Abstract
Background
Oligopeptide transporters (OPTs) play important roles in the mobilization of organic nitrogenous compounds and usually associate with tissues that show signs of rapid protein hydrolysis, such as germinating seeds and senescing leaves. This study is to investigate rice OPT genes.
Results
A total of sixteen OsOPT genes (Os for Oryza sative L.) were identified in the rice genome, which were then classified into six sections that belong to two subfamilies (the PT and YSL subfamily). The major mechanisms for evolutionary expansion of the sixteen genes during the rice genome evolution include segmental and tandem duplication. Calculation of the duplication event dates indicated that the sixteen genes originated from nine original OsOPT genes, and the duplication events could be classified into three evolutionary stages. The first evolutionary stage occurred approximately 50 million years ago (Mya) and involved the evolution of four new genes. The second evolutionary stage was approximately 20 Mya and was marked by the appearance of two new genes, and the third evolutionary stage was approximately 9 Mya when two new genes evolved. Mining of the expression database and RT-PCR analysis indicated that the expression of most duplicated OsOPT genes showed high tissue specificities. Diverse expression patterns for the sixteen genes were evaluated using both semi-quantitative RT-PCR and the MPSS data. Expression levels of some OsOPT genes were regulated by abiotic and biotic stresses suggesting the potential involvement of these gene products in rice stress adaptation. Five OsOPT gene mutants showed abnormal development and growth, the primary analysis of five OsOPT gene mutants suggested that they may be necessary for rice development.
Conclusions
These results suggested that rice-specific OsOPT genes might be potentially useful in improving rice.
Electronic supplementary material
The online version of this article (doi:10.1186/1939-8433-5-12) contains supplementary material, which is available to authorized users.
Keywords: Oligopeptide transporters, Rice, Evolutionary expansion, Expression profile
Background
The acquisition and allocation of nitrogenous compounds are essential processes in plant growth and development (Williams and Miller [2001]). Peptide transport has been demonstrated to play important roles in these processes and is a commonly observed physiological phenomenon in both prokaryotes and eukaryotes (Higgins and Payne [1981]). Peptide transporters are characterized by the ability to transport a range in different types of peptides and their derivatives across membranes in an energy-dependent manner (Lubkowitz et al. [1997]; Lubkowitz et al. [1998]). The plant peptide transporters generally belong to three different gene families: the ABC-type transporter family, the peptide transporter family (PTR) and the oligopeptide transporter family (OPT) (Ouyang et al. [2010]; Rentsch et al. [2007]; Zhao et al. [2010]), each of which recognizes peptides of a specific length. In this study, we focused on the rice OPT family. The OPT family was first characterized in yeast (Lubkowitz et al. [1997]). Several other OPT genes have also been identified from other species, such as CaOPT1 from Candida albicans SpIsp4 from Schizosaccharomyces pombe, and ScOPT2 from Schizosaccharomyces cerevisiae (Lubkowitz et al. [1998]). However, to date the OPT family has only been found in fungi, bacteria, plants and archaea, but not in animals (Lubkowitz [2006]).
The OPT genes have been classified into two subfamilies by phylogenetic analysis: the PT and the YSL subfamily (Curie et al. [2001]). Most of the information regarding the PT subfamily in plants comes from the study of Arabidopsis. Nine orthologs of the PT subfamily were identified in Arabidopsis and were named sequentially from AtOPT1 to AtOPT9. Five (AtOPT1, 4, 5, 6, and 7) of the orthologs were found to be functional tetra- and penta-peptide transporters by yeast complementation assay (Koh et al. [2002]; Osawa et al. [2006]). However, reduced AtOPT3 expression resulted in decreased accumulation of iron in seeds while high levels resulted in the accumulation of iron in other tissues, suggesting that AtOPT3 functions to maintain whole-plant iron homeostasis and iron nutrition of developing seeds (Stacey et al. [2007]). AtOPT6 proteins have the ability to transport glutathione derivatives (Cagnac et al. [2004]) and mammalian signaling peptides up to 10 amino acids in length (Pike et al. [2009]). AtOPT promoter-GUS fusion analysis revealed differential expression of 8 AtOPT genes: 6 AtOPT genes (AtOPT1 AtOPT3 AtOPT4 AtOPT6 AtOPT7 and AtOPT8) were strongly expressed in vascular tissues, 3 (AtOPT1 AtOPT3 and AtOPT8) were uniquely expressed in pollens and one (AtOPT6) was expressed in ovules (Stacey et al. [2005]). Several OPT genes were also identified from other plants. BjGT1 from B. juncea was found to transport glutathione and has been implicated in mediating heavy-metal toxicity (Bogs et al. [2003]).
The YSL subfamily is usually considered to transport the metals (Inoue et al. [2008]; Lee et al. [2009]). Iron (Fe) is an important micronutrient for living organisms (Bashir et al. [2010]). Maize yellow stripe1 (ZmYS1) from Zea mays was demonstrated as a phytosiderophore transporter (Curie et al. [2001]; Roberts et al. [2004]; Schaaf [2004]). In rice, OsYSL2 (Koike et al. [2004]), OsYSL15 (Inoue et al. [2008]; Lee et al. [2009]) and OsYSL18 (Aoyama et al. [2009]) are similar to orthologs of ZmYSL1 in that they all have the ability to transport metal-nicotianamine. However, some members (OsOPT1 OsOPT3 OsOPT4 OsOPT5 and OsOPT7) of the PT subfamily in rice have been demonstrated to transport nicotianamine-bound iron (Vasconcelos et al. [2008]).
The roles of the OPT genes in plants, especially in rice, are still under investigation. To the best of our knowledge, no biofunctional data regarding the OsOPT genes are currently available. In eukaryotes, functional diversification follows the evolutionary expansion of a gene family. The evolutionary expansion of a gene family follows duplication events of an individual gene, the chromosomal segment, or the whole genome (Lynch and Conery [2000]; Moore and Purugganan [2005]; Xue and Fu [2008]). In this study, we identified the evolutionary expansion of the rice OsOPT family and showed function diversification of the duplicated genes by expression analysis and primary analysis of the mutants.
Results
Identification of 16 OsOPT gene homologues in the rice genome
A total of 17 putative OsOPT proteins have been identified from the rice genome database by BLASTP searching. However, one locus (LOC_Os02g46860) encodes a short protein (245aa) that was not found in NCBI; therefore, we considered it to be a pseudogene and did not analyze it in this study. The predicated OsOPT proteins were analyzed for Pfam matches in the Pfam database to confirm the 16 putative OsOPT genes (Table 1) as oligopeptide transporter-like homologues and to identify their domains. The analysis showed that all 16 putative OsOPT genes were homologous to the OPT family and their predicated proteins contained the SPYxEVRxxVxxxDDP domain, which is also found in other OPT proteins (Lubkowitz et al. [1998]; Wiles et al. [2006]). Next, we analyzed the parameters and the number of transmembrane helices of OsOPT proteins and found that the OsOPT proteins usually contain 12-16 transmembrane helices with molecular weights that range from 50 to 90 KDa. These parameters were similar to the description of the OPT family in the Pfam database.
Table 1.
