Abstract
Limitless cell proliferation, evasion from apoptosis, dedifferentiation, metastatic spread and therapy resistance: all these properties of a cancer cell contribute to its malignant phenotype and affect patient outcome. MYBL2 (alias B-Myb) is a transcription factor of the MYB transcription factor family and a physiological regulator of cell cycle progression, cell survival and cell differentiation. When deregulated in cancer cells, MYBL2 mediates the deregulation of these properties. In fact, MYBL2 is overexpressed and associated with poor patient outcome in numerous cancer entities. MYBL2 and players of its downstream transcriptional network can be used as prognostic and/or predictive biomarkers as well as potential therapeutic targets to offer less toxic and more specific anti-cancer therapies in future. In this review, we summarize current knowledge on the physiological roles of MYBL2 and highlight the impact of its deregulation on cancer initiation and progression.
Facts
MYBL2 is a highly conserved member of the MYB family of transcription factors.
MYBL2 is an important physiological regulator of cell cycle progression, cell survival and cell differentiation.
Deregulation of MYBL2 expression is involved in cancer initiation and progression.
High MYBL2 expression is significantly correlated with poor patient outcome in numerous cancer entities.
Open questions
What are further players of the MYBL2 downstream transcriptional network mediating its cancer-promoting properties?
How can MYBL2 and players of its downstream transcriptional network be exploited as therapeutic targets to improve patient outcome?
Which additional cancer entities are also affected by MYBL2 deregulation and which patients could specifically benefit from using MYBL2 as a biomarker or therapeutic target?
Limitless replicative potential, evading apoptosis, tissue invasion and metastasis: these classical hallmarks of cancer, as originally proposed by Hanahan and Weinberg,1 characterize the malignant phenotype of a cancer cell. MYBL2 (V-Myb avian myeloblastosis viral oncogene homolog-like 2), a transcription factor of the MYB family of transcription factors, contributes to these properties of a cancer cell. MYBL2 is a physiological regulator of cell cycle progression, cell survival and cell differentiation, but due to its frequently found deregulation in cancer, it significantly drives cancer initiation and/or progression.
The MYB family of transcription factors comprises three members: MYB (c-Myb), MYBL1 (A-Myb) and MYBL2 (B-Myb). MYB was the first discovered family member and is the mammalian homolog of the retroviral v-Myb oncogene that causes acute leukemia in birds and can transform hematopoietic cells.2, 3 MYBL1 and MYBL2 have been cloned based on the homology to MYB.4 In mammals, MYB expression is mainly restricted to hematopoietic cells, colonic crypts and brain,5, 6 whereas MYBL1 is expressed in several regions of the developing central nervous system, germinal B-lymphocytes and reproductive systems of both genders.7, 8 In contrast, MYBL2 is expressed in basically all proliferating cells,3 which is a possible explanation for the lethal phenotype of MYBL2 knockout mice showing early embryonal death as a result of impaired inner cell mass formation,9 whereas MYBL1 deletion results in viable mice and MYB deletion leads to late embryonal death by cause of lacking erythropoiesis.7, 10
According to their tissue-specific expression, MYB and MYBL1 deregulations have been associated with certain specific cancer entities: MYB was shown to be involved in several types of leukemia, colon and breast cancer,11 whereas MYBL1 has been associated with Burkitt’s lymphoma and several types of leukemia.12 In contrast, MYBL2 deregulations occur in a broad spectrum of cancer entities as it is a central regulator of cell cycle progression, cell survival and cell differentiation in many tissue types (see ‘MYBL2 in cancer’ section). In this review, we summarize the physiological roles of MYBL2 in cell cycle regulation, cell survival and cell differentiation, and describe its deregulation as well as the resulting functional and clinical implications in cancer.
MYBL2 in Cell Cycle Regulation
MYBL2 is a cell cycle regulated and a cell cycle regulating gene. Its expression is controlled by the DREAM multiprotein complex (Dimerization partner, RB-like proteins, E2Fs and MuvB core), which is crucial in coordinating cell cycle-dependent gene expression and represses most cell cycle genes during cellular quiescence.13 This complex consists of the dimerization partner (DP1, -2, -3), the RB-like proteins p130 or p107, E2F (E2F4 or E2F5) and the multi-vulval class B core (MuvB, itself consisting of LIN9, LIN37, LIN52, LIN54 and RBBP4).13 Upon cell cycle entry, p130 or p107 dissociate from the MuvB core and from repressor E2Fs (E2F4, E2F5) due to loss of DYRK1A-dependent phosphorylation of LIN52, allowing activator E2Fs (E2F1 or E2F2 or E2F3) to transactivate early G1/S cell cycle genes, including MYBL2.13 Accordingly, MYBL2 is repressed by the DREAM complex during cellular quiescence and becomes subsequently expressed in late G1 and in S phase.13 Additionally, at a post-transcriptional level, MYBL2 expression can be suppressed by microRNAs.14, 15, 16, 17, 18, 19
Apart from MYBL2 expression, the transcriptional activity of MYBL2 is regulated by posttranslational modifications and protein–protein interactions. During late G1 and S phase, MYBL2 is phosphorylated by Cyclin A/E-CDK2, which enhances its transactivation activity, probably by releasing it from the nuclear receptor co-repressors N-CoR and SMRT, which maintain MYBL2 in an inhibited state when non-phosphorylated.20, 21, 22 Interestingly, Cyclin A-dependent phosphorylation simultaneously reduces MYBL2 protein expression by facilitating ubiquitin-mediated proteolysis of MYBL2, probably to regulate the proper alternation of events during cell cycle progression.23 p300, a transcriptional co-activating protein, is also able to stimulate MYBL2 activity by MYBL2 acetylation, depending on its phosphorylation by Cyclin A.24 However, contrary to Cyclin A, Cyclin D1 strongly inhibits transactivation activity of MYBL2 through direct interaction.25 Mechanistically, it was proposed that Cyclin D1 abolishes the activating function of p300.24 Several other co-activators, such as PARP1, ZPR9, TAF(II)250, or co-repressors, such as p107, p57 or CDK9, were further shown to modulate transactivation properties of MYBL2.26, 27, 28, 29, 30, 31, 32, 33
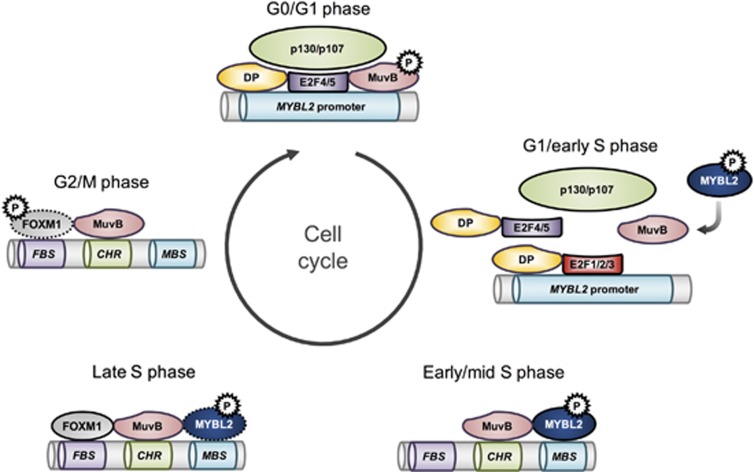
Early results already showed that MYBL2, when expressed and activated in late G1 and S phase, directly binds to the promoters and transactivates genes expressed in G2/M phase, such as Cyclin B1 (CCNB1),34 CDK1,34 Cyclin A2 (CCNA2)34 (a list of selected MYBL2 target genes34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53 is given in Table 1). Recently it was shown that the MuvB core, which dissociates from the DREAM complex upon cell cycle entry, and FOXM1 cooperate with MYBL2 to transactivate these late cell cycle genes.13, 36 Accordingly, MYB binding sites (MBS), cell cycle genes homology region (CHR, bound by LIN54 of the MuvB core) elements and FOXM1 binding motifs co-occur in the promoters of these genes.13, 36 A model emerged over the years: When DREAM dissociates upon cell cycle entry, MYBL2 becomes increasingly expressed and activated, and forms a complex with MuvB in early and mid S phase. RNAi experiments showed that knockdown of either MYBL2 or components of the MuvB core complex inhibits target gene promoter binding of the other.36 Consistently, depletion of either MBS or CHR motifs in the promoters of these target genes independently prohibits target gene promoter binding.49, 54 These results clearly indicate a dependency of both factors to one another in transactivating late cell cycle genes in early and mid S phase. Afterwards, the MYBL2–MuvB complex recruits FOXM1 in late S phase, forming a MYBL2–MuvB–FOXM1 complex.13 MYBL2 increasingly undergoes phosphorylation-dependent proteasomal degradation in late S phase,23 leading to predominant MuvB–FOXM1 complex in G2/M phase, whereby FOXM1 is increasingly activated by phosphorylation.55 Depletion of FOXM1 did not affect MYBL2 target gene promoter binding, but conversely, MYBL2 or LIN9 depletion reduced FOXM1 target gene promoter binding, indicating that the MYBL2–MuvB complex is required for FOXM1 target gene promoter binding, but not vice versa.36 As the FOXM1 DNA-binding domain has an extraordinarily low binding affinity to its target sequence and MBS and CHR motifs are necessary for FOXM1 to bind the target promoters, it was proposed that the MYBL2–MuvB complex is needed to increase target specificity for FOXM1 binding.13 The residual MuvB–FOXM1 complex dissociates due to increasing APC/C-CDH1-dependent FOXM1 degradation during M phase56 (Figure 1).
