bicaudatus provided support for both K=4 and K=5 (Figure 4 and Supplementary Figure S3). For K=5, the distribution of population clusters across the landscape according to Structure was in agreement with the discriminant analysis of principal component results (Supplementary File 3), showing a separation of the high-elevation site in the Big Thompson drainage (B3051, Figure 4) from all other sites. In the remaining sites, a north–south split was apparent in the center of the sampling region along the ridge separating the Poudre and Big Thompson drainages. Further separation divided the sites north of the Poudre River valley and those from the southern St. Vrain drainage.
Testing models of isolation: distance, resistance or environment?
The null expectation of IBD was detected only at lower elevations. Matrix regression using only Euclidean distance as a predictor of genetic distance (pairwise FST; Supplementary Tables S3 and S4) was highly significant for B. tricaudatus range-wide (F[1,26]=27.6; r2=0.52; P=0.002; Supplementary Figure S4). The same comparison for B. bicaudatus was nonsignificant when all populations were included (F[1,134]=3.6; r2=0.03; P=0.325). To disentangle species differences from landscape effects, we also tested for a relationship between Euclidian and genetic distance in the lower portion of the elevational range of B. bicaudatus where it overlaps with B. tricaudatus (that is, sites below 3000 m). When only B. bicaudatus samples from lower-elevation sites were considered, the relationship became significant (F[1,43]=19.2; r2=0.31; P=0.004).
To contrast with the null expectation of IBD tested above, regression models were used to test for IBR due to each of the landscape factors of interest. Models that considered the landscape resistance variables while accounting for Euclidean distance were all significant for B. tricaudatus, but in each case only the Euclidean contribution was significant and not the landscape resistance factors of interest (Supplementary Table S5). However, the best of the reduced models (obtained after removing variables with high VIF) revealed significant relationships between both elevation differences among sites, and elevation resistance (alt100; Supplementary Figure S5) with pairwise FST after accounting for Euclidean distance (F[3,24]=17.9; r2=0.69; P=0.001). For B. bicaudatus, slope resistance (slope100; Supplementary Figure S5) was a significant predictor of pairwise FST after accounting for Euclidean distance (Supplementary Table S5), and when considered alone (F[1,134]=28.9; r2=0.18; P=0.001). Neither the full model (F[6,129]=10.1, r2=0.32; P=0.225) nor the best of the reduced models were statistically significant (F[5,130]=11.8, r2=0.31; P=0.149) for B. bicaudatus.
The MEMGENE results (Supplementary Table S6) for B. tricaudatus were in agreement with the MRDM analysis in that a relatively high proportion of genetic distance was explained by the Euclidean resistance surface (r2adj=0.75). But for this species, none of the four resistance models explained the data better than the Euclidean distances alone did. The MEMGENE results for B. bicaudatus largely agreed with the MRDM results as well, but a lower proportion of genetic distance was explained by Euclidean distance (r2adj=0.15). In B. bicaudatus, resistance distances associated with both elevation (alt100) and slope (slope100) explained the observed genetic distances among samples better than the Euclidean models.
FST outlier loci correlate with environmental variation
Evidence for IBE was detected in the form of FST outlier loci that were correlated with environmental variables. By comparing the proportion of loci with more extreme FST values, we can learn about the relative strength of directional selection in these two species in the sampled range. In B. tricaudatus, 81 outlier loci were detected with fdist, and four with BayeScan, all of which were in agreement with fdist (Supplementary Figure S2). In B. bicaudatus, fdist detected 116 outliers that were potentially under selection, and BayeScan found 79, 69 of which were in agreement between the two methods. For B. tricaudatus and B. bicaudatus, respectively, 0.5% and 4.8% of total loci were determined to be outliers.
BayeScEnv was used to detect loci exhibiting signatures of selection that matched with expectations from a model of IBE. BayeScEnv identified a number of loci responding to each of the environmental variables tested (Table 2). In B. tricaudatus, we detected nine of 665 loci (1.4%) with significant associations with the environmental parameters, and those included all four of the outlier loci identified by BayeScan and fdist. Although there were fewer overall FST outliers in B. tricaudatus, all four were associated with elevation, and all but one were associated with temperature of the driest quarter and stream gradient. For B. bicaudatus, a total of 52 out of 1433 loci (3.6%) were significantly associated with at least one of the environmental variables, and 41 of these were in agreement with the set of FST outliers detected with BayeScan and fdist. The environmental variable associated with the greatest number of loci (n=20) was the mean temperature of the driest quarter. Stream gradient (n=12) and site elevation (n=11) were also associated with a relatively large number of outlier loci in this species.
