Abstract
Background
Although the genetic structure of rice germplasm has been characterized worldwide, few studies investigated germplasm from Thailand, the world’s largest exporter of rice. Thailand and the International Rice Research Institute (IRRI) have diverse collections of rice germplasm, which could be used to develop breeding lines with desirable traits. This study aimed to investigate the level of genetic diversity and structures of Thai and selected IRRI germplasm. Understanding the genetic structure and relationships among these germplasm will be useful for parent selection used in rice breeding programs.
Results
From the 98 InDel markers tested for single copy and polymorphism, 19 markers were used to evaluate 43 Thai and 57 IRRI germplasm, including improved cultivars, breeding lines, landraces, and 5 other Oryza species. The Thai accessions were selected from all rice ecologies such as irrigated, deep water, upland, and rainfed lowland ecosystems. The IRRI accessions were groups of germplasm having agronomic desirable traits, including temperature-sensitive genetic male sterility (TGMS), new plant type, early flowering, and biotic and abiotic stress resistances. Most of the InDel markers were genes with diverse functions. These markers produced the total of 127 alleles for all loci, with a mean of 6.68 alleles per locus, and a mean Polymorphic Information Content (PIC) of 0.440. Genetic diversity of Thai rice were 0.3665, 0.4479 and 0.3972 for improved cultivars, breeding lines, and landraces, respectively, while genetic diversity of IRRI improved and breeding lines were 0.3272 and 0.2970, respectively. Cluster, structure, and differentiation analyses showed six distinct groups: japonica, TGMS, deep-water, IRRI germplasm, Thai landraces and breeding lines, and other Oryza species.
Conclusions
Thai and IRRI germplasm were significantly different. Thus, they can be used to broaden the genetic base and trait improvements. Cluster, structure, and differentiation analyses showed concordant results having six distinct groups, in agreement with their development, and ecologies.
Electronic supplementary material
The online version of this article (doi:10.1186/1939-8433-5-19) contains supplementary material, which is available to authorized users.
Keywords: Genetic diversity, Germplasm evaluation, InDel marker, Oryza sativa, Population structure, Temperature sensitive genetic male-sterile
Background
Genetic diversity and population structure of cultivated rice (Oryza sativa L.) have been studied worldwide (Garris et al. [2005]; Yu et al., [2003]; Zhao et al. [2010]; Ali et al. [2011]; Chen et al., [2011]). However, only few Thai rice germplasm has been included in these studies (Garris et al. [2005]; Yu et al., [2003]), and to our knowledge, there is no report on genetic structure of Thai commercial cultivars grown in different ecologies in the country. Thus, there is a lack of information on genetic structure of Thai rice. Thailand is the world’s largest exporter of rice, and is famous for high-quality, long-grain white rice, because the breeding of Thai rice has been focused on maintaining good grain characteristics and quality. Thailand has a large collection of diverse rice germplasm, including the famous Thai jasmine rice (Chitrakon and Somrith [2003]).
In Thailand, rice ecologies can be classified as irrigated, rainfed lowland, deep water, and upland ecosystems. Rainfed lowland is the majority of rice growing area, followed by irrigated, deepwater and upland. Different ecologies also results in different amount of rice production. The highest average rice yield was from irrigated ecosystem, followed by deep water, rainfed lowland, and upland ecosystems. The average rice yield in the wet season is constant at about 2 t/ha (http://www.fao.org/). By its shape and geography, Thailand can be divided into four regions: the mountains and forests of the North; the vast rice fields of the Central Plains; the semi-arid farm lands of the Northeast plateau; and the tropical islands and long coastline of the peninsula South. Each region has different rice-growing environments. The Northern region produces 25% of the total rice production where upland rice is grown on hilly areas, while lowland rice is grown in lower valleys and some terraced fields. The Central region produces 30% of the total rice production where rice is planted almost everywhere across the area in wet season, and in dry season rice is planted to irrigated area of about 450,000 ha. The Northeastern region produces 41% of the total rice mainly rainfed lowland rice. Only a small portion of the total rice is produced from the Southern region where rice is planted in the west and east coasts of the peninsula (http://www.irri.org).
Oryza sativa is composed of two major subspecies, Indica and Japonica (both tropical and temperate) and several ecotypes. Several efforts have been made to assess the genetic diversity within Oryza sativa at both phenotypic and molecular levels. To estimate genetic diversity among Oryza species, several types of molecular markers, particularly simple sequence repeats (SSR), have been used (Yu et al. [2003]; Hashimoto et al. [2004]; Garris et al. [2005]; Thomson et al. [2007]; Wen et al. [2009]; Ishii et al. [2011], Zhang et al., [2011]). Polymorphisms in the SSR region are considered the results of different replications of repeated sequences, resulting in different sizes of the PCR products. However, alleles with different sequences but having the same length may yield ambiguous results of the phylogenetic analysis. Sequencing SSR products can provide clear information on the evolutionary history of these loci (Sunnucks et al. [2000]; Provan et al. [2004]). Alternatively, single-stranded conformation polymorphism (SSCP), a simple and rapid method to determine sequence variation in a large number of samples without expensive direct sequencing, was proposed to use for genotyping and mapping genetic diversity in crop plants (Kuhn et al., [2008]). SSCP is a very sensitive technique for the detection of single point mutations between different DNA fragments (Grieu et al. [2004]; Muangprom et al. [2005]). Recently, SSCP has been used in crop studies, such as marker assisted selection (Borchert and Hohe [2009]), comparative genomics (Castelblanco and Fregene [2006]), phylogenetics (Rousseau-Gueutin et al. [2009]) and fitness effects of crop QTLs (Baack et al. [2008]). Furthermore, the recent availability of rice genome sequences provides the opportunity to select genes/sequences distributed in the genome as SSCP markers.
The International Rice Research Institute (IRRI) has large collections of characterized rice germplasm. These germplasm could be used to develop breeding lines with desirable traits, and they are available to other countries. Several of IRRI germplasm have been used for improvement of rice breeding programs in Thailand. Understanding the genetic diversity and genetic relationships among Thai and IRRI germplasm is useful for parent selection to produce hybrids or to improve rice population (Moose and Mumm [2008]). Although Thailand is famous for its rice, genetic characterizations of Thai rice at the molecular level are very limited. Therefore, the aims of this study were to evaluate the level of genetic diversity and to assess genetic relationships of Thai rice germplasm, and germplasm with desirable traits obtained from IRRI.
Results
InDel marker development and polymorphisms of the SSCP markers
By testing the 4 selected rice accessions with the 98 InDel markers, only markers that were presented as single copy and showed polymorphism in at least 3 out of the 4 rice accessions were selected for genetic analysis. A total of 19 InDel markers were used to evaluate genetic diversity in 101 rice accessions (Table 1). These InDel markers were chosen from 9 out of 12 rice chromosomes and most of them were genes annotated with diverse functions, as listed in Table 2.
Table 1.