Table 1. Selected target genes transactivated by MYBL2.
| Gene symbol | Protein name | Reference(s) |
|---|---|---|
| Cell cycle regulation | ||
| AURKA | Aurora A kinase | Sadasivam et al.36 |
| CCNA1 (Sp1-mediated) | Cyclin A1 | Bartusel et al.35 |
| CCNA2 | Cyclin A2 | Zhu et al.34; Osterloh et al.37 |
| CCNB1 | Cyclin B1 | Zhu et al.34; Sadasivam et al.36; Osterloh et al.37 |
| CCND1 (Sp1-mediated) | Cyclin D1 | Bartusel et al.35 |
| CCND2 (repression) | Cyclin D2 | Papetti et al.38 |
| CDK1 | Cyclin-dependent kinase 1 | Zhu et al.34; Osterloh et al.37 |
| CDK2 (repression) | Cyclin-dependent kinase 2 | Papetti et al.38 |
| CDKN2A (repression) | p16INK4A | Huang et al.110 |
| CENPF | Centromere protein F | Iltzsche et al.42 |
| CEP55 | Centrosomal protein 55 | Wolter et al.43 |
| FGF4 | Fibroblast growth factor 4 | Johnson et al.40 |
| FOXM1 | Forkhead box M1 | Lorvellec et al.41 |
| KIFC1; KIF2C; KIF4A; KIF14; KIF20A; KIF23 | Mitotic kinesins | Wolter et al.43; Iltzsche et al.42 |
| MYB (repression) | c-Myb | Guerra et al.44 |
| MYBL2 (Sp1-mediated) | B-Myb | Sala et al.45 |
| MYC | c-Myc | Nakagoshi et al.46 (activation); Lorvellec et al.41 (activation); Papetti et al.38 (repression) |
| NUSAP1 | Nuclear- and spindle-associated protein 1 | Iltzsche et al.42 |
| PLK1 | Polo-like kinase 1 | Sadasivam et al.36; Osterloh et al.37 |
| PRC1 | Protein regulator of cytokinesis 1 | Wolter et al.43 |
| TOP2A | DNA topoisomerase IIα | Brandt et al.39 |
| Cell survival | ||
| BCL2 | Bcl-2 | Grassilli et al.47 |
| BCL2L11 | Bim | Greene et al.48 |
| BIRC5 | Survivin | Knight et al.49 |
| CLU | ApolipoproteinJ/Clusterin | Cervellera et al.50 |
| FGF4 | Fibroblast growth factor 4 | Johnson et al.40 |
| MYB (repression) | c-Myb | Guerra et al.44 |
| MYBL2 (Sp1-mediated) | B-Myb | Sala et al.45 |
| MYC | c-Myc | Nakagoshi et al.46 (activation); Lorvellec et al.41 (activation); Papetti et al.38 (repression) |
| PLK1 | Polo-like kinase 1 | Sadasivam et al.36; Osterloh et al.37 |
| VDAC2 | Voltage-dependent anion channel 2 | Yuan et al.51 |
| Differentiation | ||
| NANOG | Homeobox protein NANOG | Zhan et al.52 |
| POU5F1 | Oct-4 | Tarasov et al.53 |
| SOX2 | Sox2 | Zhan et al.52 |
| Invasion/metastasis | ||
| SNAI1 | Snail (Zinc-finger protein SNAI1) | Tao et al.120 |
Figure 1.
Regulation of MYBL2 expression and subsequent association of MYBL2 with its functional binding partners MuvB and FOXM1 during the cell cycle. In G0/G1 phase, the DREAM complex binds the MYBL2 promoter and suppresses MYBL2 expression. In late G1/early S phase, the DREAM complex dissociates due to loss of DYRK1A-dependent phosphorylation of LIN52 (part of the MuvB core), MYBL2 becomes increasingly expressed (and activated by Cyclin A/E-CDK2-dependent phosphorylation) and associates with the MuvB core to cooperatively transactivate G2/M genes in early and mid S phase. In late S phase, FOXM1 additionally associates with the MuvB–MYBL2 complex and cooperates in transactivation of these late cell cycle genes. MYBL2 increasingly becomes degraded in late S phase, leading to predominantly persisting FOXM1–MuvB complexes during G2/M phase, whereby FOXM1 is increasingly activated by phosphorylation. The residual MuvB–FOXM1 complex dissociates due to increasing APC/C-CDH1-dependent FOXM1 degradation during M phase
The association between MYBL2 and cell proliferation has already been described early by Arsura et al.57 Although some of the pioneering studies indicated a role for MYBL2 in G1/S progression,58, 59 the major role of MYBL2 in G2/M progression became increasingly clear over the recent years: RNAi-mediated MYBL2 knockdown in human cell lines and experiments in Drosophila with knockout of the MYBL2 Drosophila homolog dMyb reduces cell proliferation, expression of G2/M genes and decreases the amount of cells in G2/M phase.60, 61, 37, 38, 53, 62, 63 Although dMyb is the only gene of the MYB transcription factor family in Drosophila, it is functionally and phylogenetically equivalent to vertebrate MYBL2 and can therefore be seen as a suitable model.64 The results from Drosophila experiments are remarkable, as they indicate that an adequate proliferative capacity mediated by MYBL2 is necessary to maintain genomic stability.53, 65, 66, 67 Loss-of-function mutation of dMyb causes abnormal mitoses that are associated with multiple functional centrosomes, unequal chromosome segregation, micronuclei formation and failure to complete cell division.65 These are frequent in the later cell cycles with resulting nuclei that often show aneuploidy and/or polyploidy.65 It was also shown that MYBL2 can contribute to genomic stability by forming complexes with Clathrin and Filamin.68 This is required for proper localization of Clathrin at the mitotic spindle and is thereby stabilizing kinetochore fibers.68 Consistently, in embryonic stem cells (ESC) MYBL2 depletion leads to stalling of replication forks, disorganization of the replication program and an increase in double-strand breaks.41 It has been shown that these effects are, at least in part, mediated by deregulation of MYC and FOXM1 transcription, which are important for normal S phase progression, indicating that MYBL2 protects cells from genomic damage during S phase by promoting proper cell cycle progression.41 Chromosomal fragmentation, shorter and thicker chromatids, end-to-end fusion and chromatid loss upon MYBL2 knockdown indicates that reduced activity of MYBL2 is also associated with structural chromosomal instability.69
MYBL2 in Cell Survival
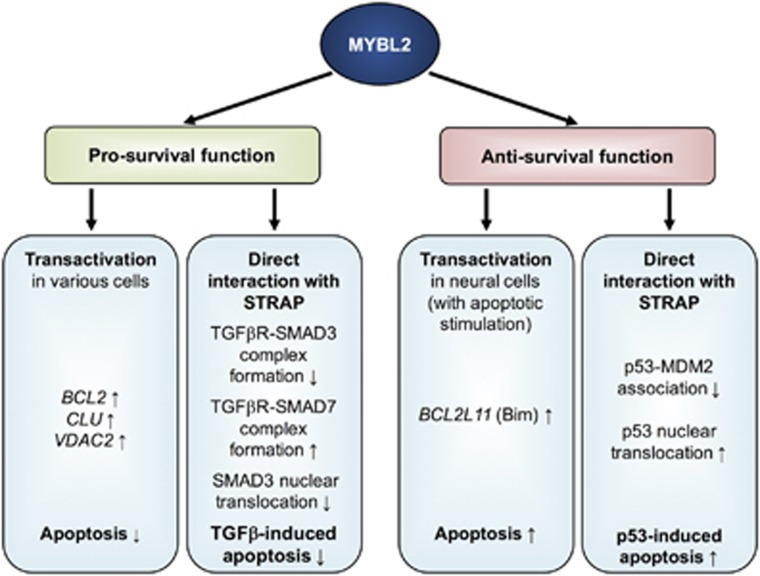
An association between MYBL2 and cell survival has already been reported in early studies. However, over the years, the role of MYBL2 in the regulation of cell survival became increasingly clear and is mainly mediated via transcriptional regulation of specific target genes, but can also be mediated by direct protein–protein interaction. The transcriptional regulation by MYBL2 seems to depend on the cell type: In most cell types MYBL2 appears to have pro-survival functions, whereas it mainly has anti-survival functions in cells of neural origin when exposed to apoptotic stimuli (Figure 2).