Basic information of sixteen rice OsOPT genes
| Locus IDa | Protein | FL-cDNAc | Identity/similarity (E-value) to SpISP4 | Putative Substrated | Phenotypes of the mutante | ||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Length (aa) |
MW (KDa) | TMb | pI | |||||
| LOC_Os01g43940 | OsOPT1 | 755 | 83.60 | 16 | 8.92 | NA | 37/57%(3.1e-140) | Iron-chelate | |
| LOC_Os02g43370 | OsYSL2 | 674 | 73.34 | 15 | 9.03 | AB126253 | 19/36%(0.00099) | Iron-chelate | Dwarf, palegreen leaf, low tillerring, long clum, late heading |
| LOC_Os02g43410 | OsYSL15 | 672 | 73.15 | 14 | 9.20 | AB190923 | 22/41%(0.00019) | Iron-chelate | |
| LOC_Os02g46850 | OsOPT8 | 559 | 56.23 | 11 | 8.59 | AK071310 | 39/58%(1.7e-107) | ||
| LOC_Os03g54000 | OsOPT7 | 757 | 83.45 | 16 | 9.18 | AK102404 | 37/57%(7.3e-123) | Iron-chelate | |
| LOC_Os04g44300 | OsYSL13 | 688 | 78.77 | 14 | 9.10 | AK067235 | 21/37%(0.0083) | ||
| LOC_Os04g44320 | OsYSL12 | 717 | 77.69 | 12 | 8.96 | AK069437 | 21/37%(0.019) | Dwarf, short panicle, lesion leaf, low tillering | |
| LOC_Os04g45860 | OsYSL9 | 657 | 74.26 | 14 | 8.50 | AK120923 | 21/38%(0.0023) | ||
| LOC_Os04g45900 | OsYSL16 | 675 | 73.81 | 14 | 9.30 | AK070304 | 21/39%(0.013) | ||
| LOC_Os04g50820 | OsOPT6 | 737 | 82.25 | 15 | 8.86 | AK070801 | 35/57%(3.5e-139) | ||
| LOC_Os04g57840 | OsYSL10 | 686 | 74.65 | 14 | 9.06 | AK069645 | 21/39%(0.011) | ||
| LOC_Os06g03540 | OsOPT2 | 761 | 85.82 | 14 | 5.95 | NA | 37/37%(2.0e-145) | ||
| LOC_Os06g03560 | OsOPT3 | 766 | 86.14 | 12 | 6.53 | AF393848 | 36/55%(1.7e-139) | Iron-chelate/GSH | Dwarf, glabrous leaf, semi-short clum, white belly rice kernel |
| LOC_Os06g03700 | OsOPT4 | 760 | 85.10 | 12 | 7.87 | AK072617 | 38/58%(2.0e-145) | Iron-chelate | Yellow-green plant, late heading, semi-dwarf, low tillerring |
| LOC_Os08g23130 | OsOPT5 | 733 | 81.63 | 12 | 9.15 | AK121257 | 35/55%(5.4e-132) | Iron-chelate | |
| LOC_Os08g38400 | OsOPT9 | 752 | 82.82 | 14 | 9.17 | AK100814 | 37/55%(5.1e-131) | Semi-dwarf, low tillering, semi-sterile pollen | |
aLocus ID was adopted from the MSU Rice Genome Annotation Project Database; bNumber of transmembrane domains; cCorresponding full-length cDNA in GenBank; dPutative substrate come from former publications. The NA indicates that this gene has no mRNA record in GenBank.eThe phenotypes of the inserted mutants of OsOPTs except of OsOPT9 were from different mutant database (OsYSL2, OsYSL12, and OsOPT3 from Rice Tos17 Insertion Mutant Database: http://tos.nias.affrc.go.jp/index.html.en, OsOPT4 from Oryza Tag Line: http://urgi.versailles.inra.fr/OryzaTagLine/cgi-bin/focus_mutant.pl).
Phylogenetic relationship, chromosomal location and gene structures of the OsOPT genes
To further confirm the phylogenetic relationship of the OPT genes, we compared the rice OsOPT genes with OPT genes from other species. Some plant OPT genes had been experimentally demonstrated to have transportation functions for oligopeptides or metal chelates (Figure 1). These comparisons also suggested that the 16 genes could be classified into the same group as the OPT family homologues. By examining the phylogenetic relationship, nine of the 16 OsOPT genes were clustered into the PT subfamily and seven were clustered into the YSL subfamily (Lubkowitz [2006]; Yen et al. [2001]). Classification of the PT subfamily was in accordance with the peptide transport (PT) clade, which was identified and studied by (Lubkowitz [2006]). The nine OsOPT genes within the PT subfamily could be further classified into four sections (A1, A2, A3 and A4), and the seven genes within the YSL subfamily were further grouped into two sections (B1 and B2) (Figure 2a).
Figure 1.

Phylogenetic tree of the oligopeptide transporter (OPT) genes among rice and other species, based on amino acid sequences. The tree was drawn according to results generated by MEGA4.0 analysis using the neighbor-joining method with an amino acid and Poisson correction model. Bootstrap values calculated for 1,000 replicates are indicated at corresponding nodes. The groups with the cycles were functionally demonstrated to transport oligopeptides and the groups with the triangles were experimentally demonstrated to transport metal compounds. Locus IDs of OsOPT genes from rice are given in Table 1, and gi numbers of OPTs from other species are given as: AfOPT1, gi|20145239|; AtOPT1, gi|9758213|; AtOPT2, gi|15218331|; AtOPT3, gi|18414644|; AtOPT4, gi|9759417|; AtOPT5, gi|4938497|; AtOPT6, gi|4469024|; AtOPT7, gi|3600039|; AtOPT8, gi|9759191|; AtOPT9, gi|9759190|; AtYSL1, gi|41352037|; AtYSL2, gi|41352039|; AtYSL3, gi|9759194|; AtYSL4, gi|41352041|; AtYSL5, gi|41352043|; AtYSL6, gi|41352045|; AtYSL7, gi|41352047|; AtYSL8, gi|41352049|; BjGT1, gi|30722286|; NcHYPO1, gi|9368956|; ZmYSL1, gi|10770865|; CaOPT1, gi|2367386|; ScOPT2, gi|6325452|; SpISP4, |19112445|.
Figure 2.

Phylogenetic relationships (a) and gene structure schematic diagram (b) for OsOPT genes. The phylogenetic tree was drawn as described in Figure 1 using SpISP4 as an outgroup. The 16 OsOPT genes from rice were clustered into six sections. The identical exon formation of OsOPT genes is connected with slashes.
The physical location of the 16 OsOPT genes could be assigned to the six rice chromosomes (Figure 3). Three genes found in section A1, OsOPT1, OsOPT6 and OsOPT8, were localized to chromosome 1, 2 and 4, respectively. All three genes found in section A2 (OsOPT2, OsOPT3 and OsOPT4) were localized to chromosome 6. Both OsOPT5 and OsOPT9 within section A3 were localized to chromosome 8. Section A4 only contained one gene, OsOPT7, which was localized to chromosome 3. All three genes found in section B1: OsYSL10, OsYSL12 and OsYSL13 were localized to chromosome 4. Among the four genes found in section B2, two genes (OsYSL9 and OsYSL16) were localized to chromosome 4 and the other two (OsYSL2 and OsYSL15) were localized to chromosome 2.
Figure 3.