Characteristics of rice accession used in this study including their ecologies, special traits and methods for their improvement
| No.a | Rice Accession | Ecologies and Special traits | Methods used for their improvementb |
|---|---|---|---|
| 1 | RD7 | Irrigated, Resistance to Bacterial Blight, and Moderated resistance to Yellow Orange Leaf virus | C4-63/Gow Ruang 88//Sigadis |
| 2 | RD23 | Irrigated, High yielding, Resistance to Bacterial Blight and Ragged stun | RD7/IR32//RD1 |
| 3 | SPR90 | Irrigated, High yielding, Resistance to Blast, Bacterial Blight, Ragged stun and Brown planthopper | RD21/IR4422-98-3-6-1//RD11/RD23 |
| 4 | CNT1 | Irrigated, High yielding, Resistance to Ragged stun, Brown planthopper, White-backed planthopper, and Blast | IR13146-158-1/IR15314-43-2-3-3//BKN6995-16-1-1-2 |
| 5 | SPR1 | Irrigated, High yielding, Resistant to Ragged stun, Brown planthopper, White-backed planthopper, Blast, Bacterial Blight, Yellow Orange Leaf virus | IR25393-57-2-3/RD23//IR27316-96-3-2-2///SPRLR77205-3-2-1-1/SPRLR79134-51-2-2 |
| 6 | PTT1 | Irrigated, Fragrance, High yielding, Resistance to Brown planthopper, White-backed planthopper, Blast, Bacterial Blight | BKNA6-18-3-2/PTT85061-86-3-2-1 |
| 7 | PSL2 | Irrigated, High yielding, Resistance to Brown planthopper and White-backed planthopper | CNTLR81122-PSL-37-2-1/SPRLR81041-194-2-1//IR56 |
| 8 | SRN1 | Rainfed for North-East, Resistance to Blast, Bacterial Blight, Drought and Salt | IR61078/IR46329-SRN-18-2-2-2 |
| 9 | NSG19 | Rainfed for North-East, Early flowering, Resistance to Brown planthopper | Breeding line |
| 10 | RD15 | Rainfed for North-East, Fragrance, Good cooking quality, Moderated resistance to Drought, Early flowering, Resistance to Brown spot | Irradiated KDML105 |
| 11 | KTH17 | Rainfed for Central plain, Good cooking quality, Moderated resistance to Gall midge | Breeding line |
| 12 | RD27 | Rainfed for Central plain, Resistance to Ragged stun, sheat blight, Blast | Khao Tah Oo/Khao Tah Haeng17 |
| 13 | LPT123 | Rainfed for Central plain, Resistance to acidic soil, Bacterial Blight and Ragged stun | Breeding line |
| 14 | GJ | Rainfed for South, Resistance to Ragged stun, and Narrow brown spot | Breeding line |
| 15 | LDP | Rainfed for South, Resistance to salt and acidic soil, and moderated resistant to Blast | Breeding line |
| 16 | LNP | Rainfed for South, Late flowering, Good cooking quality | Breeding line |
| 17 | CPL | Rainfed for South, Good milling quality | Breeding line |
| 18 | PG56 | Deep water rice, Growing well up to 5 meters deep water, Good milling quality | Breeding line |
| 19 | LMN111 | Deep water rice, Growing well up to 4 meters deep water, Resistance to drought, acidic soil, and Brown spot | Breeding line |
| 20 | HT60 | Deep water rice, Growing well in Central plain area where level of water is not higher than 1 meters high, Resistance to Blast and drought | Khao Nahng Nuey11/C4-63 |
| 21 | PNg1 | Deep water rice, Resistance to Blast in seedling stage and stem border, and Good milling quality | Composite Crosses |
| 22 | PNg | Deep water rice, Resistance to Blast and Drought | Breeding line |
| 23 | DPY | Upland rice, South, Resistance to Blast, Brown spot, Narrow brown spot, Good cooking quality | Breeding line |
| 24 | GML | Upland rice, South, Resistance to Drought, Blast and Brown spot, Narrow brown spot | Breeding line |
| 25 | JH | Upland rice, Central and North < 1000 meters above sea level, Resistance to Blast and Gall Dwarf virus | Breeding line |
| 26 | NR | Upland rice, Growing well in 1000–1400 meters above sea level, Resistance to Blast and Cold | Breeding line |
| 27 | c20878 | Upland, Primitive | Landrace |
| 28 | 23006 | Upland, Primitive | Landrace |
| 29 | c21486 | Upland, Cold resistance | Landrace |
| 30 | 23702 | Upland, Cold resistance | Landrace |
| 31 | KGH | Upland, Salt resistance | Landrace |
| 32 | TM | Resistance to drought | Landrace |
| 33 | KS | Resistance to flooding | Landrace |
| 34 | KH | Resistance to Blast | Landrace |
| 35 | KK | Resistance to Brown planthopper | Landrace |
| 36 | NG | Specialty type | Landrace |
| 37 | KD | Specialty type | Landrace |
| 38 | O. rufipogon | Oryza species, Progenitor of Asian rice | |
| 39 | O.glaberrimma | Oryza species, African cultivated rice | |
| 40 | O.brachyantha | Oryza species, FF genome | |
| 41 | O. latifolia | Oryza species, CCDD genome | |
| 42 | O. officinalis | Oryza species, CC genome | |
| 43 | IR 68301-11-6-4-4-3-6-6 | TGMS, tms3 | Breeding line |
| 44 | IR 77271-42-5-4-36 | TGMS | ID24/I 69736-175-2-2-1-1 |
| 45 | IR 76753-41-6-34-13 | TGMS | ID24/PSB RC 64 |
| 46 | IR 76761-4-3-17-34-25 | TGMS | IR 68935 S/IR32364-20-1-3-2 |
| 47 | IR 73834-21-26-15-25-4 | TGMS | ID24/IR58025B |
| 48 | IR 75589-31-27-8-33 | TGMS | ID24/IR65469-2-3-2-3-2-2 |
| 49 | IR 73827-23-26-15-7 | TGMS | ID24/IR64 |
| 50 | tms2 KDML105 | TGMS | tms2/KDML105 |
| 51 | IR 1820-52-2 | Resistance to Stem borer | Breeding line |
| 52 | IR4227-28-3-2 | Resistance to Stem borer, alkaline | Breeding line |
| 53 | IR 1539-823-1-4 | Resistance to Brown planthopper | Breeding line |
| 54 | IR 9-60 | Resistance to Brown planthopper | PETA/I-GEO-TZE |
| 55 | IR 4819-77-3-2 | Resistance to Brown planthopper | Breeding line |
| 56 | IR 13146-45-2-3 | Resistance to Brown planthopper | Breeding line |
| 57 | IR 13564-95-1 | Resistance to Brown planthopper, Bacterial Blight | Breeding line |
| 58 | IR 2035-117-3 | Resistance to white-backed planthopper, Drought | Breeding line |
| 59 | IR 36 | Resistance to Gall midge, Earliness, Multiple resistance | Siam 29/Chianan 8 |
| 60 | IR 14632-2-3 | Resistance to Blast | Breeding line |
| 61 | IR 5533-PP854-1 | Resistance to Blast | Breeding line |
| 62 | IR1905-PP11-29-4-61 | Resistance to Blast | IR 8/Tetep |
| 63 | IR 54 | Resistance to Bacterial blight | Tangkai rotan/IR 19 |