Figure 2.
MYBL2 in regulation of cell survival. MYBL2 is primarily described to have pro-survival functions, but also anti-survival functions of MYBL2 were shown. These effects are mainly mediated by MYBL2 transactivating target genes regulating cell survival (pro-survival: BCL2, CLU, VDAC; anti-survival: BCL2L11 (Bim)). Pro-survival functions were described in various cell types, whereas anti-survival functions were mainly described in cells of neural origin when exposed to apoptotic stimuli. Apart from transactivation of its target genes, MYBL2 can also regulate cell survival by direct protein–protein interaction with STRAP. On the one hand, this can lead to inhibition of TGF-β signaling pathways by inhibiting TGF-β receptor association with SMAD3 and enhancing TGF-β receptor association with SMAD7, and thereby prevent translocation of SMAD3 in the nucleus in response to TGF-β1 (pro-survival function). On the other hand, it can result in increased STRAP-mediated stimulation of p53 nuclear translocation, p53-induced apoptosis and cell cycle arrest via reduction of p53-MDM2 association
Pro-survival function via transcriptional regulation
Grassilli et al.47 showed that MYBL2 overexpression in interleukin 2-dependent murine T cells is associated with enhanced transactivation of the anti-apoptotic Bcl-2, and hence diminished cytokine dependence and enhanced resistance to apoptosis induced by doxorubicin, ceramide and dexamethasone. Consistently, the transfection of a Bcl-2-non-expressing human B-cell line with a MYBL2 expression vector induced the expression of Bcl-2 and vice versa, antisense depletion of MYBL2 decreases Bcl-2 levels and enhances apoptosis.70 Furthermore, results of Cervellera et al.50 indicate that ApolipoproteinJ/Clusterin is a MYBL2 target gene, whose expression mediates resistance to apoptosis induced by the chemotherapeutic drug doxorubicin in neuroblastoma. Santilli et al.62 further confirm these results: under conditions of thermal stress, MYBL2-dependent ApolipoproteinJ/Clusterin expression is enhanced due to redox modification of MYBL2 and constitutes a protective response mechanism to thermal injury in MEFs. MYBL2 has also been shown to suppress autophagy and to promote cell survival of ovarian oocytes by binding the promoter and directly activating the transcription of VDAC2.51
Anti-survival function via transcriptional regulation
However, in contrast to these findings, MYBL2 seems to have a contrary role concerning cell survival predominantly in neural cells. MYBL2 knockdown protects pheochromocytoma cells, sympathetic neurons and cortical neurons against cell death elicited by NGF withdrawal or DNA damage.71 This indicates a required role for MYBL2 in neuronal apoptosis after E2F de-repression due to apoptotic stimuli.71, 72 A model has been proposed by which E2F4–p130 protein complexes protect neurons from cell death by occupying the MYBL2 promoter under basal conditions, whereas under conditions of cell stress these complexes are lost and replaced by E2F1 transactivating MYBL2 and thus promoting cell death.73 In neurons, MYBL2 is able to bind the promoter and to transactivate the pro-apoptotic gene BCL2L11 (Bim).48 The following interaction of Bim with the cellular apoptotic machinery leads to caspase activation and apoptotic cell death.48 MYBL2 was also shown to be required for beta-amyloid-dependent induction of Bim and cell death, relevant in Alzheimer’s disease.74 Also in Drosophila, dMyb promotes the programmed cell death of specified sensory organ precursor daughter cells.75 However, not only in neural cells, but also in TGF-β1-treated M1 myeloid leukemia cell lines overexpressing MYBL2, TGF-β1-induced apoptosis was found to be accelerated.76
Direct protein–protein interactions
Independent of the transactivation capabilities of MYBL2, it is further able to regulate cell survival by direct interaction with the serine–threonine kinase receptor-associated protein (STRAP), for which MYBL2 can serve as a positive regulator.77 On the one hand, MYBL2 can enhance STRAP-mediated inhibition of TGF-β signaling pathways, such as apoptosis and growth inhibition, by inhibiting TGF-β receptor association with SMAD3 and enhancing TGF-β receptor association with SMAD7, and thereby prevent translocation of SMAD3 in the nucleus in response to TGF-β1 (pro-survival function).77 On the other hand, co-expression of MYBL2 results in increased STRAP-mediated stimulation of p53 nuclear translocation, p53-induced apoptosis and cell cycle arrest via reduction of p53–MDM2 association (anti-survival function).77
MYBL2 in differentiation and maintenance of stem cell properties
Several lines of evidence indicate that MYBL2 contributes to the maintenance of an undifferentiated and/or stem cell-like phenotype of a cell. Especially in stem cells, the balance between cellular quiescence on the one hand, and cell division in order to generate more stem cells (self-renewal) or to give rise to mature cells (differentiation) on the other hand, is important for the maintenance of the stem cell pool.78
Early results showed that MYBL2 protein levels decrease upon differentiation of human myeloid cell lines.79 Later on, in neuroblastoma cells, MYBL2 expression was found to be downregulated during retinoic acid-induced neural and glial differentiation and conversely, constitutive expression of MYBL2 prevents retinoic acid-induced neural differentiation.80 Compatible with this, levels of p130, a member of the DREAM complex (see ‘MYBL2 in cell cycle regulation’ section) that is able to suppress the MYBL2 promoter upon transfection, was shown to be strongly upregulated during mid/late differentiation stages, whereas MYBL2 levels decrease.81 Comparable results indicating a role for MYBL2 to maintain cells in an undifferentiated state have also been shown for several different cell types, such as leukemic cell lines,82 male gonocytes,83 intestinal epithelial cells38 and keratinocytes.84
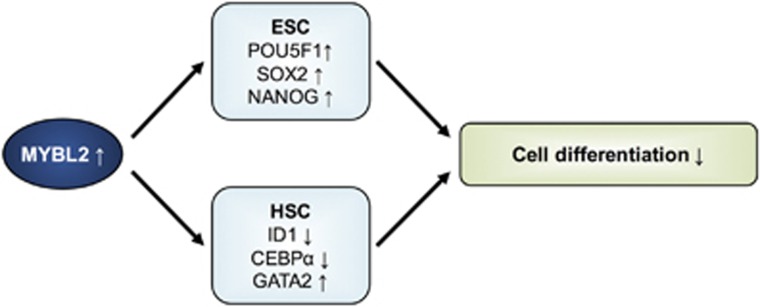
Mechanistically, for the maintenance of a pluripotent and undifferentiated phenotype of ESC, it was proposed that MYBL2 may regulate a transcriptional network that controls cell cycle progression and cell fate to sustain self-renewal and pluripotency.52 Especially for the maintenance of pluripotency, MYBL2 directly regulates the expression of POU5F1, SOX2 and NANOG, which are critical mediators of differentiation and pluripotency in ESC.52, 53, 85 Similarly, MYBL2 was shown to control self-renewal and differentiation of hematopoietic stem cells, possibly by downregulating ID1 and CEBPα, which promote cellular differentiation, while upregulating GATA2, a transcription factor shown to promote proliferation at the expense of differentiation78, 86(Figure 3).