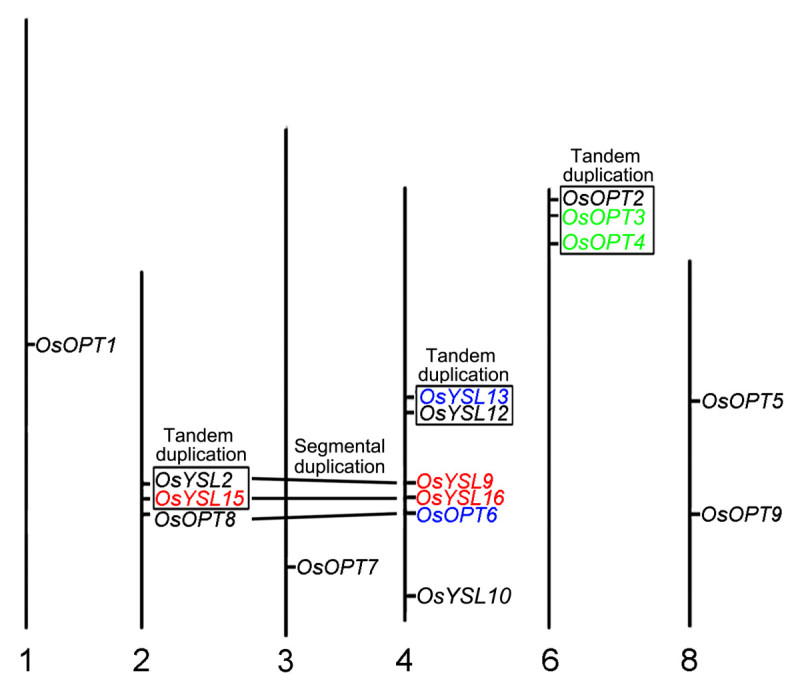
Chromosomal location, duplication event, and evolutionary stage schematic for OsOPT genes. Chromosome numbers are indicated at the bottom of each bar. The OsOPT genes represented by duplicated chromosomal segments between two chromosomes were connected by lines and the OsOPT genes that resulted from tandem duplication were contained in boxes. The OsOPT genes originating from the first evolutionary stage were shown in red, the OsOPT genes derived from the second evolutionary stage were shown in green, and the OsOPT genes derived from the third evolution stage were shown in blue.
We also determined the intron-exon boundaries for the 16 OsOPT genes to obtain more information on the genomic organization of rice OsOPT genes (Figure 2b). According to the annotation of the rice genome, three of the six sections (A2, B1 and B2) had similar intron-exon boundaries. All three genes found in section A2 (OsOPT2, OsOPT3 and OsOPT4) contain six introns and seven exons, but the length from the fourth to the sixth exon were identical. The genes in section B1 (OsYSL10, OsYSL12 and OsYSL13) had four identical exons. All four genes found in section B2 (OsYSL9, OsYSL16, OsYSL2 and OsYSL15) contain six introns and seven exons, but the length from the third to the fifth exon were identical. The identical exons were connected by slashes (Figure 2b). In addition, a 98 bp-exon was found in most of the 13 OsOPT genes except OsOPT1, OsOPT3 and OsOPT8.
Duplication and evolution analysis of the OsOPT genes
To reconstruct the expansion history of the 16 OsOPT genes in the rice genome, duplication and evolution analysis was performed. According to the duplication regions of rice chromosomes, we found that seven genes (OsOPT2, OsOPT3 and OsOPT4 in section A2, OsYSL12 and OsYSL13 in section B1, OsYSL2 and OsYSL15 in section B2) came from gene tandem duplication, which were shown in the box in Figure 3. However, three pairs (OsYSL9/OsYSL16, OsYSL2/OsYSL15 and OsOPT8/OsOPT6) were located in duplicated chromosomal regions, suggesting that these six genes came from segmental duplication.
The nonsynonymous substitution rate (Ka), the synonymous substitution rate (Ks), and their ratio (Ka/Ks) are commonly used to aid in understanding the direction of evolution and its selective strength in a coding sequence (Li et al. [2009]). The Ka/Ks ratio is also a measure to explore the mechanism of gene divergence after duplication. Ka/Ks < 1 is termed “purifying selection”, which means natural selection eliminates deleterious mutations, thus maintaining the protein as is. Alternatively, a value where Ka/Ks > 1 indicates that selection has accelerated evolution by acting to change the protein, which is termed “positive selection”. Ka/Ks = 1 is referred to as “neutral selection”, meaning that natural selection does not have any forces acting on the protein (Hurst [2002]). To analyze the evolutionary selection after the duplication events of the five pairs of OsOPT genes, we calculated Ka, Ks and Ka/Ks ratios for the seven duplicated pairs in the OsOPT family (Table 2). The Ka/Ks ratio of five pairs (OsYSL15/OsYSL2 OsYSL16/OsYSL15 OsOPT4/OsOPT3 OsOPT4/OsOPT2 and OsYSL9/OsYSL2) was less than 1, suggesting that purifying selection was acting on these five duplicated pairs. These data indicated that the mutation of the duplicated OsOPT protein is much less likely to differ between the two homologues; that is, most of the time selection eliminates deleterious mutations. However, the Ka/Ks ratio of OsYSL12/OsYSL13 and OsOPT6/OsOPT8 were more than 1, suggesting that positive selection acts on the two duplicated pairs. These data provide strong evidence to suggest that selection between these two gene pairs has acted to change the OsOPT protein (positive selection).
Table 2.
Ka/Ks analysis and estimate of the absolute dates for the duplication events between the duplicated OsOPT genes
| Duplicated pair | Ka | Ks | Ka/Ks | Date (million years) | Duplicate type | Purifying selection |
|---|---|---|---|---|---|---|
| OsYSL15/OsYSL2 | 0.1546 | 0.9061 | 0.1706 | 69.70 | Tandem | Yes |
| OsYSL16/OsYSL15 | 0.1679 | 0.8115 | 0.2069 | 62.42 | Segmental | Yes |
| OsYSL9/OsYSL2 | 0.1955 | 0.9902 | 0.1974 | 76.16 | Segmental | Yes |
| OsYSL12/OsYSL13 | 0.3304 | 0.1368 | 2.4152 | 10.52 | Tandem | No |
| OsOPT4/OsOPT3 | 0.1139 | 0.6271 | 0.1816 | 48.23 | Tandem | Yes |
| OsOPT4/OsOPT2 | 0.0791 | 0.5929 | 0.1334 | 45.60 | Tandem | Yes |
| OsOPT6/OsOPT8 | 0.5302 | 0.2936 | 1.8058 | 22.58 | Segmental | No |
We also calculated the dates of the duplication events (Table 2) by molecular clock theory (Yu et al. [2005]) using the nucleotide substitution rate (Ks) and the neutral evolutionary rate (λ) of 6.5 × 10−9 substitutions per silent site per year for rice genome (Yu et al. [2005]). According to the first whole-genome duplication event of grass genomes (Gaut [2002]; Salse et al. [2008]), duplication events for the three pairs (OsYSL15/OsYSL2, OsYSL16/OsYSL15 and OsYSL9/OsYSL2) occurred within the last 70 to 50 million years, after the origin of grasses (Gaut [2002]; Kellogg [2001]) and before the divergence of rice and maize (Gaut [2002]; Stebbins [1981]). Duplication events for the two pairs (OsOPT4/OsOPT3 and OsOPT4/OsOPT2) occurred within the last 50 to 20 million years, after the divergence of rice and maize, but before Zizaniinae and Oryzinae were diverged from one another (Guo and Ge [2005]). Duplication events for the other two pairs (OsYSL12/OsYSL13 and OsOPT6/OsOPT8) occurred within the last 20 to 9 million years, after Zizaniinae and Oryzinae were separated and before the Oryza genus branched off from the remaining genera of Oryzeae (Guo and Ge [2005]). Therefore, the evolutionary origin of the 16 OsOPT genes may have occurred in three evolutionary stages. The genes that came from the three evolutionary stages were shown in three different colors (Figure 3).