| 64 | IR 8608-298-3-1 | Resistance to Bacterial blight, Tungro virus | Breeding line |
| 65 | IR 10176-24-6-2 | Earliness | Breeding line |
| 66 | IR 9202-25-1-3 | Earliness | Breeding line |
| 67 | IR 50 | Earliness | I-geo-tze/IR 49 a |
| 68 | IR 58 | Earliness | Fb 24/IR 57 a |
| 69 | IR 72 | Earliness | Taichung native 1/Chianun 242 |
| 70 | IR 2153--338-3 | Grain Quality | Breeding line |
| 71 | IR 12-178-2-3 | Grain Quality | MONG CHIM VANG A/I-GEO-TZE |
| 72 | IR 30 | Multiple resistance | Fb 24/Chianung yu 280 |
| 73 | IR 32 | Multiple resistance | Bpi 76/Tainan 3 |
| 74 | IR 29 | Multiple resistance | Fb 24/Kaohsiung 68 |
| 75 | IR 28 | Multiple resistance | Fb 24/Taichung 172 |
| 76 | IR 26 | Multiple resistance | Tangkai rotan/Kaohsiung 68 |
| 77 | IR 4570-83-3-3-2 (IR 48) | Multiple resistance | Breeding line |
| 78 | IR 442-2-58 | Resistance to Submergence | Breeding line |
| 79 | IR 1529-430-3 (IR 43) | Resistance to Drought | Breeding line |
| 80 | IR 5853-118-5 (IR 52) | Resistance to Drought | Breeding line |
| 81 | IR5178-1-1-4 | Resistance to Drought | Breeding line |
| 82 | Azucena | Resistance to Drought | Breeding line |
| 83 | Pokkali | Resistance to Salt | Unknown derivative method |
| 84 | IR 3941-14-2-2-3 | Resistance to Cold | Breeding line |
| 85 | IR 32429-122-3-1-2 | Resistance to Cold | Breeding line |
| 86 | IR 42015-83-3-2-2 | Resistance to Cold | Breeding line |
| 87 | IR 10206-29-2 | Resistance to Salt | Breeding line |
| 88 | IR 17494-32-3-1-1-3 | Resistance to Salt | Breeding line |
| 89 | IR 5657-33-2 | Resistance to Salt | Breeding line |
| 90 | IR 4422-98-3-6-1 | Resistance to Salt | Breeding line |
| 91 | IR 4630-22-2-17 | Resistance to Salt | Breeding line |
| 92 | IR 29725-21-1-3-2 | Resistance to Salt | Breeding line |
| 93 | IR 13540-56-3-2-1 | Resistance to Alkaline | Breeding line |
| 94 | IR 9764-45-2-2 | Resistance to Alkaline | Breeding line |
| 95 | IR 8 | Plant Type | Peta/Dee-geo-woo-gen |
| 96 | IR 24 | Plant Type | Chianan 8/Tangkai rotan |
| 97 | IR 5 | Plant Type | Peta/Tangkai rotan |
| 98 | IR 22 | Plant Type | Taichung 172/Tangkai rotan |
| 99 | IR6023-10-1-1 | Plant Type | Breeding line |
| 100 | KDML105 | Rainfed for North and North East, Fragrance, Good cooking quality and Resistant to Salt, Drought, Acidic soil | Breeding line |
| 101 | Nipponbare | Japonica | Yamabiko/Sachikaze |
a Number 1–26 and 100, Commercial Thai rice lines; 27–37, Landraces with special trait; 38–42,The other Oryza species; 43–99, Germplasm with desirable traits from IRRI.
b Sources : http://iris.irri.org/; Chitrakon & Somrith [2003].
Table 2.
Characteristics of the InDel markers including their chromosome locations and their annotations
| Genes | Sequence | Size | Tm | Position | Annotation |
|---|---|---|---|---|---|
| (Chromosome; Mb) | |||||
| 1gAP006530 | F: ACCCCCAGCATCTCCTCGTC | 261 | 55 | 1; 15.0 | intergenics |
| R: ACTGGGCCAGGGCTGAGTCT | (close to OsS01g26950) | ||||
| 1gAP003855 | F: CTTGCGCGGTCGAGTAGACG | 289 | 55 | 1; 25.7 | intergenics |
| R: AGCGGTGTGATCCCCAAAGG | (close to Os01g45310) | ||||
| Os01g48270 | F: AGGCAAGCATTGGAAATAGG | 210 | 55 | 1; 27.6 | AAA-type ATPase family |
| R: TGGTAACAATCGCACCTTGA | protein, putative, expressed | ||||
| Os01g57310 | F: CGATGAACTGGAACACCATGA | 290 | 57 | 1; 33.1 | rp1-like protein, putative, expressed |
| R: GGCAACAGAGCCATACTTTGA | |||||
| Os01g72550 | F: CCG AGT TCA GGC GAG TGT TC | 382 | 55 | 1; 42.4 | OsCML19-Calmodulin- |
| R: TCA TTG TTT GGC ACT CCT CG | Related calcium sensor protein | ||||
| Os02g48500 | F: AGGCAATGGAGCACCAAGTT | 277 | 57 | 2; 29.6 | hypothetical protein |
| R:TTGTACTGTTGGGGTTGGCA | |||||
| Os04g22440 | F: ATCCTCGATGACACCGACCT | 352 | 57 | 4; 12.6 | hypothetical protein |
| R: TGCCCTTGGTAACTTGCTTCT | |||||
| Os04g30430 | F: GCTTCTCCTGGTTGTATGC | 163 | 52 | 4: 18.0 | nuclear transport factor 2, |
| R:AAAATAGGGAGGCAGATAGAC | putative, expressed | ||||
| 4gAL606639 | F: TTTTGTGAAACTTGACCCTC | 112 | 52 | 4; 28.8 | intergenics |
| R: GCGTCCATGTCTTTATTGTG | (close to OS04g48750) | ||||
| 5gAC137622 | F: CTCGCTGTTTACTGACTGG | 155 | 52 | 5; 13.8 | intergenics |
| R: TTTGATGTACTGCCTGCTCT | |||||
| Os06g11600 | F: TGCTGTGGGGCCTCTAATGA | 211 | 55 | 6; 6.1 | growth regulator related |
| R: TGAGACAACACCCACCCACC | protein, putative, expressed | ||||
| Os06g51110 | F: GAT GGC AAA CAC CAA CAG GA | 340 | 55 | 6; 30.9 | cyclin, putative, |
| R: GAG GGT TGG TTT GCC AGT GT | expressed | ||||
| Os07g26740 | F: GGGGAAGCGTCGTTATGACC | 259 | 55 | 7; 15.4 | 60 S ribosomal protein L44, |
| R: CCTTGATCGGGTGCTGAGAG | putative, expressed | ||||
| Os07g027000 | F: GCGCACTGTGATGCAAGATG | 358 | 55 | 7; 15.6 | retrotransposon protein |
| R: GACCTTGTCGGGATGTGCAG | putative, unclassified | ||||
| Os07g27590 | F: CTGTTGAAGGGGAGGAGCGT | 244 | 55 | 7;16.1 | retrotransposon protein, |
| R: TACGGTGCACTTCGGTCGTC | putative, unclassified | ||||
| Os08g41690 | F: TGCGAGGATGGAGTTCTTGA | 250 | 55 | 8; 26.3 | expressed protein |
| R: CAATCCCTTCACCAGAAGGAC | |||||
| Os08g41950 | F: GTC AGC CTG AAG TGC AGC AG | 418 | 55 | 8; 26.5 | OsMADS7 - MADS-box |
| R: CGG CAC CAC ATA TAT GCC AC | family with MIKCc type- | ||||
| box, expressed | |||||
| Os09g08960 | F: GGACTGAAAACACGATCGCA | 280 | 55 | 9; 4.7 | retrotransposon protein, |
| R: TTTTGGGGATCATCATCGACT | putative, unclassified | ||||
| Os11g41390 | F: AAGAAAAATATCTATTGAGGAGTG | 178 | 52 | 11; 24.3 | hypothetical protein |
| R: GGAGGACCATAAATGACGG |
The 19 InDel markers produced the total of 127 alleles for all loci, ranging from 4 alleles (Os06g11600 and 4gAL606639) to 12 alleles (Os07g27590), with the average of 6.68 (Table 3). Genetic diversity values ranged from 0.149 (Os06g51110) to 0.768 (Os04g22440), with the average of 0.476. Polymorphism information content (PIC) values ranged from 0.145 (Os06g51110) to 0.737 (Os04g22440), with the average of 0.440.
Table 3.