Figure 3.
MYBL2 in differentiation and maintenance of stem cell properties. In various studies, high MYBL2 levels were shown to be associated with cell dedifferentiation (see ‘MYBL2 in differentiation and maintenance of stem cell properties’ section). Mechanistically, the role of MYBL2 in regulation of differentiation was mainly investigated in embryonic stem cells (ESC) and hematopoietic stem cells (HSC): In ESC, MYBL2 was shown to directly control the expression of POU5F1, SOX2 and NANOG, which are critical regulators of differentiation and maintenance of pluripotency. In HSC, MYBL2 was shown to downregulate ID1 and CEBPα, which promote cellular differentiation, and to upregulate GATA2, a transcription factor shown to promote proliferation at the expense of differentiation. Thus, MYBL2 helps the cell to maintain in an undifferentiated, pluripotent, but proliferative state
In summary, these studies indicate that MYBL2 helps the cell to maintain an undifferentiated, pluripotent, but proliferative state.
MYBL2 in Cancer
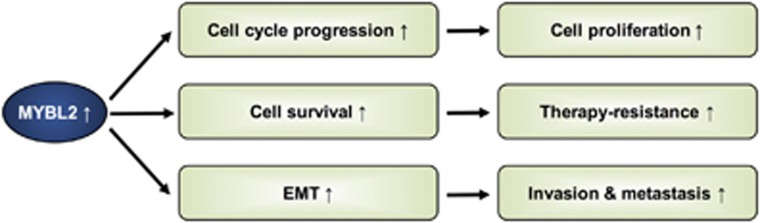
The roles of MYBL2 in cell cycle progression, cell survival and cell differentiation suggest that deregulation of MYBL2 may has an oncogenic potential. It can contribute significantly to cancer progression by promoting cancer cell proliferation, therapy resistance and metastatic spread (Figure 4). Indeed, MYBL2 is frequently found to be overexpressed in several cancer entities and associated with poor patient outcome87, 88, 89, 90, 91, 92, 93 (Table 2).
Figure 4.
MYBL2 deregulation in promotion of cancer initiation and/or progression. Upregulation of MYBL2 is described in numerous cancer entities and is associated with poor patient outcome (Table 2). It leads to an increase in cell cycle progression, cell survival and epithelial-to-mesenchymal transition (EMT), thus promoting cell proliferation, therapy resistance, invasion and metastatic spread
Table 2. MYBL2 deregulation associated with patient outcome in different cancer entities.
| Tumor entity | MYBL2 deregulation | Association with patient survival | References |
|---|---|---|---|
| Acute myeloid leukemia | Overexpression | MYBL2 expression is an independent prognostic factor for disease-free survival and cumulative incidence of relapse | Fuster et al.17 |
| Bladder carcinoma | Overexpression (amplification) | Overrepresentation of amplicons in high-grade and recurrent cases | Nord et al.87 |
| Breast cancer | Overexpression | Overexpression is associated with short overall patient survival and short disease-free survival | Inoue and Fry88 |
| Colorectal cancer | Overexpression | Overexpression is correlated with worse disease-free survival and MYBL2 is an independent prognostic factor for poor patient survival | Ren et al.89 |
| Esophageal squamous cell carcinoma | Overexpression (amplification) | High MYBL2 expression and high MYBL2 copy-number are associated with poor patient survival | Qin et al.90 |
| Hepatocellular carcinoma | Overexpression, high phosphorylation levels | High levels of total and phosphorylated MYBL2 and high levels of LIN9–MYBL2 complex in HCC with poorer outcome | Calvisi et al.91 |
| Neuroblastoma | Overexpression | MYBL2 expression is associated with increased risk of death and worse overall survival | Raschellà et al.92 |
| Renal cell carcinoma | Positivity | Positivity for MYBL2 expression is significantly correlated with clinical stage III and IV | Sakai et al.93 |
Mechanisms of MYBL2 deregulation in cancer
On the one hand, altered MYBL2 expression can arise from chr20q13 amplification, which is described for several cancer entities, for example, breast cancer, colorectal cancer and ovarian cancer.94, 95, 96 On the other hand, it can be caused by deregulation of DREAM complex assembly, for example, due to p53 mutation or transformation by the HPV16 E7 oncogene and thereby uncoupling MYBL2 expression from negative transcriptional regulation and enabling MYBL2 to increasingly bind to MuvB and FOXM1 (see below). Additionally, MYBL2 expression can be deregulated at a post-transcriptional level via deregulation of microRNAs, a class of small non-coding RNAs often found to be deregulated in cancer97 and of which some were shown to suppress MYBL2 mRNA expression.14, 15, 16, 17, 18, 19
p53 signaling is frequently altered in cancers.98 Physiologically, p53 directly activates p21, which prevents following p130 phosphorylation by cyclin-dependent kinases, leading to a switch from MYBL2–MuvB to DREAM complex by shifting MuvB-associated proteins from MYBL2 to E2F4/DP1/p130.99 It has been described that the p53-p21-DREAM pathway represses MYBL2 expression, especially under conditions of cellular stress, such as DNA damage, which mechanistically explains MYBL2 deregulation, and thus deregulation of cell cycle progression and cell survival in p53-mutated cancers.100, 101 This is in accordance with results from Parikh et al.102 showing that MYBL2 is disproportionately upregulated in many p53 mutant cancers. MYBL2 has even been shown to overcome DNA damage-induced G2 checkpoint arrest in p53 mutant cells103 and constitutive expression of MYBL2 has been shown to overcome p53-induced G1 checkpoint arrest.59 Furthermore, the oncoviral HPV16 E7 protein is able to deregulate DREAM complex assembly and to thereby drive MYBL2 expression.104 Consistent to this, HPV16-immortalized cells show upregulated expression of MYBL2.105 Mechanistically, the HPV16 E7 oncogene can bind to p130, promote its proteasomal degradation and thereby disassemble the DREAM complex.106 E7 in addition directly binds to the MYBL2–MuvB–FoxM1 complex, leading to cooperative transcriptional activation of mitotic genes.106 MYBL2 moreover mediates abrogation of DNA damage-induced G1 checkpoint arrest, via regulation of CDK1 expression in E7 transformed cells107 and was shown to rescue oncogene-induced cellular senescence,14, 108 a permanent cell cycle arrest that cells must bypass during cancer development,14, 109 probably by suppressing p16INK4A expression.14, 108, 110
MYBL2 in deregulation of proliferation