Table 2. Results from BayeScEnv revealed loci responding to each of the five environmental variables tested.
| B. tricaudatus (n=665 loci) | B. bicaudatus (n=1433 loci) | |
|---|---|---|
| Mean temperature of the driest quarter | 3 | 20 (23) |
| Stream gradient | 3 | 12 (14) |
| Elevation | 3 (5) | 11 (14) |
| Canopy cover | 0 | 9 (10) |
| Annual range in temperature | 2 | 6 (8) |
The number of FST outliers associated with each environmental parameter (described in Supplementary Table S2) that were also detected with BayeScan is presented. In cases where additional outliers were detected with BayScEnv but not observed with BayeScan, the total number is in parentheses. Some loci showed correlations with more than one environmental factor.
Discussion
Our data show that limited gene flow reduces genetic variation and connectivity in populations at higher sites along an elevation gradient. By comparing genetic structure and diversity among samples from different elevations, with expectations based on specific models of IBD, resistance (IBR) or environment (IBE), we show that the effects of landscape barriers on dispersal are stronger at higher elevations. Our findings also show that populations at higher elevations are more genetically differentiated than those at lower elevations, and this holds true for comparisons between closely related species at different elevations, as well as comparisons among sites along the gradient within the higher elevation species. In addition, we found significant differences in the number of outlier loci with elevation. The correlation of outlier loci with specific environmental variables supports the hypothesis that high-elevation populations may show more local adaptation. Local adaptation is generally hampered by extensive gene flow because locally adapted alleles that do not convey an advantage in other locations can be swamped out from the surrounding population matrix. However, studies of natural populations provide evidence that local adaptation can occur even in the presence of ongoing gene flow (Gonzalo‐Turpin and Hazard, 2009; Sarup et al., 2009). The larger number of outliers at higher elevations suggest that limited gene flow combined with natural selection promote divergence among high-elevation populations. These observations advance our understanding of the drivers of population structure in complex montane habitats, with important implications for the conservation of stream ecosystems.
Genetic diversity and population structure in montane stream insects
Lower levels of heterozygosity and smaller effective population sizes were observed at higher elevation sites relative to lower elevations. Specifically, Ho declined at high elevations in B. bicaudatus and Ne declined in high-elevation populations of B. tricaudatus. Reduced diversity at high elevations can result from smaller population sizes, population bottlenecks due to fluctuations in population sizes, or from founder effects as organisms colonize new habitats. Moreover, genetic drift from these three factors will be exacerbated by reduced gene flow in isolated alpine taxa (DeChaine and Martin, 2004). Other surveys of elevation gradients in aquatic insects have demonstrated that diversity at multiple scales (from genetic to taxonomic) is linked to their position along the elevation gradients (Clarke et al., 2008; Múrria et al., 2013), especially as organisms reach the edge of their elevational ranges (Hodkinson, 2005; Gill et al., 2014).
Demographic history and effective population sizes undoubtedly have a key role in shaping population structure, by modulating the number of breeding individuals at a given location and thus influencing the rate of adaptive evolution (Gossmann et al., 2012). However, the amount of gene flow (or lack thereof) among sites is likely to be the primary factor structuring populations in species with large populations and similar life histories (Hughes, 2007; Finn et al., 2011). Despite expectations of similar dispersal potential, our results reveal important differences between the species in terms of the number of subpopulation lineages and the distribution of genetic variation across the landscape. Gene flow was more extensive overall in B. tricaudatus and extended to the largest spatial scale (that is, among drainages). For this lower-elevation species, analysis with Structure indicated only two genetic clusters (Figure 4). However, the underlying model used by this software does not perform well when a pattern of IBD is present (as is the case here). Thus it is likely that B. tricaudatus is in fact a single panmictic population with the only clear obstacle to gene flow being geographic distance among sites.