Number of unique alleles, Nei genetic diversity, heterozygosity and PIC values at each of InDel markers testing 101 rice accessions
| Markers | No. of allele | Nei Genetic diversity | Heterozygosity | PIC |
|---|---|---|---|---|
| 1gAP006530 | 6 | 0.629 | 0.598 | 0.573 |
| 1gAP003855 | 5 | 0.506 | 0.409 | 0.468 |
| Os01g48270 | 6 | 0.373 | 0.351 | 0.354 |
| Os01g57310 | 8 | 0.426 | 0.385 | 0.387 |
| Os01g72550 | 6 | 0.512 | 0.338 | 0.470 |
| Os02g48500 | 7 | 0.251 | 0.207 | 0.240 |
| Os04g22440 | 11 | 0.768 | 0.756 | 0.737 |
| Os04g30430 | 5 | 0.439 | 0.399 | 0.399 |
| 4gAL606639 | 4 | 0.583 | 0.531 | 0.533 |
| 5gAC137622 | 7 | 0.692 | 0.662 | 0.650 |
| Os06g11600 | 4 | 0.512 | 0.506 | 0.400 |
| Os06g51110 | 5 | 0.149 | 0.062 | 0.145 |
| Os07g26740 | 7 | 0.267 | 0.126 | 0.256 |
| Os07g27000 | 6 | 0.343 | 0.187 | 0.325 |
| Os07g27590 | 12 | 0.711 | 0.711 | 0.680 |
| Os08g41690 | 8 | 0.404 | 0.395 | 0.370 |
| Os08g41950 | 8 | 0.619 | 0.599 | 0.554 |
| Os09g08960 | 5 | 0.343 | 0.291 | 0.324 |
| Os11g41390 | 7 | 0.517 | 0.484 | 0.489 |
| Mean | 6.68 | 0.476 | 0.421 | 0.440 |
| Total | 127 | - | - | - |
Genetic diversity and genetic difference among groups of populations
Using 19 SSCP InDel markers, genetic diversity of Thai rice, IRRI germplasm, and other Oryza species were 0.436, 0.322, and 0.547, respectively (Table 4). To determine genetic difference among the three groups, we performed AMOVA and pairwise analyses. The AMOVA results showed that 15.06% of the variation was caused by differences among groups, while the remaining 84.94% was caused by differences within groups. The pairwise Fst estimates among these three groups indicated that all the three groups were significantly different from each other.
Table 4.
Summary of polymorphisms according to groups of populations
| Samples | Sample size | Mean no. of allele | Effective number of alleles | Mean genetic diversity | Exp. Heterozygosity | No. of polymorphic loci |
|---|---|---|---|---|---|---|
| All | 100 | 5.8 | 1.9 | 0.419 | 0.421 | 19 |
| 1. Thai rice lines | 38 | 3.2 | 2.0 | 0.436 | 0.441 | 17 |
| -Thai improved cultivars | 11 | 2.3 | 1.8 | 0.367 | 0.383 | 15 |
| -Thai breeding lines | 16 | 2.7 | 2.0 | 0.448 | 0.464 | 17 |
| -Thai landraces | 11 | 2.4 | 1.9 | 0.398 | 0.417 | 17 |
| 2. IRRI rice lines | 57 | 3.7 | 1.7 | 0.322 | 0.324 | 18 |
| -IRRI improved cultivars | 24 | 2.6 | 1.7 | 0.328 | 0.334 | 16 |
| - IRRI breeding lines | 33 | 3.2 | 1.6 | 0.298 | 0.302 | 18 |
| 3. The other Oryza species | 5 | 3.1 | 2.9 | 0.547 | 0.638 | 16 |
Because IRRI germplasm was used to improve rice breeding in Thailand, in this study we tested for genetic diversity and genetic difference among the groups of Thai and IRRI rice samples to determine the effects of each classification, such as improved cultivars, breeding lines, and landraces. The results showed that genetic diversity of Thai rice were 0.367, 0.448 and 0.398 for improved cultivars, breeding lines, and landraces, respectively. On the other hand, genetic diversity of IRRI improved and breeding lines were 0.327 and 0.297, respectively (Table 4). To determine genetic difference among the six groups, which are Thai improved cultivars, Thai breeding lines, Thai landraces, IRRI improved cultivars, IRRI breeding lines, and the other Oryza species, the AMOVA and pairwise analyses were performed. The AMOVA results showed that 13.04% of the variations were caused by differences among groups, while the remaining 86.96% were caused by differences within groups. Pairwise Fst estimates among groups ranged from 0.029 to 0.349. There were no significant difference among Thai improved cultivars, Thai local breeding lines and Thai landraces. Similarly, there were no significant difference between IRRI improved and IRRI breeding lines. However, all Thai groups, classified as Thai improved cultivars, Thai local breeding lines and Thai landraces were significantly different from IRRI improved and IRRI breeding lines (Table 5).
Table 5.
Pairwise population differentiation according to groups of populations as measured by F st
| Populationsa | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 0.000 | |||||
| 2 | 0.029 ns | 0.000 | ||||
| 3 | 0.049 ns | 0.038 ns | 0.000 | |||
| 4 | 0.254** | 0.233* | 0.258** | 0.000 | ||
| 5 | 0.089* | 0.153** | 0.142** | 0.349** | 0.000 | |
| 6 | 0.093* | 0.143** | 0.180** | 0.346** | 0.035 ns | 0.000 |
a 1)Thai improved cultivars, 2)Thai breeding lines, 3)Thai landraces, 4) The other Oryz a species 5)IRRI improved cultivars, and 6) IRRI breeding lines.
** Significant at P < 0.001, * Significant at P < 0.05, ns Not Significant.
Clustering of rice accessions using SSCP InDel markers
The UPGMA cluster diagram differentiated 4 species of other Oryza species and showed two major groups that correspond to the Indica and Japonica subspecies (Figure 1). Group (G) I was Japonica rice. Using Nipponbare as a representative for the temperate Japonica and Azucena (IRRI breeding line) as a representative for the tropical Japonica, five Thai and one IRRI accessions were grouped with Japonica rice lines by clustering closer to Azucena. All the Thai accessions in this group are upland rice. Group (G) II were clusters of Indica, which includes four sub-groups: GII-1, GII-2, GII-3, and GII-4, and additional two isolated single accessions. The GII-1 had 11 accessions which were all Thai landraces and Thai local breeding lines, except O. rufipogon, and one improved cultivars (No. 8, SRN1). KDML105 (No. 100), which is the famous Thai jasmine rice, was also sorted into this group by clustering with its derivative (No. 10, RD15) and a Thai landrace (No. 37, KD). The GII-2 had eight accessions that were all TGMS types, with the exception of No. 29. The GII-3 was the largest group with 60 accessions, most of which were IRRI germplasm. It should be noted that seven of the Thai improved lines and three Thai breeding lines were in this group (Figure 1). The GII-4 had eight accessions, which majority were deep water lines. The two isolated single accessions were Thai irrigated line (improved, No. 6, PTT1) and Thai rainfed central plain (breeding, No. 13, LPT123).
Figure 1.