As described for non-malignant cells (see ‘MYBL2 in cell cycle regulation’ section), MYBL2 has also been shown to drive cell proliferation and/or cell cycle progression in cancer cells, such as breast cancer,111 cervical cancer,112 colorectal cancer,89 hepatocellular carcinoma,91 leukemic cells,15 lung adenocarcinoma42 and neuroblastoma (in MYCN-amplified cell lines).113
MYBL2 in cancer therapy resistance
Resistance to chemo- and radiotherapy is one of the main properties of a cancer that determines cancer progression and patient outcome. MYBL2 overexpression in interleukin 2-dependent murine T cells was shown to be associated with enhanced resistance to drug-induced apoptosis by doxorubicin, ceramide and dexamethasone, due to increased transactivation of the anti-apoptotic Bcl-2 by MYBL2.47 These results from Grassilli et al. are in accordance with results from Levenson et al.,114 showing that MYBL2 is upregulated upon genetic suppressor element-induced drug resistance to DNA-interactive agents, such as aphidicolin, hydroxyurea, cytarabine, etoposide, doxorubicin and mafosfamide in fibrosarcoma cells. In neuroblastoma, MYBL2 directly regulates expression of ApolipoproteinJ/Clusterin and thereby mediates resistance to apoptosis induced by doxorubicin.50
However, MYBL2 was not only shown to mediate resistance to chemotherapeutic agents, but also resistance to DNA damage, as, for example, caused by radiation. Under physiological conditions, such as in p53 wild-type cells, DNA damage results in p53-dependent binding of p130 and E2F4 to MuvB and the dissociation of the MYBL2–MuvB complex.99, 100, 101 Also, upon ionizing radiation, Cyclin F suppresses the MYBL2-regulated transcriptional program by directly interacting with MYBL2 and thereby suppressing Cyclin A-mediated phosphorylation of MYBL2.115 On the contrary, under non-physiological conditions, such as in p53 mutant cells, MYBL2 fails to dissociate from MuvB, which contributes to increased G2/M gene expression in response to DNA damage.103 In accordance, DT40 chicken B cells lacking MYBL2 show increased sensitivity to DNA damage elicited by UV irradiation and alkylation.116 Consistently, in Ewing sarcoma cells, MYBL2 can be destroyed quickly upon UV irradiation, leading to induction of apoptosis,117 whereas this is not the case in neuroblastoma, where MYBL2 levels do not change upon irradiation, making the cells resistant to UV-induced apoptosis.117 Interestingly, in neuroblastoma cells MYBL2 is found to be hypophosphorylated and overexpression of a non-phosphorylatable MYBL2 mutant in HEK 293 cells can protect cells from UV-induced apoptotic cell death, suggesting that decreased Cyclin A-dependent phosphorylation, accompanied by decreased activation but also decreased proteasomal degradation, can facilitate the survival promoting activity of MYBL2.117
Consistent with these results, a pro-survival role for MYBL2 has also been shown in several cancer cell lines, such as colorectal cancer,89 hepatocellular carcinoma91 and leukemia cells.118
MYBL2 in invasion and metastasis
Early results of Iwai et al.58 have shown that the introduction of an inducible dominant interfering Myb protein into ESC lead to dissociation of ESC colonies into dispersed single cells and to reduced adhesion of the ESC to the culture dish. Cell adhesion analyses have shown that MYBL2 suppression decreased the adhesion with extracellular matrix proteins, such as Laminin, Collagen and Fibronectin, probably due to reduced cell surface expression of Beta1 Integrin.58
However, in contrast to these early findings, a role for MYBL2 in epithelial-to-mesenchymal transition (EMT), a process in which epithelial cells lose their polarity and gain migratory and invasive properties,119 has been proposed: In breast cancer cells, MYBL2 knockdown is able to restore the expression of the epithelial marker E-Cadherin, the formation of cell–cell junctions and to suppress cell invasion, anchorage-independent growth and tumor formation.120 Conversely, MYBL2 overexpression decreased the expression of the E-Cadherin, but increased expression of mesenchymal markers.120 Mechanistically, it was proposed that MYBL2 upregulates the expression of the major EMT regulator SNAIL, thereby mediating activation of EMT and cancer cell invasion.120
In accordance with this, MYBL2 protein levels have been shown to be significantly upregulated in matched breast cancer metastases compared to the primary tumor.111 Similar results were shown for prostate cancer and renal cell carcinoma: MYBL2 is overexpressed in prostate cancer (xenograft) metastases,121 whereas in renal cell carcinoma MYBL2 was found to be expressed in metastases from primary tumors being MYBL2 negative.93
Conclusions
MYBL2 is a central regulator of cell cycle progression, cell survival and cell differentiation. Deregulation of MYBL2 expression can contribute significantly to cancer progression by promoting cancer cell proliferation, therapy resistance, metastatic spread and is correlated with poor patient outcome in several cancer entities. Therefore, MYBL2 and/or players of its downstream transcriptional network could serve as effective targets for cancer treatment. Although no direct MYBL2 inhibitor is available yet, CDK2 inhibition could be used to reduce MYBL2 activity in MYBL2 high-expressing cancers. Yet, highly specific CDK2 inhibitors are lacking, but several more or less specific CDK inhibitors have already been in clinical trials for cancer treatment.122, 123 Also, several inhibitors interfering with players of the downstream transcriptional network of MYBL2, such as inhibitors against Aurora kinases,124 FGF receptors,125 Kinesins,126 Bcl-2127 and BIRC5 (Survivin)128 have already been in clinical trials and may serve as an effective, more specific and less toxic future anti-cancer therapy in cancers highly expressing MYBL2.
Acknowledgments
JM acknowledges support from the Kind-Philipp-Foundation. The laboratory of TGPG is supported by grants from the ‘Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed)’, the Daimler and Benz Foundation in cooperation with the Reinhard Frank Foundation, by LMU Munich’s Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative, the ‘Mehr LEBEN für krebskranke Kinder – Bettina-Bräu-Stiftung’, the Walter Schulz Foundation, the Friedrich-Baur Foundation, the Wilhelm Sander-Foundation (2016.167.1) and by the German Cancer Aid (DKH-111886 and DKH-70112257).