The higher elevation species, B. bicaudatus, did not conform to a model of IBD, and a greater proportion of the genetic variance was partitioned among sites within drainages compared to the larger spatial scale of the drainages themselves. High divergence at small spatial scales has been observed in other studies of ribosomal and mitochondrial sequence variation in stream invertebrates, where adjacent sites in a single stream showed greater differentiation than sites kilometers apart (Monaghan et al., 2005; Schultheis and Hughes, 2005). As a consequence, no clear signal of IBD has been observed in many of these studies despite apparently extensive dispersal. Structure divided B. bicaudatus into multiple subpopulations that showed differentiation both among drainages, and between high- and low-elevation sites within drainages (Figure 4).
Models of isolation along elevation gradients
In this study, sites at higher elevations separated by steep topographic slopes were more genetically differentiated than would be expected based on Euclidean distance alone. Thus, a model of IBR is most appropriate when considering higher elevation sites because of the constraints on dispersal imposed by elevation and slope. Topographic effects such as slope have been found to act as significant barriers to dispersal in other landscape genetic studies of species with an aquatic life history stage and overland dispersal (Murphy et al., 2010). This is likely due to the energetic cost associated with traversing steep rugged areas, and may be especially so in species such as mayflies with very short adult life spans. Overall, the patterns of diversity and population structure observed here fit well with a valley-mountain model of population structure (Funk et al., 2005). Under this model, low-elevation sites are characterized by higher effective population sizes and levels of connectivity than their high-elevation counterparts, and gene flow between low and high elevations is limited, often resulting in diversification of high-elevation sites (Clarke et al., 2008; Múrria et al., 2013).
In contrast to observations at higher elevations, the lower-elevation sites in this study showed greater genetic connectivity, presumably due to regular exchange of migrants in a relatively flat topographic landscape. Increased genetic differentiation among high-elevation sites could be explained by reduced gene flow among sites separated by topographical barriers and/or genetic drift at isolated sites resulting in reduced genetic diversity and smaller effective population sizes. Regardless of the exact mechanism, baetid populations at high-elevation sites are expected to be more vulnerable to local extinction due to lower effective population sizes and limited amounts of standing genetic variation for natural selection to act upon.
Evidence for local adaptation in montane mayflies
Hierarchical dendritic stream networks in mountain ranges can provide environmental settings conducive to local adaptation and diversification. This is because elevation is often correlated with environmental gradients in a number of factors including temperature, precipitation, flow regime, as well as habitat and community composition (Lytle and Poff, 2004; Múrria et al., 2013). These selection gradients overlaid on a stream system create conditions whereby standing genetic variation can be divided and subsequently shaped into locally adapted lineages (Funk et al., 2005; Clarke et al., 2008; Hughes et al., 2009). The evolution of local adaptation depends on both the strength of local selective pressures and the homogenizing effect of immigration from other sites. In this context, demographic isolation can be beneficial to some extent by allowing populations to undergo an adaptive response to local conditions, as has been observed in numerous studies of organisms inhabiting the edges of their latitudinal or elevational ranges (Bridle and Vines, 2007; Halbritter et al., 2015).
Many more putative targets of natural selection, as well as an overall higher proportion of FST outliers, were detected in the higher elevation species B. bicaudatus. Adaptation to strong selective forces imposed by high-elevation environments have been documented in a number of montane species (Hodkinson, 2005; Storz et al., 2009; Cheviron and Brumfield, 2012). Reduced gene flow due to landscape effects, combined with selection at high elevations, appears to underlie adaptive divergence at many B. bicaudatus loci. This is consistent with a model of IBE in this species and supports the idea that isolated, high-elevation sites are an incubator for local adaptation in mayflies. Although genetic drift due to isolation and small effective population sizes are likely to accelerate diversification, the effect of drift alone would result in genome-wide diversification, rather than a subset of outlier loci. Further, many of the FST outliers detected in this species were significantly associated with the environmental parameters of elevation, temperature of the driest quarter and stream gradient. This reveals a link between genetic differentiation and elevation-related variation. Our observations of more extensive population structure and increased resistance from landscape features provide a parsimonious explanation for how such diversification could develop.