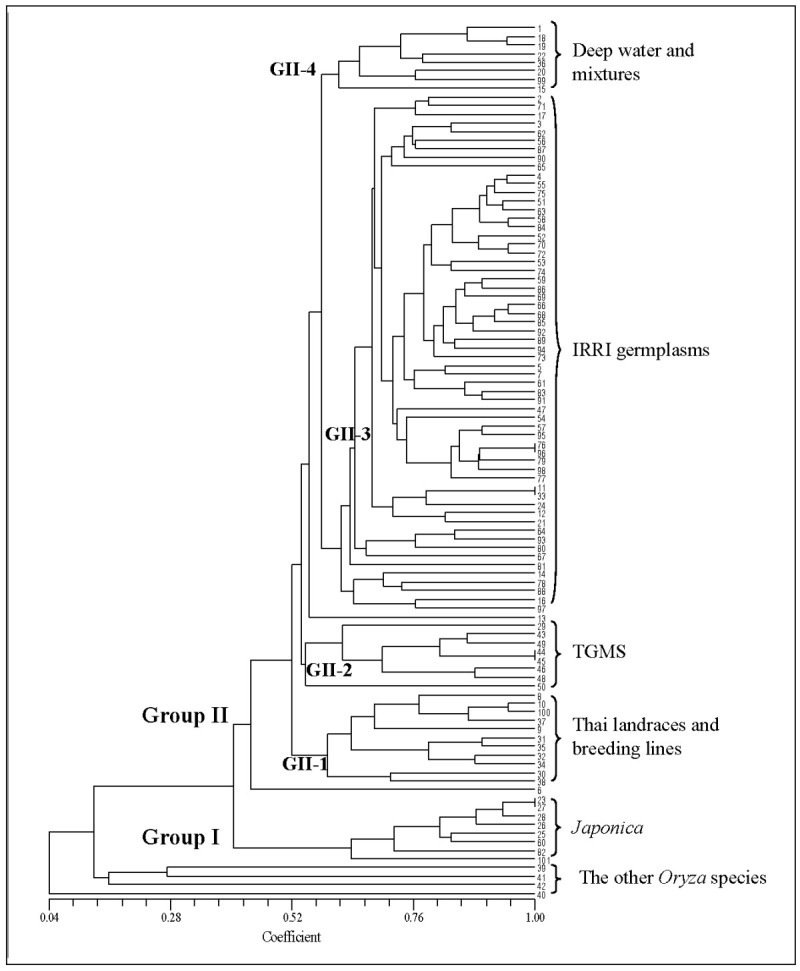
Cluster diagram based on the Dice genetic similarity matrix by UPGMA analysis calculated from alleles of 101 rice accessions detected by 19 SSCP InDel markers; Number followed details in Table 1 ; 1-26 and 100 are commercial Thai rice lines; 27-37 are landraces with selected traits; 38-42 are the other Oryza speciese; 43-99 are germplasm having desirable traits obtained from IRRI; 101 is Nipponbare.
Genetic structure and differentiation
Using the data of 19 polymorphic InDel markers, the model-based method was performed to determine the genetic structure among all 101 samples. The Bayesian-based clustering method demonstrated that the highest log likelihood score was obtained when the number of populations (k) was equal to six. The population structure based on the k = 6 showed similar results to the UPGMA tree (Figure 1) by sorting rice into 6 different color-coded groups, which are Japonica (red), TGMS (green), deep water and mixtures (blue), IRRI germplasm (yellow), the other Orzya species (pink), and Thai landraces and breeding lines (light blue) (Figure 2). The first group in red, Japonica, had the Q values ranging from 0.91 to 0.989, except Azucena, No. 82 (Q = 0.795). All Thai rice in this group was upland rice. The second group in green contained all TGMS lines with the Q-values ranged from 0.87 to 0.985. The TGMS line, tms2 KDML105 (No. 50), was not in this group but it had the highest Q value (Q = 0.369) with group of the other Oryza species. The third group in blue had the Q values ranging from 0.808 to 0.985. The majority of accessions in this group were Thai deep water rice. The fourth group was the largest group (shown in yellow) with Q values ranging from 0.812 to 0.984, most of which were IRRI germplasm, except three Thai improved cultivars from irrigated area(No. 2, 4 and 5). The fifth group in pink contained all the other Oryza species with their Q-values higher than 0.98, except for the O. rufipogon (No. 38) that was grouped with Thai landraces. The last group in light blue had the Q values ranging from 0.811 to 0.987. Four out of the eight accessions in this group were Thai landraces (No. 30, 31, 35, 37), while the remaining accessions were all Thai commercial lines from the Northeast.
Figure 2.
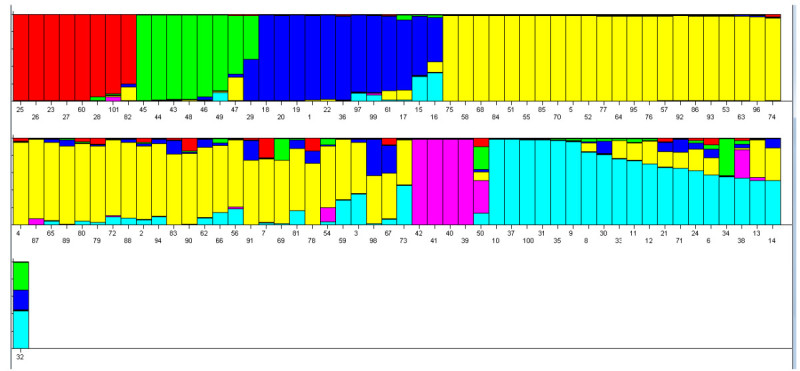
Estimated population structure using k = 6; individual rice line is represented by a vertical bar broken into colored segments, with lengths in proportion to Q values: red, Japonica ; green, TGMS; dark blue, Deep water rice and mixtures; yellow, IRRI germplasms; pink, the other Oryza species; light blue, Thai landraces and breeding lines. The numbers marked below each line indicate the rice accession numbers as shown in details in Table 1; 1-26 and 100 are commercial Thai rice lines; 27-37 are landraces with selected traits; 38-42 are the other Oryza species; 43-99 are germplasm with desirable traits from IRRI; 101 is Nipponbare.
To determine genetic diversity and genetic difference in the rice samples according to population structure, we re-analyzed the data according to the population structure in Figure 2. The results showed that genetic diversity of Japonica, TGMS, deep water and mixtures, IRRI germplasm, the other Oryza species, and Thai landraces and breeding lines were 0.256, 0.239, 0.303, 0.255, 0.565, and 0.355, respectively (Table 6). The AMOVA results showed that 35.28% of the variations were caused by differences among groups, while the remaining 64.72% were caused by differences within groups. Pairwise Fst estimates among groups ranged from 0.204 to 0.680, and indicated that these groups were significantly different from each other.
Table 6.
Summary of polymorphisms for 6 groups of populations according to population structure in Figure 2
| Samples | Sample size | Mean no. of allele | Effective number of alleles | Mean genetic diversity | Exp. Heterozygosity | No. of polymorphic loci |
|---|---|---|---|---|---|---|
| All | 100 | 5.8 | 1.9 | 0.419 | 0.421 | 19 |
| 1) Japonica ( red) | 8 | 2.1 | 1.5 | 0.256 | 0.273 | 14 |
| 2) TGMS ( green) | 8 | 1.7 | 1.5 | 0.239 | 0.257 | 11 |
| 3) Deep water ( deep blue) | 12 | 2.2 | 1.7 | 0.303 | 0.316 | 13 |
| 4) IRRI germplasms ( Yellow) | 48 | 2.9 | 1.5 | 0.255 | 0.258 | 15 |
| 5) The other Oryza species ( Pink) | 5 | 3.1 | 3.0 | 0.565 | 0.661 | 16 |
| 6) Thai landraces and breeding lines (blue) | 19 | 2.6 | 1.8 | 0.355 | 0.365 | 14 |
Discussion
Molecular characterization is the alternative approach to overcome several limitations of morphological characterization, which are high experimental cost, long evaluation time, and environmental effects. We reported genetic analysis in rice using groups of SSCP InDel markers, most of which were developed from putative rice genes containing short InDel. The SSCP technique can overcome the limitation of the SSR technique, which could not distinguish different DNA sequences when the DNA fragments are of the same length. DNA separation on SSCP gels is based on both size and conformation, which is determined by the DNA primary structure. Typically, single-copy amplifications showed the two-band SSCP profile, indicating a separation of sense and anti-sense strands.