Footnotes
Edited by G Raschellà
The authors declare no conflict of interest.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100: 57–70. [DOI] [PubMed] [Google Scholar]
- Roussel M, Saule S, Lagrou C, Rommens C, Beug H, Graf T et al. Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature 1979; 281: 452–455. [DOI] [PubMed] [Google Scholar]
- Ness SA. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol Dis 2003; 31: 192–200. [DOI] [PubMed] [Google Scholar]
- Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S et al. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res 1988; 16: 11075–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Lee HW, Jeon GS, Lee HY, Lee KH, Cho SS. Constitutive expression of c-myb mRNA in the adult rat brain. Brain Res 2001; 892: 203–207. [DOI] [PubMed] [Google Scholar]
- Zorbas M, Sicurella C, Bertoncello I, Venter D, Ellis S, Mucenski ML et al. c-Myb is critical for murine colon development. Oncogene 1999; 18: 5821–5830. [DOI] [PubMed] [Google Scholar]
- Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, Orth J et al. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature 1997; 386: 713–717. [DOI] [PubMed] [Google Scholar]
- Trauth K, Mutschler B, Jenkins NA, Gilbert DJ, Copeland NG, Klempnauer KH. Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. EMBO J 1994; 13: 5994–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Patestos NP, Maekawa T, Ishii S. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem 1999; 274: 28067–28070. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 1991; 65: 677–689. [DOI] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer 2008; 8: 523–534. [DOI] [PubMed] [Google Scholar]
- Golay J, Luppi M, Songia S, Palvarini C, Lombardi L, Aiello A et al. Expression of A-myb, but not c-myb and B-myb, is restricted to Burkitt’s lymphoma, sIg+ B-acute lymphoblastic leukemia, and a subset of chronic lymphocytic leukemias. Blood 1996; 87: 1900–1911. [PubMed] [Google Scholar]
- Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 2013; 13: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, D Dimaio. B-Myb, cancer, senescence, and microRNAs. Cancer Res 2011; 71: 5370–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauli G, Voltan R, di Iasio MG, Bosco R, Melloni E, Sana ME et al. miR-34a induces the downregulation of both E2F1 and B-Myb oncogenes in leukemic cells. Clin Cancer Res 2011; 17: 2712–2724. [DOI] [PubMed] [Google Scholar]
- Papetti M, LH Augenlicht. Mybl2 downregulated during colon epithelial cell maturation, is suppressed by miR-365. Am J Physiol Gastrointest Liver Physiol 2011; 301: G508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster O, Llop M, Dolz S, García P, Such E, Ibáñez M et al. Adverse prognostic value of MYBL2 overexpression and association with microRNA-30 family in acute myeloid leukemia patients. Leuk Res 2013; 37: 1690–1696. [DOI] [PubMed] [Google Scholar]
- Wu Z, Huang X, Huang X, Zou Q, Guo Y. The inhibitory role of Mir-29 in growth of breast cancer cells. J Exp Clin Cancer Res 2013; 32: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Kim J, He L, Creighton CJ, Gunaratne PH, Hawkins SM et al. Functional analysis of miR-34c as a putative tumor suppressor in high-grade serous ovarian cancer. Biol Reprod 2014; 91: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A, Kundu M, Casella I, Engelhard A, Calabretta B, Grasso L et al. Activation of human B-MYB by cyclins. Proc Natl Acad Sci USA 1997; 94: 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TK, Schweppe RE, Septer J, Lewis RE. Phosphorylation of B-Myb regulates its transactivation potential and DNA binding. J Biol Chem 1999; 274: 36741–36749. [DOI] [PubMed] [Google Scholar]
- Li X, McDonnell DP. The transcription factor B-Myb is maintained in an inhibited state in target cells through its interaction with the nuclear corepressors N-CoR and SMRT. Mol Cell Biol 2002; 22: 3663–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S, Carena I, Brondani V, Klempnauer KH, Ferrari S. Degradation of B-Myb by ubiquitin-mediated proteolysis: involvement of the Cdc34-SCF(p45Skp2) pathway. Oncogene 2000; 19: 2986–2995. [DOI] [PubMed] [Google Scholar]
- Schubert S, Horstmann S, Bartusel T, Klempnauer K-H. The cooperation of B-Myb with the coactivator p300 is orchestrated by cyclins A and D1. Oncogene 2004; 23: 1392–1404. [DOI] [PubMed] [Google Scholar]
- Horstmann S, Ferrari S, Klempnauer KH. Regulation of B-Myb activity by cyclin D1. Oncogene 2000; 19: 298–306. [DOI] [PubMed] [Google Scholar]
- Cervellera MN, Sala A. Poly(ADP-ribose) polymerase is a B-MYB coactivator. J Biol Chem 2000; 275: 10692–10696. [DOI] [PubMed] [Google Scholar]
- Santilli G, Cervellera MN, Johnson TK, Lewis RE, Iacobelli S, Sala A. PARP co-activates B-MYB through enhanced phosphorylation at cyclin/cdk2 sites. Oncogene 2001; 20: 8167–8174. [DOI] [PubMed] [Google Scholar]
- Seong H-A, Kim K-T, Ha H. Enhancement of B-MYB transcriptional activity by ZPR9, a novel zinc finger protein. J Biol Chem 2003; 278: 9655–9662. [DOI] [PubMed] [Google Scholar]
- Bartusel T, Klempnauer K-H. Transactivation mediated by B-Myb is dependent on TAF(II)250. Oncogene 2003; 22: 2932–2941. [DOI] [PubMed] [Google Scholar]
- Sala A, De Luca A, Giordano A, Peschle C. The retinoblastoma family member p107 binds to B-MYB and suppresses its autoregulatory activity. J Biol Chem 1996; 271: 28738–28740. [DOI] [PubMed] [Google Scholar]
- Joaquin M, Bessa M, Saville MK, Watson RJ. B-Myb overcomes a p107-mediated cell proliferation block by interacting with an N-terminal domain of p107. Oncogene 2002; 21: 7923–7932. [DOI] [PubMed] [Google Scholar]
- Joaquin M, Watson RJ. The cell cycle-regulated B-Myb transcription factor overcomes cyclin-dependent kinase inhibitory activity of p57(KIP2) by interacting with its cyclin-binding domain. J Biol Chem 2003; 278: 44255–44264. [DOI] [PubMed] [Google Scholar]
- De Falco G, Bagella L, Claudio PP, De Luca A, Fu Y, Calabretta B et al. Physical interaction between CDK9 and B-Myb results in suppression of B-Myb gene autoregulation. Oncogene 2000; 19: 373–379. [DOI] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. EMBO J 2004; 23: 4615–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartusel T, Schubert S, Klempnauer K-H. Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene 2005; 351: 171–180. [DOI] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev 2012; 26: 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh L, von Eyss B, Schmit F, Rein L, Hübner D, Samans B et al. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J 2007; 26: 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papetti M, Augenlicht LH. MYBL2, a link between proliferation and differentiation in maturing colon epithelial cells. J Cell Physiol 2011; 226: 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt TL, Fraser DJ, Leal S, Halandras PM, Kroll AR, Kroll DJ. c-Myb trans-activates the human DNA topoisomerase IIalpha gene promoter. J Biol Chem 1997; 272: 6278–6284. [DOI] [PubMed] [Google Scholar]
- Johnson LR, Johnson TK, Desler M, Luster TA, Nowling T, Lewis RE et al. Effects of B-Myb on gene transcription: phosphorylation-dependent activity and acetylation by p300. J Biol Chem 2002; 277: 4088–4097. [DOI] [PubMed] [Google Scholar]
- Lorvellec M, Dumon S, Maya-Mendoza A, Jackson D, Frampton J, García P. B-Myb is critical for proper DNA duplication during an unperturbed S phase in mouse embryonic stem cells. Stem Cells 2010; 28: 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iltzsche F, Simon K, Stopp S, Pattschull G, Francke S, Wolter P et al. An important role for Myb-MuvB and its target gene KIF23 in a mouse model of lung adenocarcinoma. Oncogene 2017; 36: 110–121. [DOI] [PubMed] [Google Scholar]
- Wolter P, Hanselmann S, Pattschull G, Schruf E, Gaubatz S. Central spindle proteins and mitotic kinesins are direct transcriptional targets of MuvB, B-MYB and FOXM1 in breast cancer cell lines and are potential targets for therapy. Oncotarget 2017; 8: 11160–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra J, Withers DA, Boxer LM. Myb binding sites mediate negative regulation of c-myb expression in T-cell lines. Blood 1995; 86: 1873–1880. [PubMed] [Google Scholar]