Outlier loci that are significantly correlated with abiotic factors can be an indication of selection; however, the SNP markers are unlikely to be the direct targets of selection. Further sequencing and annotation of the genomes of these taxa will be required before we can establish direct functional associations. Genetic linkage with sites under selection could explain the observed associations with these environmental variables, and would be in agreement with a model of IBE at higher elevation sites
Conclusions
The higher elevation sites in this study were isolated by topographical barriers and a lack of suitable intervening habitat, resulting in apparent adaptive divergence in the higher elevation species across its elevational range. At elevations below 3000 m, however, both species exhibited substantial gene flow among populations, indicating that dispersal among lower-elevation sites is less restricted than at higher elevations. In some cases, complex topography or other landscape features could explain observations of more extensive population structure at higher elevations independent of an adaptive response to natural selection. However, reduced gene flow among high-elevation sites creates conditions whereby adaptive divergence is more likely to occur. Indeed, the finding of many more FST outliers in the high-elevation species provides support for this hypothesis, as other mechanisms such as drift would be more likely to result in genome-wide effects. Furthermore, the finding that many of the FST outlier loci were also associated with environmental factors related to elevation provides further support that elevational change (or its covariates) are driving genetic differentiation at these loci.
Although the higher elevation species are likely to possess novel genetic diversity due to local adaptation associated with topographic and climatic factors, they are also likely to be more vulnerable to shifting disturbance regimes and environmental changes due to lack of genetic connectivity and lower overall genetic diversity. Future studies measuring physiological tolerance of mayflies from different elevations, and how population differences relate to patterns of population genetic structure, will be an important next step in confirming landscape effects on diversification and vulnerability in montane taxa.
Data Archiving
RAD-tag sequences are available from the NCBI SRA database under project accession ID: PRJNA377573. SNP data and landscape resistance data are available from the DRYAD data depository under accession number: doi:10.5061/dryad.02j5s.
Acknowledgments
This paper was supported by the U.S. National Science Foundation through a collaborative Dimensions of Biodiversity grant, awards DEB-1046408, DEB-1045960 and DEB-1045991. We thank Alisha A. Shah, Cameron Ghalambor, Erin Larson, Keeley McNeil, Steve Thomas, Juan Guayasamin, Rachel Harrington and the rest of the EVOTRAC team for their input on project design and analysis. We also thank Peter A. Schweitzer and the staff of the Cornell Biotechnology Resource Center and Genomics Facility for sequencing work, as well as Steven M. Bogdanowicz of the Cornell Evolutionary Genetics Core Facility for advice and support with library preparation. Erika Mudrak at the Cornell Statistical Consulting Unit provided valuable advice regarding statistical analysis, and Paul Galpern and Pedro Peres-Neto were instrumental in modifying and implementing Memgene.
Author contributions
WCF, KRZ, ACE and NRP designed the project; BAG, KLC and BCK collected and identified the specimens; NRP, MMG and CGB analyzed the data; ACE, BCK, NLP, BAG, NRP and CGB contributed to the landscape analysis; and NRP and MMG wrote the paper with input from all other authors.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Alp M, Keller I, Westram AM, Robinson CT. (2012). How river structure and biological traits influence gene flow: a population genetic study of two stream invertebrates with differing dispersal abilities. Freshwater Biol 57: 969–981. [Google Scholar]
- Baguette M, Blanchet S, Legrand D, Stevens VM, Turlure C. (2013). Individual dispersal, landscape connectivity and ecological networks. Biol Rev 88: 310–326. [DOI] [PubMed] [Google Scholar]
- Bálint M, Domisch S, Engelhardt C, Haase P, Lehrian S, Sauer J et al. (2011). Cryptic biodiversity loss linked to global climate change. Nat Clim Change 1: 313–318. [Google Scholar]
- Bauernfeind E, Moog O. (2000). Mayflies (Insecta : Ephemeroptera) and the assessment of ecological integrity: a methodological approach. Hydrobiologia 422: 71–83. [Google Scholar]