Differential expression of the OsOPT genes under normal growth conditions
Organ-specific and developmental expression of the 16 OsOPT genes under normal growth conditions were obtained from the rice MPSS database and RT-PCR (Figure 4). The data showed that expression profiles of the OsOPT genes were different during various developmental stages and in organs. The expression profiles for most OsOPT genes from the MPSS database (Figure 4A) matched well with the results from RT-PCR (Figure 4B). At the booting stage (Figure 4B), results from the RT-PCR showed that eleven of the 16 OsOPT genes were constitutively expressed in all four tested organs, three genes (OsOPT5 OsYSL10 and OsYSL15) were expressed in at least one organ and two genes (OsOPT1 and OsOPT6) were not expressed. However, when comparing the expression of OsOPT genes from the Rice Expression Profile Database (RiceXPro: http://ricexpro.dna.affrc.go.jp/index.html) (Sato et al. [2010]), we found that OsOPT6 was specifically expressed in the 1.2-2.0 mm portion of the anthers (data not shown). Therefore, we confirmed that genes were expressed under normal growth conditions except OsOPT1, which also did not have a corresponding mRNA sequence in GenBank (Table 1).
Figure 4.

Expression profiles of OsOPT genes in various rice organs. A. Transcript abundance of OsOPT genes in tissue-specific libraries from the MPSS database. The transcript abundance of OsOPT genes were normalized to transcripts per million. B. Organ-specific expression analysis of OsOPT genes at booting stages by RT-PCR. The rice plants were grown in a normal, natural field.
Data from the MPSS database suggested that some genes showed differential expression profiles in the developmental organs (Figure 4A). For the vegetative organs, four genes (OsOPT4, OsOPT7, OsYSL13 and OsYSL12) were differentially expressed among the roots of 14-day-old seedlings and 60-day-old seedlings; however, three genes (OsOPT4, OsOPT7, OsYSL13) were expressed at a higher level and one gene (OsYSL12) was expressed at a lower level in the 60-day-old roots than the 14-day-old roots. Three genes (OsOPT2, OsOPT7 and OsOPT9) had lower expression in the 14-day-old leaves than the 60-day-old leaves. For the reproductive organs, only five genes (OsOPT5, OsOPT8, OsOPT9, OsOPT12 and OsYSL16) were expressed in the male organs (mature pollen); whereas, six genes (OsOPT4, OsOPT8, OsOPT9, OsYSL12, OsYSL13 and OsYSL16) were expressed in the female organs (ovary and mature stigma). In addition, seven genes were expressed in the 35-day-old callus.
Expression of the OsOPT genes under abiotic and biotic stresses
To gain insight into the comprehensive roles of the OsOPT family members in response to various stresses, their expression patterns were investigated in rice seedlings subjected to abiotic (salt, drought and cold) (Figure 5) and biotic (two rice diseases) stresses (Figure 6) by mining the MPSS database. Only the genes, whose expression have at least a two-fold increase or decrease, were considered to have responded to stresses. Among the 16 OsOPT genes, nine genes showed differential expression in at least one tissue or at least one stress treatment compared to the control.
Figure 5.

Expression profiles of OsOPT genes under abiotic stress treatments. The data were from the MPSS database. Roots and leaves from 14-d- seedlings stressed in 250 mM NaCl (salt) for 24 h, lack of water (drought) for 5d, and in 4 °C cold for 24 h. Roots and leaves from 14-d- seedlings were used as a control.
Figure 6.

Differential expression of OsOPT genes in response to Magnaporthe grisea , and Xanthomonas oryzae. These data were from the MPSS database. A. Expression after X. oryzae inoculation. The resistant reaction group was leaves from a 2-month-old Nipponbare-Xa21 treated with X. oryzae (resistant variety) and the susceptible reaction group was leaves from a 2-month-old Nipponbare-Xa21 treated with X. oryzae (susceptible variety). The mock treatment group was leaves from a 2-month-old Nipponbare-Xa21 treated with water. B. Expression after M. grisea inoculation. The resistant reaction group was leaves from a 3-week-old Nipponbare-Pi9 treated with M. grisea (resistant variety) and the susceptible reaction group was leaves from a 3-week-old Nipponbare treated with M. grisea (susceptible variety). The mock treatment group was leaves from a 3-week-old Nipponbare treated with water.
In response to salt (250 mM NaCl) treatment, four genes (OsOPT4, OsOPT7, OsYSL13 and OsYSL16) were upregulated in the roots; however, five genes (OsOPT2, OsOPT7, OsOPT9, OsYSL13 and OsYSL12) were downregulated and one gene (OsOPT3) was upregulated in the leaves. In response to drought treatment, seven genes (OsOPT2, OsOPT4, OsOPT7, OsOPT8, OsYSL13 OsYSL15 and OsYSL16) were upregulated and one gene (OsYSL9) was downregulated in the roots; however, only two genes (OsYSL12 and OsYSL13) were upregulated and six genes (OsOPT2, OsOPT3, OsOPT4, OsOPT7, OsOPT9 and OsYSL16) were downregulated in the leaves. In response to cold (4oC) treatment, three genes (OsOPT8, OsYSL9 and OsYSL12) in the roots and seven genes (OsOPT2, OsOPT3, OsOPT4, OsOPT7, OsOPT9, OsYSL13 and OsYSL16) were downregulated in the leaves, and only one gene (OsYSL9) was upregulated in the leaves.
We also analyzed the responses of the OsOPT genes to infection with a fungus, Magnaporthe grisea, or a bacteria, Xanthomonas oryzae (Figure 6), which are known to cause rice blast and rice bacterial streak, two main rice diseases that cause severe decreases in rice yield. The expression changes of OsPTRs were determined after the inoculation. The microarray data and the infection with M. grisea and X. oryzae were published previously (Nobuta et al. [2007]). We found that four genes (OsOPT8 OsYSL12 OsYSL13 and OsYSL16) were upregulated and three genes (OsOPT3 OsOPT5 and OsYSL9) were downregulated by M. grisea treatment (Figure 6A), and there was no response difference among these seven genes between the resistance reaction and the susceptible reaction of the rice variety Nipponbare. In response to X. oryzae treatment (Figure 6B), upregulated expression of two genes (OsOPT8 and OsYSL12) were observed only during the early inoculation, and two genes (OsYSL13 and OsYSL16) were upregulated during the whole inoculation; and two genes (OsOPT2 and OsOPT5) showed downregulation.
Expression response of the OsOPT genes to hormone treatments
We used semi-quantitative RT-PCR to monitor the effect various hormones would have on the expression of OsOPT genes. A total of fourteen genes were amplified in three-week-old rice seedlings subjected to various hormone treatments, including 2,4-dichlorophenoxyacetic acid (2,4-D, an auxin), kinetin (KT, a cytokinin), abscisic acid (ABA) and gibberellic acid (GA3) (Figure 7). The results indicated that the expression of most (11) OsOPT genes were not regulated by the four phytohormone treatments. However, the expression of OsYSL15 was downregulated by the four hormones in the roots and the leaves, OsOPT7 was dowregulated by GA3 and ABA in the roots, and OsOPT2 was upregulated by ABA in the roots.
Figure 7.