- Sala A, Saitta B, De Luca P, Cervellera MN, Casella I, Lewis RE et al. B-MYB transactivates its own promoter through SP1-binding sites. Oncogene 1999; 18: 1333–1339. [DOI] [PubMed] [Google Scholar]
- Nakagoshi H, Kanei-Ishii C, Sawazaki T, Mizuguchi G, Ishii S. Transcriptional activation of the c-myc gene by the c-myb and B-myb gene products. Oncogene 1992; 7: 1233–1240. [PubMed] [Google Scholar]
- Grassilli E, Salomoni P, Perrotti D, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells overexpressing B-Myb is associated with B-Myb-dependent bcl-2 induction. Cancer Res 1999; 59: 2451–2456. [PubMed] [Google Scholar]
- Greene LA, Liu DX, Troy CM, Biswas SC. Cell cycle molecules define a pathway required for neuron death in development and disease. Biochim Biophys Acta 2007; 1772: 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene 2009; 28: 1737–1747. [DOI] [PubMed] [Google Scholar]
- Cervellera M, Raschella G, Santilli G, Tanno B, Ventura A, Mancini C et al. Direct transactivation of the anti-apoptotic gene apolipoprotein J (clusterin) by B-MYB. J Biol Chem 2000; 275: 21055–21060. [DOI] [PubMed] [Google Scholar]
- Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H, Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy 2015; 11: 1081–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan M, Riordon DR, Yan B, Tarasova YS, Bruweleit S, Tarasov KV et al. The B-MYB transcriptional network guides cell cycle progression and fate decisions to sustain self-renewal and the identity of pluripotent stem cells. PLoS ONE 2012; 7: e42350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov KV, Tarasova YS, Tam WL, Riordon DR, Elliott ST, Kania G et al. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS ONE 2008; 3: e2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller GA, Quaas M, Schümann M, Krause E, Padi M, Fischer M et al. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res 2012; 40: 1561–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol 2008; 10: 1076–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol 2008; 28: 5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsura M, Introna M, Passerini F, Mantovani A, Golay J. B-myb antisense oligonucleotides inhibit proliferation of human hematopoietic cell lines. Blood 1992; 79: 2708–2716. [PubMed] [Google Scholar]
- Iwai N, Kitajima K, Sakai K, Kimura T, Nakano T. Alteration of cell adhesion and cell cycle properties of ES cells by an inducible dominant interfering Myb mutant. Oncogene 2001; 20: 1425–1434. [DOI] [PubMed] [Google Scholar]
- Lin D, Fiscella M, O’Connor PM, Jackman J, Chen M, Luo LL et al. Constitutive expression of B-myb can bypass p53-induced Waf1/Cip1-mediated G1 arrest. Proc Natl Acad Sci USA 1994; 91: 10079–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen AL, Jackson J, Harmon BP, Fung SM, Ramsay G, Bishop JM. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev 1998; 12: 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Akimaru H, Hou D-X, Takahashi T, Ishii S. Myb controls G(2)/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J 2002; 21: 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santilli G, Schwab R, Watson R, Ebert C, Aronow BJ, Sala A. Temperature-dependent modification and activation of B-MYB: implications for cell survival. J Biol Chem 2005; 280: 15628–15634. [DOI] [PubMed] [Google Scholar]
- Pilkinton M, Sandoval R, Colamonici OR. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene 2007; 26: 7535–7543. [DOI] [PubMed] [Google Scholar]
- Davidson CJ, Tirouvanziam R, Herzenberg LA, Lipsick JS. Functional evolution of the vertebrate Myb gene family: B-Myb, but neither A-Myb nor c-Myb, complements Drosophila Myb in hemocytes. Genetics 2005; 169: 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S-M, Ramsay G, Katzen AL. Mutations in Drosophila myb lead to centrosome amplification and genomic instability. Development 2002; 129: 347–359. [DOI] [PubMed] [Google Scholar]
- Manak JR, Mitiku N, Lipsick JS. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc Natl Acad Sci USA 2002; 99: 7438–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard JL, Amatruda JF, Stern HM, Subramanian A, Finkelstein D, Ziai J et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc Natl Acad Sci USA 2005; 102: 13194–13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Ishidao T, Nomura T, Shinagawa T, Tanaka Y, Yonemura S et al. A B-Myb complex containing clathrin and filamin is required for mitotic spindle function. EMBO J 2008; 27: 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García P, Frampton J. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. J Cell Sci 2006; 119: 1483–1493. [DOI] [PubMed] [Google Scholar]
- Lang G, Gombert WM, Gould HJ. A transcriptional regulatory element in the coding sequence of the human Bcl-2 gene. Immunology 2005; 114: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Biswas SC, Greene LA. B-myb and C-myb play required roles in neuronal apoptosis evoked by nerve growth factor deprivation and DNA damage. J Neurosci 2004; 24: 8720–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Greene LA. Regulation of neuronal survival and death by E2F-dependent gene repression and derepression. Neuron 2001; 32: 425–438. [DOI] [PubMed] [Google Scholar]
- Iyirhiaro GO, Zhang Y, Estey C, O’Hare MJ, Safarpour F, Parsanejad M et al. Regulation of ischemic neuronal death by E2F4-p130 protein complexes. J Biol Chem 2014; 289: 18202–18213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SC, Shi Y, Vonsattel J-PG, Leung CL, Troy CM, Greene LA. Bim is elevated in Alzheimer’s disease neurons and is required for beta-amyloid-induced neuronal apoptosis. J Neurosci 2007; 27: 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovani MK, Brachmann CB, Ramsay G, Katzen AL. The dREAM/Myb-MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin. Dev Biol 2012; 372: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies J, Wolff L. Acceleration of apoptosis in transforming growth factor beta 1-treated M1 cells ectopically expressing B-myb. Cancer Res 1995; 55: 501–504. [PubMed] [Google Scholar]
- Seong H-A, Manoharan R, Ha H. B-MYB positively regulates serine-threonine kinase receptor-associated protein (STRAP) activity through direct interaction. J Biol Chem 2011; 286: 7439–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ, Ma’ayan A, Lieu YK, John P, Reddy MVR, Chen EY et al. B-myb is an essential regulator of hematopoietic stem cell and myeloid progenitor cell development. Proc Natl Acad Sci USA 2014; 111: 3122–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsura M, Luchetti MM, Erba E, Golay J, Rambaldi A, Introna M. Dissociation between p93B-myb and p75c-myb expression during the proliferation and differentiation of human myeloid cell lines. Blood 1994; 83: 1778–1790. [PubMed] [Google Scholar]
- Raschellà G, Negroni A, Sala A, Pucci S, Romeo A, Calabretta B. Requirement of b-myb function for survival and differentiative potential of human neuroblastoma cells. J Biol Chem 1995; 270: 8540–8545. [DOI] [PubMed] [Google Scholar]
- Raschellà G, Tanno B, Bonetto F, Negroni A, Claudio PP, Baldi A et al. The RB-related gene Rb2/p130 in neuroblastoma differentiation and in B-myb promoter down-regulation. Cell Death Differ 1998; 5: 401–407. [DOI] [PubMed] [Google Scholar]
- Bies J, Hoffman B, Amanullah A, Giese T, Wolff L. B-Myb prevents growth arrest associated with terminal differentiation of monocytic cells. Oncogene 1996; 12: 355–363. [PubMed] [Google Scholar]
- Latham KE, Litvin J, Orth JM, Patel B, Mettus R, Reddy EP. Temporal patterns of A-myb and B-myb gene expression during testis development. Oncogene 1996; 13: 1161–1168. [PubMed] [Google Scholar]
- Maruyama H, Ishitsuka Y, Fujisawa Y, Furuta J, Sekido M, Kawachi Y. B-Myb enhances proliferation and suppresses differentiation of keratinocytes in three-dimensional cell culture. Arch Dermatol Res 2014; 306: 375–384. [DOI] [PubMed] [Google Scholar]
- Tarasov KV, Testa G, Tarasova YS, Kania G, Riordon DR, Volkova M et al. Linkage of pluripotent stem cell-associated transcripts to regulatory gene networks. Cells Tissues Organs 2008; 188: 31–45. [DOI] [PubMed] [Google Scholar]