Previously, SSR have been used to determine genetic variation in rice. The reported number of allele per locus, genetic diversity, and Polymorphism information content (PIC) were ranged from 4.8-14.0, 6.2-6.8 and 0.63-0.70, respectively (Ni et al. [2002]; Garris et al. [2005]; Pessoa-Filho et al. [2007]; Ram et al. [2007]). Very recenly, Ali et al. ([2011]) genotyped 409 Asian rice accessions originated from 79 countries representing all the major rice growing regions of the world using 36 SSR markers. They reported an average of 9.17 alleles per marker (range from 2 to 24), a mean genetic diversity of 0.68, and an average PIC of 0.63. In addition, Chen et al. ([2011]) studies genetic diversity of 300 rice accessions representing major geographic areas of rice growing countries in the world using 372 SNP markers. They detected 744 alleles at 372 markers, an average gene diversity of 0.358, and an average PIC of 0.285. Using 19 SSCP InDel markers to determine genetic variation in 101 rice accessions, we found that the 19 markers produced the average number of allele of 6.68, and the average PIC of 0.44. Our results on average allele per locus, genetic diversity, and PIC were higher than that reported by the study using SNP marekers (Chen et al., [2011]). Comparing to other studies using SSR, our result on the average number of allele per locus was comparable to several studies (Ni et al. [2002]; Yu et al. [2003]). However, the average PIC produced by our method was lower than that reported by the other studies using SSR, concordant with the earlier study in pearl millet, which reported the average PIC value of 0.49 by SSCP relative to the SSR value of 0.72 tested on the same genotype panel (Bertin et al. [2005]).
The subset of 19 markers selected from the total of 98 markers provided the resulting groups corresponding to the Indica and Japonica subspecies, and correlated quite well with their ecologies and their known information on their development. Previous study showed that genetic diversity was also determined in genus Orysza using 11 ISSR markers selected from 30 ISSR markers (Joshi et al. [2000]). Small numbers of markers can be used to estimate genetic diversity as shown in the earlier study when a subset of 30 markers provided the same results as using all 111 markers with the same genetic distance matrices and dendrograms (Ni et al. [2002]). Similarly, Ali et al. ([2011]) also showed that a subset of 36 SSR markers gave nearly the same results as using 169 SSR markers for population structure analysis.
Here we showed that SSCP InDel markers can be used to study plant breeding. SSCP Indel gene-based markers are very specific and can utilize their known positions in the rice genome. In addition, SNP and InDel are abundant in rice (Feltus et al. [2004]; Shen et al. [2004]; Chen et al., [2011]), which allows for the development of InDel markers even in small target areas.
Our results from the analysis of the 3 main rice groups showed that the other Oryza species had the highest genetic diversity, followed by Thai rice lines and IRRI germplasm. Similar to the study using Indian germplasm, the genetic diversity of the other Oryza species in this report is higher than that of the cultivated rice (Ram et al. [2007]). However, the genetic diversity of the other Oryza species used in this report (0.55) is higher than the genetic diversity of the 7 wild rice species (0.436) reported by Ram et al. ([2007]). Our results indicated that Oryza brachyantha, No. 40, (FF genome) is the most divergent species among the other Oryza species, which is also in agreement with reports from previous studies (Joshi et al. [2000], Jacquemin et al. [2009]; Lu et al. [2009]). Similarly, Oryza officinalis No. 42, (CC genome) and Oryza latifolia No. 41, (CCDD genome) are grouped together, and separated from Oryza brachyantha, supporting that they are in the officinalis complex, including diploid CC and tetraploid CCDD genomes(Joshi et al. [2000]).
The selected IRRI germplasm (including 57 rice accessions from several countries) showed lower genetic diversity than that of the Thai commercial cultivars (improved and breeding lines) and Thai landraces. The Thai breeding lines had the highest genetic diversity while the IRRI breeding lines had the lowest. Interestingly, genetic diversity of Thai landraces was lower than that of Thai breeding lines, possibly because the Thai landraces were selected only from the North and the Central parts of the country, while the breeding lines were from all over Thailand.
Our cluster analysis showed two major groups of Oryza sativa corresponding to the Indica and Japonica subspecies, similar to other studies (Wen et al. [2009]; Chen et al., [2011]). Both cluster and structure analyses separated a group of other Oryza species and showed that O. rufipogon (No. 38), which is considered the progenitor of Oryza sativa, was most related to Oryza sativa. Interestingly, the famous Thai jasmine rice, KDML105 (No. 100) was grouped with Thai landraces and breeding lines, and O. rufipogon, which indicates that the strain is native to Thailand. Our analyses support the existences of five subpopulations of Oryza sativa, similar to earlier studies (Garris et al. [2005]; Zhao et al. [2010], Ali et al., [2011]; Chen et al., [2011]). Several rice accessions included in our study were used in the report by Zhao et al. ([2010]), and classifications of their subpopulations were concordant. However, we did not have known samples of aus and Group V subpopulations (Zhao et al. [2010]) in this study, thus we can not indicate if some of our subpopulations were aus and Group V. Some of Thai upland rice lines and one IRRI germplasm were grouped with a well known tropical Japonica, Azuzena, which further suggests that these upland rice lines are also tropical Japonica. All irrigated rice lines, which were improved cultivars, have genetic backgrounds of IRRI germplasm supporting the results of cluster and structure analyses. The results from diversity analysis showed that the Thai germplasm were more diverse than the tested IRRI germplasm, concordant with the results from structure analysis.
Although Thailand is quite small, 513115 sq. km. (approximately the same size as France), different ecologies and geography in distinct parts of the country could have some effects on our rice diversity. The first three of the four rice ecologies: irrigated, deep water, upland, and rainfed lowland ecosystems, showed distinct groups for both cluster and structure analyses. The irrigated rice lines, planted in irrigated areas in the Central plain, were grouped together and showed similar genetic structure to IRRI germplasm. Most of the tested upland rice lines were planted in hilly areas in the North, and they were grouped with Japonica rice. Deep water rice lines were planted in specific areas having high levels of water in the Central plain, and they were clustered together in a distinct group. Interestingly, three out of the four rainfed lowland rice lines from the South showed genetic structure that is similar to the deep water rice. In addition, groups that were sorted by population structure also displayed significant genetic difference among them.
All tested TGMS lines controlled by 3 tgms genes were grouped together by cluster analysis into one sub-group of Indica. Results from structure analysis also supported this information, with the exception of tms2 KDML105, which showed genetic structure similar to the group of the other Oryza species. Results from cluster and structure analyses indicated that tms2 KDML105 (No. 50) was quite distinctive from the other TGMS lines, and it contained several different genetic fragments, including the other Oryza species (Q = 0.369), TGMS (Q = 0.266), Thai landraces and breeding lines (Q = 0.141), and IRRI germplasm (Q = 0.101), in agreement with its genetic background containing some part of Japonica genome (Pitnjam et al. [2008]), and its development (Lopez et al. [2003]). C21489 (No. 29), a cold-resistance, Thai landrace with no available information on sterility, is also clustered with the TGMS sub-group. However, the structure analysis showed that it had lower Q value.
Conclusions
Our study showed the utility of SSCP InDel markers for genetic analysis of Thai and IRRI rice germplasm, as alternative to SSR markers. The resulting genetic structure and differentiation of these samples were in agreement with their ecologies and their known information on their development. The results indicated that genetic diversity of Thai commercial rice lines (improved cultivars and local breeding lines) and Thai landraces were higher than that of the tested rice germplasms obtained from IRRI. Our molecular analysis indicated that some of our cultivars were japonica rice, and genetic diversity is present in this set of the available germplasms. Differentiation analysis indicated that groups of IRRI germplasm were significantly different from Thai groups. Thus, these germplasm can be used to broaden the genetic base and trait improvements in rice breeding programs. Cluster and structure analyses showed concordant results having six distinct groups, and differentiation analysis supported that they were significantly different from each other. The results also indicate that TGMS lines which could be used as female parents were different from the other groups making them good candidates used to create rice hybrids having high yields. Genetic diversity and genetic relationship among these germplasms will be useful for parental line selection used in rice breeding programs and in hybrid production.