Expression profiles of OsOPT genes under various hormone treatments. Three-week-old seedlings were irrigated with Hoagland's Solution containing 5 μM gibberellic acid (GA3), 5 μM 2,4-dichlorophenoxyacetic acid (2,4-D), 5 μM kinetin (KT), or 25 μM abscisic acid (ABA) for 24 h. Control (CK) was from the water-treated seedlings. Actin1 was used as an internal control.
Primary phenotypes of the inserted mutants for five OsOPT genes
To acquire more clues regarding the biological function of OsOPT genes, we searched main rice inserted mutant databases. Inserted mutants of all sixteen OsOPT genes were found in different mutant databases (Additional file 1: Table S1). However, the phenotype records of mutants of only five OsOPT genes were found in two mutant databases and their observed phenotypes were integrated into Table 1. The segregation rate and the plant numbers recorded in the mutant database can be found in Additional file 1: Table S1. From the records of the database, we found that some plants of the five OsOPT mutants displayed abnormal phenotypes under normal growth conditions (Table 1, Additional file 1: Table S1). Of course, it should be further investigated whether the phenotypes of the mutants from the database are linked with the five OsOPT genes.
From National Institute of Agrobiological Sciences (NIAS) of Japan (Miyao et al. [2003]), we obtained the Tos17-insertion mutants of OsOPT3 (OsGT1) and OsOPT9. The inserted loci for OsOPT3 (OsGT1) and OsOPT9 were confirmed by flanking sequence method. Then the osptr3 and osptr9 homozygote mutants were identified from the offspring by PCR method and were use for phenotype observation. The homozygote osptr3 mutant plant showed signs of serious lesion mimic symptom in the leaves even though they were grown under natural field (Figure 8A) and this symptom was not observed when the homozygote osptr3 mutant plant was grown in shade (data not shown). In addition, the homozygote osptr9 mutant mainly had semi-sterile pollen (Figure 8B) and low seed setting (data not shown). However, we did not observe the semi-dwarf and low tillering phenotype in the osopt9 mutants, which were observed in the mutant database as shown in Table 1.
Figure 8.
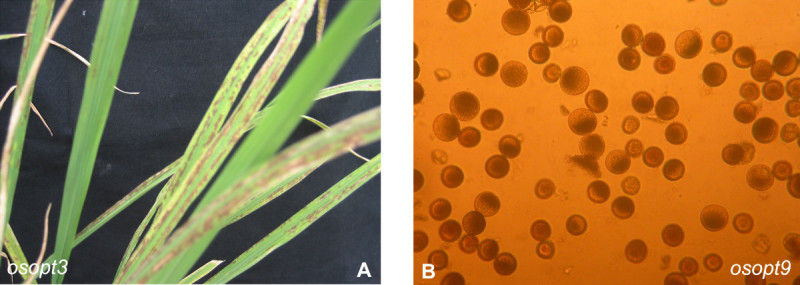
Main phenotypes of two OsOPT inserted mutants ( osopt3 and osopt9 ). A. Leaf lesions from a homozygote osopt3 mutant. B. Semi-sterile pollens from a homozygote osopt9 mutant. The homozygote plants (osopt3 and osopt9) were grown in natural fields during normal rice season in Guangzhou, China.
Discussion
Evolutionary expansion of the OsOPT gene family in rice
Gene duplication plays an important role in the expansion of a gene family (Adams and Wendel [2005]; Hughes [1994]). Several mechanisms have been suggested to drive the expansion of a gene family, including segmental duplication, tandem duplication and retroposition (Cannon et al. [2004]; Kong et al. [2007]). In this study, we identified sixteen members of the OsOPT gene family from the rice genome. We found that both the segmental duplication and the tandem duplication contributed to the evolutionary expansion of the OsOPT gene family in rice.
To speculate on the evolutionary orders of the rice OsOPT family, we considered protein sequence similarities with SpISP4 (Table 1), dates of the gene appearance (Table 2), rice genome evolutionary events (Krom and Ramakrishna [2010]), etc. The rice OsOPT members, which are more similar to SpISP4 from S. pombe, might have evolved earlier in the rice genome (Krom and Ramakrishna [2010]). The molecular clock was also used to compute the date of duplication by calculating the number of substitutions per silent site (Ks). Yu et al. ([2005]) identified an evolutionary rate of 6.5 × 10−9 substitutions per silent site per year by analyzing the duplication history of the rice genome, dating the duplication event to 53 million years ago (Mya). Based on the dates of the duplication events (Table 2) and similar E-value to SpISP4 (Table 1), we speculated that the evolutionary expansion of the OsOPT gene family in the rice genome underwent three stages. The three evolutionary stages of the OsOPT genes were shown with three colors in Figure 3. Before the evolutionary expansion of the OsOPT gene family in the rice genome, nine OsOPT genes (gene name in black) were distributed on six chromosomes (Figure 3). The first evolutionary stage was from the origin of the grasses (70 million years ago) to the divergence of rice and maize (approximately 50 million years ago). The original nine OsOPT gene homologues were also found in the maize genome (data not shown). During the first stage, OsYSL15 arose from OsYSL2 located on chromosome 2 after one tandem duplication event. Next, through one segmental duplication event between chromosome 2 and chromosome 4, OsYSL9 and OsYSL16 arose from OsYSL2 and OsYSL15 on chromosome 4. The four newly evolved genes were shown in red (Figure 3). The second evolutionary stage occurred at the point of divergence between rice and maize (approximately 50 million years ago) to the separation of Zizaniinae and Oryzinae (approximately 20 million years ago). In the second stage, OsOPT4 and OsOPT3 arose from OsOPT2 on chromosome 6 by two tandem duplication events. The two newly evolved genes were shown in green (Figure 3). The third evolutionary stage was from the separation of Zizaniinae and Oryzinae (approximately 20 million years ago) to the point where Oryza branched-off from the remaining Oryzeae genera (approximately 9 million years ago). In the third stage, OsYSL13 arose from OsYSL12 on chromosome 4 after one tandem duplication event; OsOPT6 arose from OsOPT8 on chromosome 4 following one segmental duplication event. The two newly evolved genes were shown in blue (Figure 3). After the three evolutionary stages, the number of the rice OsOPT gene family increased to sixteen genes. Notably, purifying selection occurred at stage 1 and stage 2, but positive selection occurred at stage 3.
Different functions and multiple roles of the OsOPT genes in rice development
Some plant OPT genes have been characterized in embryo development (Stacey et al. [2005]), glutathione transport (Zhang et al. [2004]), heavy metal detoxification (Cagnac et al. [2004]), seed germination (Muntz [1998]), and long distance nitrogen allocation (Williams and Miller [2001]). The genome-wide expression analysis of the OsOPT gene family showed that eleven genes (OsOPT2 OsOPT3 OsOPT 4 OsOPT7 OsOPT8 OsOPT9 OsYSL2 OsYSL9 OsYSL12 OsYSL13 and OsYSL16) had transcript accumulation in all tissues, and some in germinating seeds, roots, panicles and callus (Figure 4). These data suggested multiple functions for these oligopeptide transporters in rice and they might be involved in housekeeping functions during rice growth and development. On the other hand, some genes may play roles in specific tissues. The expression of OsOPT6 was not detectable in most of the tested tissues (Figure 4), but it was specifically expressed in the 1.2-2.0 mm portion of the anthers (Data from RiceXpro expression database).