- Briegel K, Lim KC, Plank C, Beug H, Engel JD, Zenke M. Ectopic expression of a conditional GATA-2/estrogen receptor chimera arrests erythroid differentiation in a hormone-dependent manner. Genes Dev 1993; 7: 1097–1109. [DOI] [PubMed] [Google Scholar]
- Nord H, Segersten U, Sandgren J, Wester K, Busch C, Menzel U et al. Focal amplifications are associated with high grade and recurrences in stage Ta bladder carcinoma. Int J Cancer 2010; 126: 1390–1402. [DOI] [PubMed] [Google Scholar]
- Inoue K, Fry EA. Novel molecular markers for breast cancer. Biomark Cancer 2016; 8: 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Wang L, Shen X, Xiao X, Liu Z, Wei P et al. MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am J Cancer Res 2015; 5: 1542–1552. [PMC free article] [PubMed] [Google Scholar]
- Qin H-D, Liao X-Y, Chen Y-B, Huang S-Y, Xue W-Q, Li F-F et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet 2016; 98: 709–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvisi DF, Simile MM, Ladu S, Frau M, Evert M, Tomasi ML et al. Activation of v-Myb avian myeloblastosis viral oncogene homolog-like2 (MYBL2)-LIN9 complex contributes to human hepatocarcinogenesis and identifies a subset of hepatocellular carcinoma with mutant p53. Hepatology 2011; 53: 1226–1236. [DOI] [PubMed] [Google Scholar]
- Raschellà G, Cesi V, Amendola R, Negroni A, Tanno B, Altavista P et al. Expression of B-myb in neuroblastoma tumors is a poor prognostic factor independent from MYCN amplification. Cancer Res 1999; 59: 3365–3368. [PubMed] [Google Scholar]
- Sakai N, Kubota Y, Shuin T. Statistically significant expression of B-myb in clinically advanced human renal-cell carcinomas. Int J Oncol 1993; 2: 419–423. [DOI] [PubMed] [Google Scholar]
- Shi H, Bevier M, Johansson R, Grzybowska E, Chen B, Eyfjörd JE et al. Single nucleotide polymorphisms in the 20q13 amplicon genes in relation to breast cancer risk and clinical outcome. Breast Cancer Res Treat 2011; 130: 905–916. [DOI] [PubMed] [Google Scholar]
- Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, Hopt U et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med 2007; 85: 293–304. [DOI] [PubMed] [Google Scholar]
- Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T et al. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res 2000; 6: 1833–1839. [PubMed] [Google Scholar]
- Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle Georget Tex 2008; 7: 2643–2646. [DOI] [PubMed] [Google Scholar]
- Stracquadanio G, Wang X, Wallace MD, Grawenda AM, Zhang P, Hewitt J et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat Rev Cancer 2016; 16: 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaas M, Müller GA, Engeland K. p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle 2012; 11: 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Quaas M, Steiner L, Engeland K. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res 2016; 44: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Quaas M, Nickel A, Engeland K. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget 2015; 6: 41402–41417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh N, Hilsenbeck S, Creighton CJ, Dayaram T, Shuck R, Shinbrot E et al. Effects of TP53 mutational status on gene expression patterns across 10 human cancer types. J Pathol 2014; 232: 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannefeld M, Klassen E, Gaubatz S. B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res 2009; 69: 4073–4080. [DOI] [PubMed] [Google Scholar]
- Lam EW, Morris JD, Davies R, Crook T, Watson RJ, Vousden KH. HPV16 E7 oncoprotein deregulates B-myb expression: correlation with targeting of p107/E2F complexes. EMBO J 1994; 13: 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Nakao Y, Pater MM, Tang SC, Pater A. Expression of cellular genes in HPV16-immortalized and cigarette smoke condensate-transformed human endocervical cells. J Cell Biochem 1997; 66: 309–321. [PubMed] [Google Scholar]
- Pang CL, Toh SY, He P, Teissier S, Ben Khalifa Y, Xue Y et al. A functional interaction of E7 with B-Myb-MuvB complex promotes acute cooperative transcriptional activation of both S- and M-phase genes. (129 c). Oncogene 2014; 33: 4039–4049. [DOI] [PubMed] [Google Scholar]
- Fan X, Chen JJ. Role of Cdk1 in DNA damage-induced G1 checkpoint abrogation by the human papillomavirus E7 oncogene. Cell Cycle 2014; 13: 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowla SN, Lam EW-F, Jat PS. Cellular senescence and aging: the role of B-MYB. Aging Cell 2014; 13: 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8: 729–740. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wu J, Li R, Wang P, Han L, Zhang Z et al. B-MYB delays cell aging by repressing p16 (INK4α) transcription. Cell Mol Life Sci CMLS 2011; 68: 893–901. [DOI] [PMC free article] [PubMed]
- Tao D, Pan Y, Lu H, Zheng S, Lin H, Fang H et al. B-myb is a gene implicated in cell cycle and proliferation of breast cancer. Int J Clin Exp Pathol 2014; 7: 5819–5827. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nor Rashid N, Yusof R, Watson RJ. Disruption of repressive p130-DREAM complexes by human papillomavirus 16 E6/E7 oncoproteins is required for cell-cycle progression in cervical cancer cells. J Gen Virol 2011; 92: 2620–2627. [DOI] [PubMed] [Google Scholar]
- Gualdrini F, Corvetta D, Cantilena S, Chayka O, Tanno B, Raschellà G et al. Addiction of MYCN amplified tumours to B-MYB underscores a reciprocal regulatory loop. Oncotarget 2010; 1: 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson VV, Davidovich IA, Roninson IB. Pleiotropic resistance to DNA-interactive drugs is associated with increased expression of genes involved in DNA replication, repair, and stress response. Cancer Res 2000; 60: 5027–5030. [PubMed] [Google Scholar]
- Klein DK, Hoffmann S, Ahlskog JK, O’Hanlon K, Quaas M, Larsen BD et al. Cyclin F suppresses B-Myb activity to promote cell cycle checkpoint control. Nat Commun 2015; 6: 5800. [DOI] [PubMed] [Google Scholar]
- Ahlbory D, Appl H, Lang D, Klempnauer K-H. Disruption of B-myb in DT40 cells reveals novel function for B-Myb in the response to DNA-damage. Oncogene 2005; 24: 7127–7134. [DOI] [PubMed] [Google Scholar]
- Schwab R, Caccamo A, Bettuzzi S, Anderson J, Sala A. B-MYB is hypophosphorylated and resistant to degradation in neuroblastoma: implications for cell survival. Blood Cells Mol Dis 2007; 39: 263–271. [DOI] [PubMed] [Google Scholar]
- Vrana JA, Decker RH, Johnson CR, Wang Z, Jarvis WD, Richon VM et al. Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene 1999; 18: 7016–7025. [DOI] [PubMed] [Google Scholar]
- Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol 2016; 43: 7–13. [DOI] [PubMed] [Google Scholar]
- Tao D, Pan Y, Jiang G, Lu H, Zheng S, Lin H et al. B-Myb regulates snail expression to promote epithelial-to-mesenchymal transition and invasion of breast cancer cell. Med Oncol 2015; 32: 412. [DOI] [PubMed] [Google Scholar]
- Bar-Shira A, Pinthus JH, Rozovsky U, Goldstein M, Sellers WR, Yaron Y et al. Multiple genes in human 20q13 chromosomal region are involved in an advanced prostate cancer xenograft. Cancer Res 2002; 62: 6803–6807. [PubMed] [Google Scholar]
- Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 2015; 14: 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R. Cyclin-dependent protein kinase inhibitors including palbociclib as anticancer drugs. Pharmacol Res 2016; 107: 249–275. [DOI] [PubMed] [Google Scholar]
- Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN, Gandara DR. Aurora kinases as anticancer drug targets. Clin Cancer Res 2008; 14: 1639–1648. [DOI] [PubMed] [Google Scholar]
- Chae YK, Ranganath K, Hammerman PS, Vaklavas C, Mohindra N, Kalyan A et al. Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget 2016; 8: 16052–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran G, Tátrai P, Gergely F. Hitting the brakes: targeting microtubule motors in cancer. Br J Cancer 2015; 113: 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Huang D. Targeting BCL2 with BH3 mimetics: basic science and clinical application of venetoclax in chronic lymphocytic leukemia and related b cell malignancies. Clin Pharmacol Ther 2017; 101: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg H, Suri P, Gupta JC, Talwar GP, Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int 2016; 16: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]