Methods
Plant materials
A total of 101 rice accessions, which include 43 Thai accessions, 57 germplasm obtained from the International Rice Research Institute (IRRI), and Nipponbare were used in this study (Table 1). The Thai accessions included 27 commercial cultivars, 11 landraces, and 5 other Oryza species. The commercial cultivars were cultivars grown in all rice ecologies through out the country such as irrigated area, rain fed low land, up land, and deep water. These commercial cultivars were 11 improved and 16 local breeding lines. The improved lines were cultivars with high yielding and/or cultivars with agronomic desirable traits. The improved cultivars were classified by their development through crossing among local cultivars and/or with other genetic sources, and their recent pedigrees were known (Table 1). The local breeding cultivars had not been bred through modern breeding procedures and their precise pedigrees were unknown. The landraces were local lines, which have not been planted for commercialization, but they have different special traits (Table 1). The other Oryza species included O.glaberrimma, African cultivated rice, and four species of wild rice: O. rufipogon, O.branchyantha, O. latifolia, and O. officcinalis. The rice accessions obtained from IRRI were groups of germplasm having agronomic desirable traits, including temperature-sensitive genetic male sterility (TGMS), new plant type, early flowering, and biotic and abiotic stress resistances (Table 1). Two accessions, KDML 105 (the premium jasmine rice) and Nipponbare, which are well known Indica and Japonica rice respectively, were used as controls for genetic diversity analysis and as references for control of allele sizing variation between electrophoresis runs. Nipponbare was not included in genetic diversity and pairwise analysis aimed to compare Thai and IRRI germplasm. Genetic structure of the Thai accessions is very limited, but most of them were classified by morphology as Indica rice. Young leaves from 3–5 mature plants of each accession grown in greenhouse were collected for DNA isolation using the CTAB method (Murray and Tompson [1980]).
Marker development and SSCP analysis
A total of 98 InDel (Insertion-Deletion) markers were developed for the SSCP analysis, most of which were gene-based markers. Some of the 98 markers were reported by Pitnjam et al. ([2008]) and Shen et al. ([2004]). Other markers are from the 48 genes distributed over the entire rice genome, 4 genes per chromosome. According to information in the rice databases (GRAMENE http://www.gramene.org/; and http://shenghuan.shnu.edu.cn/ricemarker, last accessed on December, 2011), primers were designed using the Primer 3 program (http://biowb.sdsc.edu/CGI/BW.cgi) to amplify regions of the genes containing at least two SNPs. To increase the possibility of identifying polymorphisms between rice accessions and to facilitate polymorphism observation using SSCP, sizes of the PCR products amplified from these primers were about 200–300 bp. The markers were first used to test polymorphism among 4 rice accessions (KDML105, c20878, Nipponbare, and O. rufipogon) representing indica commercial Thai rice, landrace, japonica and other Oryza species. The markers show polymorphism in at least 3 rice accessions and shown the presence of only single copy gene were selected to test the whole set of rice samples. Details of the selected InDel markers used in this study were shown in Table 2. Amplification and polymorphism detection using SSCP were performed as previously described (Pitnjam et al. [2008]).
Data analysis
Data were entered in the form of single-individual genotypes. Then, Program POPGENE Version 1.32 was used to calculate number of alleles per locus, average heterozygosity, and genetic diversity (Nei genetic diversity) for each maker (Yeh et al. [1999]). The informativeness of markers was assessed by calculating polymorphic information content (PIC; Botstein et al. [1980]) using PowerMarker V3.0 (Liu and Muse [2005]). The genetic diversity level within group of rice accessions was analyzed by setting up parameters as 3 {Thai rice lines, IRRI germplasm, and other Oryza species}, and 6 {Thai landraces, Thai improved cultivars, Thai breeding lines, IRRI improved cultivars, IRRI breeding lines, and other Oryza species} sources of germplasm. In addition, pairwise genetic distances between rice accessions were calculated based on the similarity coefficients following Nei and Li ([1979]). The resulting similarity matrices were used to construct dendrogram by the unweighted pair-group method with arithmetic means (UPGMA) though NTSYS-pc version 2.0 (Rohlf [1998]). Bayesian-based clustering method of analysis using the software package STRUCTURE (Prichard et al. [2000]) was performed to infer the number of populations (k) required for accurate data interpretation without prior information on the number of groups of accessions at which the individuals were studied. The membership probabilities (Q) calculated from STRUCTURE ≥ 0.80 were used to assign rice accessions to clusters. Rice accessions with membership probabilities < 0.80 for all clusters were used to detect possible genetic exchanges among groups of rice accessions. In order to describe the population genetic structure and variability among populations, the Analysis of Molecular Variance (AMOVA) was performed using the ARLEQUIN 3.11 software (Excoffier et al. [2005]). The total variance was partitioned among individuals within the same populations as well as among different populations. The permutational procedure was then used to provide significant tests for each of the hierarchical variance components based on the original distance matrices (used 1000 permutation). Wright’s inbreeding coefficient (FST) values calculated by AMOVA were used as inter-population genetic distance measurements as described by Huff ([1997]).
Acknowledgements
We thank International Rice Research Institute (IRRI), Pathum Thani Rice Research Institute, Dr. Theeayut Toojinda, and Dr. Suriyan Chaum, for providing seeds of rice accessions used in this study. We also thank colleagues and friends for their critical reading and comments on the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/1939-8433-5-19) contains supplementary material, which is available to authorized users.
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
SC and KP performed genotyping. SC, WS and KU were involved in data analysis. AM designed the study, interpreted the results and prepared the manuscript. All authors read and approved the final manuscript.
Contributor Information
Sriprapai Chakhonkaen, Email: sriprapai.cha@biotec.or.th.
Keasinee Pitnjam, Email: keasinee.pit@biotec.or.th.
Wachira Saisuk, Email: Wachira.sas@thammasat.net.
Kittipat Ukoskit, Email: ku@tu.ac.th.
Amorntip Muangprom, Email: amorntip.mua@biotec.or.th.