AtOPT6 (Pike et al. [2009]; Cagnac et al. [2004]) and OsOPT3 (OsGT1) (Zhang et al. [2004]) have been characterized GSH transporters and have been suspected of having a role in mediating heavy metal phytotoxicity. However, little is known about the role of the OsOPT genes in response to stress. We obtained some clues regarding the role of OsOPT genes in response to stresses by examining their expression level. Nine of the 16 OsOPT genes (OsOPT2 OsOPT4 OsOPT7 OsOPT7 OsOPT9 OsYSL12 OsYSL13 OsYSL15 and OsYSL16) showed a response to at least one abiotic stress treatment (Figure 5). Of the two main rice disease treatments, the expression of seven genes (OsOPT3 OsOPT5 OsOPT8, and OsYSL9 OsYSL12 OsYSL13 and OsYSL16) changed after M. grisea treatment and the expression of six genes (OsOPT2 OsOPT5 OsOPT8 OsYSL12 OsYSL13 and OsYSL16) changed after X. oryzae treatment (Figure 6).
Functional diversification of OsOPT genes
Functional diversification is a result of the evolutionary expansion of a gene family by gene duplication and often accompanies changes in the expression profile of the gene family member (Hughes [1994]; Lynch and Conery [2000]). The OPT family has been divided into two subfamilies (the PT and YSL subfamily) (Curie et al. [2001]), where the YSL subfamily was believed to be able to transport iron compounds and the PT subfamily was believed to be able to transport oligopeptides with four-five amino acid residues (Lubkowitz [2006]). However, several members of the PT subfamily have also been identified as transporters of iron compounds, such as AtOPT3 (Stacey et al. [2007]), OsOPT1 OsOPT4 and OsOPT7 (Vasconcelos et al. [2008]). These data may support the idea that the two subfamilies originated from a common ancestor.
However, expression diversification between the duplicated pairs could be found in all seven duplicated genes (Table 3), indicating that functional diversification occurred following the duplication event. Based on the OsOPT gene expression data from the MPSS database and our experimental results, we found some characteristics of the OsOPT gene expression profiling in this study, including the following. (1) The rice OsOPT genes were differentially expressed at various developmental stages and in tissues. Eleven genes (OsOPT2, OsOPT3, OsOPT4, OsOPT7, OsOPT8, OsOPT9, OsYSL2, OsYSL9, OsYSL12, OsYSL13 and OsYSL16) showed transcript accumulation in all tissues (Figure 4B). However, subtle differences in the expression level of the eleven genes could be found in the developing plants; OsOPT4, OsOPT7, OsYSL13 and OsYSL12 were differentially expressed between the roots of 14-d -seedlings and 60-d -seedlings. (2) The expression profiles of the duplicated OsOPT genes had distinct tissue specificities and response to stimuli. (3) Fifteen OsOPT genes were expressed and the expression of one gene (OsOPT1) could not be confirmed.
Table 3.
Expression diversification between the duplicated OsOPT genes
| Duplicated pair | Tissues | Abiotic stress | Biotic stress | |||
|---|---|---|---|---|---|---|
| Salt | Drought | Cold | X. oryzae | M grisea | ||
| OsYSL15/OsYSL2 | + | - | - | - | - | - |
| OsYSL16/OsYSL15 | + | + | + | + | + | |
| OsYSL9/OsYSL2 | + | + | + | + | + | - |
| OsYSL12/OsYSL13 | + | + | + | + | + | + |
| OsOPT4/OsOPT3 | + | + | + | - | + | + |
| OsOPT4/OsOPT2 | + | + | - | - | + | + |
| OsOPT6/OsOPT8 | + | + | + | + | + | + |
+, Expression difference exists between the two members of the pair, -, No expression difference between the two members of the pair.
The transport functions and the expression patterns of the sixteen OsOPT genes have been diversified following their evolution. In the PT subfamily, three members (OsOPT1 OsOPT4, and OsOPT7) have been shown to be able to transport iron-chelates in yeast (Vasconcelos et al. [2008]). OsOPT3 (OsGT1) could also transport glutathione, some amino acids and peptides (Zhang et al. [2004]). In the YSL subfamily, OsYSL2 (Koike et al. [2004]) and OsYSL15 (Inoue et al. [2008]; Lee et al. [2009]) have been shown to be able to transport metal-nicotianamine. OsOPT6 arose from OsOPT8 by one segmental duplication event approximately 22.58 million years ago, but their expression profiles were different. OsOPT8 was expressed in all developing organs (Figure 4) and it was also induced by M. grisea; however, OsOPT6 was not expressed under normal growth conditions and during testing with biotic and abiotic stresses. OsOPT6 is not a pseudogene because it has a corresponding full-length mRNA (AK070801) that was found in the RAD-DB database (Table 1) and it might be specifically expressed in the 1.2-2.0 mm portion of the anthers.
To our knowledge there are few biofunctional data about rice OsOPT genes. Some offspring of the five inserted OsOPT mutants showed abnormal symptoms which were recorded in the mutant database and the symptoms of the five OsOPT mutants were distinct (Table 1). Mutants of OsYSL2 OsYSL12 and OsOPT3 were Tos17 insertion and mutant of OsOPT4 was T-DNA insertion. Of course, these phenotypes from the mutant database might not link with the OsPTR genes, because the mutant lines may carry multiple Tos17 or T-DNA insertions and the tissue-culture induced mutations (Hirochika et al. [1996]). By observing the phenotypes of two homozygote T-DNA inserted mutant osopt3 (osgt1) and osopt9, we found osptr3 showed a lesion mimic phenotype when the osptr3 mutant plant was grown under light of normal growth field (Figure 8A), but no this symptom under shade (data now shown). In addition, OsOPT3 (OsGT1) has been found to be a glutathione transporter (Zhang et al. [2004]). Glutathione is the major endogenous antioxidant produced by the cells, participating directly in the neutralization of free radicals and reactive oxygen compounds (Szalai et al. [2009]). These suggested that OsOPT3 (OsGT1) may play a role in tolerance to oxidative burst. The homozygote osptr9 mutant showed semi-sterile pollens (Figure 8B), which suggested that OsPTR9 may play a role in pollen development. These preliminary data could suggest that the rice OPT family may be necessary for rice growth and they may play diverse roles in rice.
The OsOPT gene family has been researched for more than ten years. In plants, it seems to have the ability to transport oligopeptide or metals, but due to the large number of potential substrates, the real roles of the OsOPT gene family are not fully understood. In this study, we described the evolutionary expansion and expression profiling of the rice OsOPT gene family and provided useful information for further study on the function of the OsOPT gene family in rice.
Conclusions
Rice OPT family contains 16 genes, and is classified into the PT and YSL subfamily with 9 and 7 genes, respectively. The 16 OsOPT genes evolved from 9 original OsOPT genes after 3 evolutionary stages. Their expression patterns of 16 OsOPT genes and phenotypes of five OsOPT mutants are different.
Methods
Identification of OsOPT gene homologues
To identify putative OPT genes in the rice genome, a reference OPT protein (SpISP4) (GI:19112445) from S. pombe was chosen as a query sequence to search in the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) by BLASTP. The protein sequences satisfied E < 0.1 were selected as the candidate proteins, which were compared with the OPT records in GenBank. Then, the physical and chemical parameters of the putative OsOPT proteins were computed by ProtParam in the ExPASy Proteomics Server (http://www.expasy.ch/tools/protparam.html). The prediction of transmembrane helices was performed by TMHMM Server v. 2.0 (http://genome.cbs.dtu.dk/services/TMHMM-2.0/). To confirm above OsOPT genes as the OPT gene homologue,we also analyzed the predicated OsOPT proteins in the Pfam database (http://www.sanger.ac.uk/Software/Pfam/search.shtml).