- Beaumont MA, Nichols RA. (1996). Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society B-Biological Sciences 263: 1619–1626. [Google Scholar]
- Bilton DT, Freeland JR, Okamura B. (2001). Dispersal in freshwater invertebrates. Annu Rev Ecol Syst 32: 159–181. [Google Scholar]
- Bridle JR, Vines TH. (2007). Limits to evolution at range margins: when and why does adaptation fail? Trends Ecol Evol 22: 140–147. [DOI] [PubMed] [Google Scholar]
- Brittain JE. (1982). Biology of mayflies. Annu Rev Entomol 27: 119–147. [Google Scholar]
- Bunn SE, Hughes JM. (1997). Dispersal and recruitment in streams: evidence from genetic studies. J N Am Benthol Soc 16: 338–346. [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. (2013). Stacks: an analysis tool set for population genomics. Mol Ecol 22: 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron Z, Brumfield R. (2012). Genomic insights into adaptation to high-altitude environments. Heredity 108: 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A, Mac Nally R, Bond N, Lake P. (2008). Macroinvertebrate diversity in headwater streams: a review. Freshwater Biol 53: 1707–1721. [Google Scholar]
- Cushman SA, McKelvey KS, Hayden J, Schwartz MK. (2006). Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168: 486–499. [DOI] [PubMed] [Google Scholar]
- DeChaine EG, Martin AP. (2004). Historic cycles of fragmentation and expansion in Parnassius smintheus (Papilionidae) inferred using mitochondrial DNA. Evolution 58: 113–127. [DOI] [PubMed] [Google Scholar]
- Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR. (2014). NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14: 209–214. [DOI] [PubMed] [Google Scholar]
- Dodds G. (1923). Mayflies from Colorado: descriptions of certain species and notes on others. Trans Am Entomol Soc 49: 93–114. [Google Scholar]
- Edmunds GF Jr, Jensen SL, Berner L. (1976) The Mayflies Of North And Central America. University of Minnesota Press. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Finn DS, Bonada N, Múrria C, Hughes JM. (2011). Small but mighty: headwaters are vital to stream network biodiversity at two levels of organization. J N Am Benthol Soc 30: 963–980. [Google Scholar]
- Foll M, Gaggiotti O. (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WC, Blouin MS, Corn PS, Maxell BA, Pilliod DS, Amish S et al. (2005). Population structure of Columbia spotted frogs (Rana luteiventris) is strongly affected by the landscape. Mol Ecol 14: 483–496. [DOI] [PubMed] [Google Scholar]
- Galpern P, Peres-Neto PR, Polfus J, Manseau M. (2014). MEMGENE: spatial pattern detection in genetic distance data. Methods Ecol Evol 5: 1116–1120. [Google Scholar]
- Gattolliat J-L. (2004). First reports of the genus Nigrobaetis Novikova & Kluge (Ephemeroptera: Baetidae) from Madagascar and La Réunion with observations on Afrotropical biogeography. Rev Suisse Zool 111: 657–669. [Google Scholar]
- Gill BA, Harrington RA, Kondratieff BC, Zamudio KR, Poff NL, Funk WC. (2014). Morphological taxonomy, DNA barcoding, and species diversity in southern Rocky Mountain headwater streams. Freshwater Sci 33: 288–301. [Google Scholar]
- Gonzalo‐Turpin H, Hazard L. (2009). Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J Ecol 97: 742–751. [Google Scholar]
- Goslee SC, Urban DL. (2007). The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22: 1–19. [Google Scholar]
- Gossmann TI, Keightley PD, Eyre-Walker A. (2012). The effect of variation in the effective population size on the rate of adaptive molecular evolution in eukaryotes. Genome Biol Evol 4: 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. (2005). HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5: 184–186. [Google Scholar]
- Gregory TR. (2002) The C-value paradox (Doctoral Dissertation). The University of Guelph. [Google Scholar]
- Halbritter AH, Billeter R, Edwards PJ, Alexander JM. (2015). Local adaptation at range edges: comparing elevation and latitudinal gradients. J Evol Biol 28: 1849–1860. [DOI] [PubMed] [Google Scholar]
- Hodkinson ID. (2005). Terrestrial insects along elevation gradients: species and community responses to altitude. Biol Rev 80: 489–513. [DOI] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. (2009). Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JM. (2007). Constraints on recovery: using molecular methods to study connectivity of aquatic biota in rivers and streams. Freshwater Biol 52: 616–631. [Google Scholar]
- Hughes JM, Mather PB, Hillyer MJ, Cleary C, Peckarsky B. (2003). Genetic structure in a montane mayfly Baetis bicaudatus (Ephemeroptera : Baetidae), from the Rocky Mountains, Colorado. Freshwater Biol 48: 2149–2162. [Google Scholar]