References
- Ali ML, McClung AM, Jia MH, Kimball JA, McCouch SR, Georgia CE. A Rice Diversity Panel Evaluated for Genetic and Agro-Morphological Diversity between Subpopulations and its Geographic Distribution. Crop Sci. 2011;51:2021–2035. doi: 10.2135/cropsci2010.06.0359. [DOI] [Google Scholar]
- Baack EJ, Sapir Y, Chapman MA, Burke JM, Rieseberg LH. Selection on domestication traits and quantitative trait loci in crop–wild sunflower hybrids. Mol Ecol. 2008;17:666–677. doi: 10.1111/j.1365-294X.2007.03596.x. [DOI] [PubMed] [Google Scholar]
- Bertin I, Zhu JH, Gale MD. SSCP-SNP in pearl millet—a new marker system for comparative genetics. Theor Appl Genet. 2005;110:1467–1472. doi: 10.1007/s00122-005-1981-0. [DOI] [PubMed] [Google Scholar]
- Borchert T, Hohe A. Identification of molecular markers for the flower type in the ornamental crop Calluna vulgaris. Euphytica. 2009;170:203–213. doi: 10.1007/s10681-009-9926-3. [DOI] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Castelblanco W, Fregene M. SSCP-SNP-based conserved ortholog set (COS) markers for comparative genomics in cassava (Manihot esculenta crantz) Plant Mol Biol Rep. 2006;24:229–236. doi: 10.1007/BF02914061. [DOI] [Google Scholar]
- Chen H, He H, Zou Y, Chen W, Yu R, Liu X, Yang Y, Gao Y, Xu J, Fan L, et al. Development and application of a set of breeder-friendly SNP markers for genetic analyses and molecular breeding of rice (Oryza sativa L.) Theor Appl Genet. 2011;123:869–879. doi: 10.1007/s00122-011-1633-5. [DOI] [PubMed] [Google Scholar]
- Chitrakon S, Somrith B. The development of Thai rice varieties. In: Lorlowhakarn S, editor. Science and technology with Thai rice. 2003. pp. 29–76. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH. An SNP Resource for Rice Genetics and Breeding Based on Subspecies Indica and Japonica Genome Alignments. Genome Res. 2004;14:1812–1819. doi: 10.1101/gr.2479404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic Structure and Diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieu F, Joseph D, Norman P, Iacopetta B. Development of a rapid genotyping method for single nucleotide polymorphisms and its application in cancer studies. Oncol Rep. 2004;11:501–504. [PubMed] [Google Scholar]
- Hashimoto Z, Mori N, Kawamura M, Ishii T, Yoshida S, Ikegami M, Takumi S, Nakamura C. Genetic diversity and phylogeny of Japanese sake-brewing rice as revealed by AFLP and nuclear and chloroplast SSR markers. Theor Appl Genet. 2004;109:1586–1596. doi: 10.1007/s00122-004-1794-6. [DOI] [PubMed] [Google Scholar]
- Huff DR. RAPD characterization of heterogeneous perennial ryegrass cultivars. Crop Sci. 1997;37:557–564. doi: 10.2135/cropsci1997.0011183X003700020041x. [DOI] [Google Scholar]
- Ishii T, Hiraoka T, Kanzaki T, Akimoto M, Shishido R, Ishikawa R. Rice. 2011. Evaluation of Genetic Variation Among Wild Populations and Local Varieties of Rice. [Google Scholar]
- Jacquemin J, Laudié M, Cooke R. A recent duplication revisited: phylogenetic analysis reveals an ancestral duplication highly-conserved throughout the Oryza genus and beyond. BMC Plant Biol. 2009;9:146. doi: 10.1186/1471-2229-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar SD. Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet. 2000;100:1311–1320. doi: 10.1007/s001220051440. [DOI] [Google Scholar]
- Kuhn DN, Motamayor JC, Meerow AW, Borrone JW, Schnell RJ. SSCP markers provide a useful alternative to microsatellites in genotyping and estimating genetic diversity in populations and germplasm collections of plant specialty crops. Electrophoresis. 2008;29:4096–4108. doi: 10.1002/elps.200700937. [DOI] [PubMed] [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lopez MT, Toojinda T, Vanavichit A, Tragoonrung S. Microsatellite markers flanking the tms2 gene facilitated tropical TGMS rice line development. Crop Sci. 2003;43:2267–2271. doi: 10.2135/cropsci2003.2267. [DOI] [Google Scholar]
- Lu Y, Curtiss J, Percy RG, Hughs SE, Yu S, Zhang J. DNA Polymorphisms of Genes Involved in Fiber Development in a Selected Set of Cultivated Tetraploid Cotton. Crop Sci. 2009;49:1695–1704. doi: 10.2135/cropsci2008.12.0693. [DOI] [Google Scholar]
- Moose SP, Mumm RH. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008;147:969–977. doi: 10.1104/pp.108.118232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muangprom A, Thomas SG, Sun TP, Osborn TC. A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol. 2005;137:931–938. doi: 10.1104/pp.104.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Tompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonuclease. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Colowit PM, Mackill DJ. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci. 2002;42:601–607. doi: 10.2135/cropsci2002.0601. [DOI] [Google Scholar]
- Pessoa-Filho M, Beló A, Alcochete AA, Rangel PH, Ferreira ME. A set of multiplex panels of microsatellite markers for rapid molecular characterization of rice accessions. BMC Plant Biol. 2007;7:23. doi: 10.1186/1471-2229-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnjam K, Chakhonkaen S, Toojinda T, Muangprom A. Identification of a deletion in tms2 and development of gene-based markers for selection. Planta. 2008;228:813–822. doi: 10.1007/s00425-008-0784-3. [DOI] [PubMed] [Google Scholar]
- Prichard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Wolters P, Caldwell KH, Powell W. High-resolution organellar genome analysis of Triticum and Aegilops sheds new light on cytoplasm evolution in wheat. Theor Appl Genet. 2004;108:1182–1190. doi: 10.1007/s00122-003-1538-z. [DOI] [PubMed] [Google Scholar]
- Ram SG, Thiruvengadam V, Vinod KK. Genetic diversity among cultivars, landraces and wild relatives of rice as revealed by microsatellite markers. J Appl Genet. 2007;48:337–345. doi: 10.1007/BF03195230. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. NTSYS- pc: Numerical taxonomy and multivariate analysis system, version 2.0,Exeter Software. New York: Setauket; 1998. [Google Scholar]
- Rousseau-Gueutin M, Gaston A, Aïnouche A, Aïnouche ML, Olbricht K, Staudt G, Richard L, Denoyes-Rothan B. Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): New insights from phylogenetic analyses of low-copy nuclear genes. Mol Phylogenet Evol. 2009;51:515–530. doi: 10.1016/j.ympev.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Shen Y, Jiang H, Jin JP, Zhang Z-B, Xi B, He Y-Y, Wang G, Wang C, Qian L, Li X, Yu QB, Liu HJ, Chen DH, Gao JH, Huang H, Shi TL, Yang ZN. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 2004;135:1198–1205. doi: 10.1104/pp.103.038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnucks P, Wilson AC, Beheregaray LB, Zenger K, French J, Taylor AC. SSCP is not so difficult: the application and utility of single-stranded conformation polymorphism in evolutionary biology and molecular ecology. Mol Ecol. 2000;9:1699–1710. doi: 10.1046/j.1365-294x.2000.01084.x. [DOI] [PubMed] [Google Scholar]
- Thomson MJ, Septiningsih EM, Suwardjo F, Santoso TJ, Silitonga TS, McCouch SR. Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers. Theor Appl Genet. 2007;114:559–568. doi: 10.1007/s00122-006-0457-1. [DOI] [PubMed] [Google Scholar]
- Wen W, Mei H, Feng F, Yu S, Huang Z, Wu J, Chen L, Xu X, Luo L. Population structure and association mapping on chromosome 7 using a diverse panel of Chinese germplasm of rice (Oryza sativa L.) Theor Appl Genet. 2009;119:459–470. doi: 10.1007/s00122-009-1052-z. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Yang RC, Boyle T. POPGENE version1.32, Microsoft window-base software for population genetic analysis: a quick user’s guide. Alberta, Canada: University of Alberta, Center for International Forestry Research; 1999. [Google Scholar]
- Yu SB, Xu WJ, Vijayakumar CH, Ali J, Fu BY, Xu JL, Jiang YZ, Marghirang R, Domingo J, Aquino C, Virmani SS, Li ZK. Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theor Appl Genet. 2003;108:131–140. doi: 10.1007/s00122-003-1400-3. [DOI] [PubMed] [Google Scholar]
- Zhang P, Li J, Li X, Liu X, Zhao X, et al. Population Structure and Genetic Diversity in a Rice Core Collection (Oryza sativa L.) Investigated with SSR Markers. PLoS One. 2011;6:e27565. doi: 10.1371/journal.pone.0027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wright M, Kimball J, Eizenga G, McClung A, Kovach M, Tyagi W, Ali ML, Tung CW, Reynolds A, Bustamante CD, McCouch SR. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS One. 2010;24(5):e10780. doi: 10.1371/journal.pone.0010780. [DOI] [PMC free article] [PubMed] [Google Scholar]