Phylogenetic analysis and gene structure
Multiple sequence alignments of amino acid sequences were performed using ClustalX (version 2.0.9) (Thompson et al. [1997]). Unrooted phylogenetic and molecular evolutionary analyses were constructed using MEGA4.0 by the neighbor-joining method with a Poisson correction model, using 1000 replicates for bootstrap analysis (Tamura et al. [2007]). Exon-intron organization was determined using the genome browser in RAP-DB (http://rapdb.dna.affrc.go.jp/). Gene structures were displayed using Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/chinese.php).
Chromosomal localization and gene duplication
The chromosomal localization of the OsOPT genes was determined by mapview in NCBI (http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=4530). The OsOPT genes present on the duplicated chromosomal segments were identified by segmental genome duplication of rice available at MSU-RGA (http://rice.plantbiology.msu.edu/analyses_search_blast.shtml) with the maximum distance permitted between collinear gene pairs of 100 kb. The OsOPT genes separated by a maximum of twenty genes were identified as tandem duplicated genes (Lynch and Conery [2000]; Moore and Purugganan [2005]).
Ka/Ks analysis and calculation of the date of duplication events
The number of nonsynonymous substitutions per nonsynonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks) of duplicated genes were calculated with DnaSP Version 5. The duplication date could be computed with the number of substitutions per silent site (Ks) (Comeron [1999], Shiu et al. [2004]; Yang et al. [2008]). Yu et al. ([2005]) calculated a neutral evolutionary rate (λ) of 6.5 × 10−9 substitutions per silent site per year to analyze the duplication history of the rice genome. Here, we calculated the dates of the duplication events by the equation T = Ks/2λ, for rice, the λ = 6.5 × 10−9 (Yang et al. [2008]).
Rice materials and treatments
The rice cultivar Nipponbare (O. sativa L. japonica) was used in this study. For expression analyzes of the OsOPT genes under normal conditions, total RNA was extracted from different tissues of the rice plants grown in the natural field of South China Botanical Garden, at the booting stage.
For phytohormone treatments, seeds were soaked in water and germinated at 28 °C for 2 days and then grown in Hoagland's Solution (Baba and Takahashi [1956]) under natural conditions for 3 weeks. Next, the seedlings were cultured in Hoagland's solution containing 5 μM gibberellic acid (GA3), 5 μM 2,4-dichlorophenoxyacetic acid (2,4-D, an auxin), 5 μM kinetin (KT, a cytokinin), or 25 μM abscisic acid (ABA), respectively, for 24 h.
For various treatments, published data were used as a reference for the growth conditions, treatments and experiments on the plant materials in the digital expression database (Nobuta et al. [2007]).
RNA extraction and semi-quantitative RT-PCR analysis
Total RNA was extracted using Trizol reagent according to the manufacturer’s instructions(Invitrogen, http://www.invitrogen.com). First-strand cDNA was synthesized from 3 μg of total RNA treated with DNase I using M-MLV reverse transcriptase (Promega, http//http://www.promega.com). The first-strand cDNA was used as a template for semi-quantitative PCR after normalizing to rice Actin1 (AB047313). Semi-quantitative RT-PCR was performed in a 20 μL reaction volume containing 1 μL cDNA solution, 1 × PCR buffer, 0.25 μM dNTPs, 1.0 μM gene-specific primers and 0.5 U Taq polymerase (Takara, Japan) using the following conditions: 94 °C for 3 min (1 cycle), 94 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min (30-35 cycles) and 72 °C for 10 min (1 cycle). All primer sequences used in this study are listed in Additional file 2: Table S2. To ensure the primers were specific for the correct OsOPT genes, primers were designed using software Primer Primier v5.00 (PREMIER Biosoft international). The amplification products obtained with the different primer pairs were purified using gel electrophoresis and sequenced. Sequences were then aligned on the RAP-DB website to determine their specificity.
Digital expression analysis using the MPSS database
The massively parallel signature sequencing (MPSS) database (http://mpss.udel.edu/rice/) was searched for 20-base signatures from 22 mRNA libraries representing 18 different tissues/organs in rice expression evidence analysis (Nakano et al. [2006]). Only the signatures uniquely identified for an individual gene and had a perfect match (100 % identity over 100 % of the length of the tag) were used for analysis. The normalized abundance (tags per million, tpm) of these signatures for a given gene in a given library represents the quantitative estimate of the expression of that gene. The 22 mRNA libraries of MPSS have been group into four groups to analyze the expression profile of the OsOPT family in various developmental stages, organs and various stress treatments.
Biofunctional analysis of OsOPTs
Two approaches were used to obtain biofunctional clues on OsOPTs. First, different rice inserted mutant databases were searched. The information on the inserted mutants of OsOPTs was retrieved from OryGenesDB (an interactive tool for rice reverse genetics, http://orygenesdb.cirad.fr/index.html), searched by their gene locus ID, and the results were listed in Additional file 1: Table S1. Then, two mutant databases (OTL: http://urgi.versailles.inra.fr/OryzaTagLine/cgi-bin/focus_mutant.pl and NIAS: http://tos.nias.affrc.go.jp/index.html.en) were further searched using the plant names of the inserted mutants in Additional file 1: Table S1 to obtain the observed phenotypes of these mutants. Five OsOPTs mutants had their observed phenotypes in two mutant databases. The second approach was analysis of two OsOPTs (OsOPT3 and OsOPT9) inserted mutants, which were from NIAS, and their main phenotypes were obtained from the homozygote osopt3 and osopt9 mutant plants.
Electronic supplementary material
Additional file 1: Table S1. Information of the inserted mutants of OsOPTs in different mutant database. (DOC 46 KB)
Additional file 2: Table S2. List of the semi-quantitative RT-PCR primers used in this study. (DOC 187 KB)
Below are the links to the authors’ original submitted files for images.
Acknowledgments
We thank to National Institute of Agrobiological Sciences (NIAS), Japan, for rice mutants (ND1017 and NG0095). This research was partly supported by NSFC grants (30900116/31071352), grant (KSCX3-EW-N-07) of the Chinese Academy of Science, Technology Innovation Program of Functional Development of Instrument and Equipment (YG2011081) of the Chinese Academy of Sciences, and grants (2011A020201003/2007A0201000001/S2011040000236) from Guangdong Province, China.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/1939-8433-5-12) contains supplementary material, which is available to authorized users.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TL, JZ and KF carried out expression and mutant studies. TF, YL and YW participated in mining digital data. XX grew plants and contributed reagents. MZ designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Tao Liu, Email: liutao6212533@163.com.
Jiqing Zeng, Email: zengjq@scbg.ac.cn.
Kuaifei Xia, Email: xiakuaifei@scbg.ac.cn.
Tian Fan, Email: ftt860630@163.com.
Yuge Li, Email: liyugezi@yahoo.com.cn.
Yaqin Wang, Email: 576653141@qq.com.
Xinlan Xu, Email: xxl@scbg.ac.cn.
Mingyong Zhang, Email: zhangmy@scbg.ac.cn.