- Hughes JM, Schmidt DJ, Finn DS. (2009). Genes in streams: using DNA to understand the movement of freshwater fauna and their riverine habitat. Bioscience 59: 573–583. [Google Scholar]
- Jombart T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. [DOI] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grunwald NJ. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2: e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. (2015). Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15: 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Fortin MJ. (2010). Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol Ecol Resour 10: 831–844. [DOI] [PubMed] [Google Scholar]
- Lytle DA, Poff NL. (2004). Adaptation to natural flow regimes. Trends Ecol Evol 19: 94–100. [DOI] [PubMed] [Google Scholar]
- McCafferty WP, Randolph RP, Jacobus LM. (2012) Mayflies of the Intermountain West. American Entomological Institute. [Google Scholar]
- Monaghan MT, Gattolliat JL, Sartori M, Elouard JM, James H, Derleth P et al. (2005). Trans-oceanic and endemic origins of the small minnow mayflies (Ephemeroptera, Baetidae) of Madagascar. Proceedings of the Royal Society B-Biological Sciences 272: 1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morihara D, McCafferty W. (1979). The Baetis larvae of North America (Ephemeroptera: Baetidae). Transactions of the American Entomological Society 105: 139–221. [Google Scholar]
- Murphy MA, Dezzani R, Pilliod DS, Storfer A. (2010). Landscape genetics of high mountain frog metapopulations. Mol Ecol 19: 3634–3649. [DOI] [PubMed] [Google Scholar]
- Múrria C, Bonada N, Arnedo MA, Prat N, Vogler AP. (2013). Higher β‐and γ‐diversity at species and genetic levels in headwaters than in mid‐order streams in Hydropsyche (Trichoptera). Freshwater Biol 58: 2226–2236. [Google Scholar]
- Nomura T. (2008). Estimation of effective number of breeders from molecular coancestry of single cohort sample. Evol Appl 1: 462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz‐Vinas I, Loot G, Stevens V, Blanchet S. (2015). Evolutionary processes driving spatial patterns of intraspecific genetic diversity in river ecosystems. Mol Ecol 24: 4586–4604. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. (2012). Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. (2006). CODA: Convergence diagnosis and output analysis for MCMC. R News 6: 7–11. [Google Scholar]
- Rutschmann S, Gattolliat JL, Hughes SJ, Báez M, Sartori M, Monaghan MT. (2014). Evolution and island endemism of morphologically cryptic Baetis and Cloeon species (Ephemeroptera, Baetidae) on the Canary Islands and Madeira. Freshwater Biol 59: 2516–2527. [Google Scholar]
- Sarup P, Frydenberg J, Loeschcke V. (2009). Local adaptation of stress related traits in Drosophila buzzatii and Drosophila simulans in spite of high gene flow. J Evol Biol 22: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Schiffers K, Bourne EC, Lavergne S, Thuiller W, Travis JM. (2013). Limited evolutionary rescue of locally adapted populations facing climate change. Philos Trans R Soc Lond B Biol Sci 368: 20120083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis AS, Hughes JM. (2005). Spatial patterns of genetic structure among populations of a stone-cased caddis (Trichoptera : Tasimiidae) in south-east Queensland, Australia. Freshwater Biol 50: 2002–2010. [Google Scholar]
- Shah V, McRae B. (2008). Circuitscape: a tool for landscape ecology. Proceedings of the 7th Python in Science Conference 7: 62–66. [Google Scholar]
- Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H et al. (2009). Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci USA 106: 14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC et al. (2004). Extinction risk from climate change. Nature 427: 145–148. [DOI] [PubMed] [Google Scholar]
- Villemereuil P, Gaggiotti OE. (2015). A new FST‐based method to uncover local adaptation using environmental variables. Methods Ecol Evol 6: 1248–1258. [Google Scholar]
- Vuataz L, Rutschmann S, Monaghan MT, Sartori M. (2016). Molecular phylogeny and timing of diversification in Alpine Rhithrogena (Ephemeroptera: Heptageniidae). BMC Evol Biol 16: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IJ, Bradburd GS. (2014). Isolation by environment. Mol Ecol 23: 5649–5662. [DOI] [PubMed] [Google Scholar]
- Ward JV, Kondratieff BC. (1992) An illustrated guide to the mountain stream insects of Colorado.
- Wright S. (1969) The theory of gene frequenciesEvolution And The Genetics Of Populations Vol. 2, University of Chicago Press: Chicago, IL, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.