References
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Kobayashi T, Takahashi M, Nagasaka S, Usuda K, Kakei Y, et al. OsYSL18 is a rice iron (III)-deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol Biol. 2009;70:681–692. doi: 10.1007/s11103-009-9500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba S, Takahashi Y. Laboratory manual in crop science: Water and sand culture. Tokyo: Nogyo-Gijitsu Kyokai; 1956. pp. 99–119. [Google Scholar]
- Bashir K, Ishimaru Y, Nishizawa NK. Iron uptake and loading into rice grains. Rice. 2010;3:122–130. doi: 10.1007/s12284-010-9042-y. [DOI] [Google Scholar]
- Bogs J, Bourbouloux A, Cagnac O, Wachter A, Rausch T, Delrot S. Functional characterization and expression analysis of a glutathione transporter, BjGT1, from Brassica juncea: evidence for regulation by heavy metal exposure. Plant Cell Environ. 2003;26:1703–1711. doi: 10.1046/j.1365-3040.2003.01088.x. [DOI] [Google Scholar]
- Cagnac O, Bourbouloux A, Chakrabarty D, Zhang MY, Delrot S. AtOPT6 transports glutathione derivatives and is induced by primisulfuron. Plant Physiol. 2004;135:1378–1387. doi: 10.1104/pp.104.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Gaut BS. Evolutionary dynamics of grass genomes. New Phytol. 2002;154:15–28. doi: 10.1046/j.1469-8137.2002.00352.x. [DOI] [Google Scholar]
- Guo YL, Ge S. Molecular phylogeny of Oryzeae (Poaceaf) based on DNA sequences from chloroplast, mitochondrial, and nuclear genomes. Am J Bot. 2005;92:1548–1558. doi: 10.3732/ajb.92.9.1548. [DOI] [PubMed] [Google Scholar]
- Higgins CF, Payne JW. The peptide pools of germinating barley grains - relation to hydrolysis and transport of storage proteins. Plant Physiol. 1981;67:785–792. doi: 10.1104/pp.67.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc R Soy B. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, et al. Rice OsYSL15 is an iron-regulated iron(iii)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem. 2008;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S, Wiles AM, Sharp JS, Naider FR, Becker JM, Stacey G. An oligopeptide transporter gene family in Arabidopsis. Plant Physiol. 2002;128:21–29. doi: 10.1104/pp.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, et al. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004;39:415–424. doi: 10.1111/j.1365-313X.2004.02146.x. [DOI] [PubMed] [Google Scholar]
- Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, DePamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873–885. doi: 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- Krom N, Ramakrishna W. Conservation, rearrangement, and deletion of gene pairs during the evolution of four grass genomes. DNA Res. 2010;17:343–352. doi: 10.1093/dnares/dsq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chiecko JC, Kim SA, Walker EL, Lee Y, Guerinot ML, et al. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 2009;150:786–800. doi: 10.1104/pp.109.135418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Z, Vang S, Yu J, Wong GKS, Wang J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J Mol Evol. 2009;68:414–423. doi: 10.1007/s00239-009-9222-9. [DOI] [PubMed] [Google Scholar]
- Lubkowitz M. The OPT family functions in long-distance peptide and metal transport in plants. Genet Eng (NY) 2006;27:35–55. doi: 10.1007/0-387-25856-6_3. [DOI] [PubMed] [Google Scholar]
- Lubkowitz MA, Hauser L, Breslav M, Naider F, Becker JM. An oligopeptide transport gene from Candida albicans. Microbiol. 1997;143:387–396. doi: 10.1099/00221287-143-2-387. [DOI] [PubMed] [Google Scholar]
- Lubkowitz MA, Barnes D, Breslav M, Burchfield A, Naider F, Becker JM. Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Mol Microbiol. 1998;28:729–741. doi: 10.1046/j.1365-2958.1998.00827.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Muntz K. Deposition of storage proteins. Plant Mol Biol. 1998;38:77–99. doi: 10.1023/A:1006020208380. [DOI] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, et al. An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- Osawa H, Stacey G, Gassmann W. ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochem J. 2006;393:267–275. doi: 10.1042/BJ20050920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Cai ZY, Xia KF, Wang YQ, Duan J, Zhang MY. Identification and analysis of eight peptide transporter homologs in rice. Plant Sci. 2010;179:374–382. doi: 10.1016/j.plantsci.2010.06.013. [DOI] [Google Scholar]
- Pike S, Patel A, Stacey G, Gassmann W. Arabidopsis OPT6 is an oligopeptide transporter with exceptionally broad substrate specificity. Plant Cell Physiol. 2009;50:1923–1932. doi: 10.1093/pcp/pcp136. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Let. 2007;581:2281–2289. doi: 10.1016/j.febslet.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Roberts LA, Pierson AJ, Panaviene Z, Walker EL. Yellow stripe1 expanded roles for the maize iron-phytosiderophore transporter. Plant Physiol. 2004;135:112–120. doi: 10.1104/pp.103.037572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, et al. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2010;39:D141–148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ludewig U, Bulent EE, Mori S, Kitahara T, von Wiren N. ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem. 2004;279:9091–9096. doi: 10.1074/jbc.M311799200. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G. Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta. 2005;223:291–305. doi: 10.1007/s00425-005-0087-x. [DOI] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, et al. The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 2007;146:589–601. doi: 10.1104/pp.107.108183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Coevolution of grasses and herbivores. Ann Missouri Bot Gard. 1981;68:75–86. doi: 10.2307/2398811. [DOI] [Google Scholar]
- Szalai G, Kellos T, Galiba G, Kocsy G. Glutathione as an antioxidant and regulatory molecule in plants. J Plant Growh Regul. 2009;28:66–80. doi: 10.1007/s00344-008-9075-2. [DOI] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos MW, Li GW, Lubkowitz MA, Grusak MA. Characterization of the PT clade of oligopeptide transporters in rice. Plant Genome. 2008;1:77–88. doi: 10.3835/plantgenome2007.10.0540. [DOI] [Google Scholar]
- Wiles AM, Naider F, Becker JM. Transmembrane domain prediction and consensus sequence identification of the oligopeptide transport family. Res Microbiol. 2006;157:395–406. doi: 10.1016/j.resmic.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Williams L, Miller A. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:659–688. doi: 10.1146/annurev.arplant.52.1.659. [DOI] [PubMed] [Google Scholar]
- Xue C, Fu Y. Preservation of duplicate genes by originalization. Genetica. 2008;136:69–78. doi: 10.1007/s10709-008-9311-5. [DOI] [PubMed] [Google Scholar]
- Yang Z, Gu S, Wang X, Li W, Tang Z, Xu C. Molecular evolution of the cpp-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol. 2008;67:266–277. doi: 10.1007/s00239-008-9143-z. [DOI] [PubMed] [Google Scholar]
- Yen MR, Tseng YH, Saier MH. Maize yellow Stripe1, an iron-phytosiderophore uptake transporter, is a member of the oligopeptide transporter (OPT) family. Microbiol. 2001;147:2881–2883. doi: 10.1099/00221287-147-11-2881. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Bourbouloux A, Cagnac O, Srikanth CV, Rentsch D, Bachhawat AK. A novel namily of transporters mediating the transport of glutathione derivatives in plants. Plant Physiol. 2004;134:482–491. doi: 10.1104/pp.103.030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Huang J, Yu H, Wang L, Xie W. Genomic survey, characterization and expression profile analysis of the peptide transporter family in rice (Oryza sativa L.) BMC Plant Biol. 2010;10:92. doi: 10.1186/1471-2229-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Information of the inserted mutants of OsOPTs in different mutant database. (DOC 46 KB)
Additional file 2: Table S2. List of the semi-quantitative RT-PCR primers used in this study. (DOC 187 KB)


