Dear Editor,
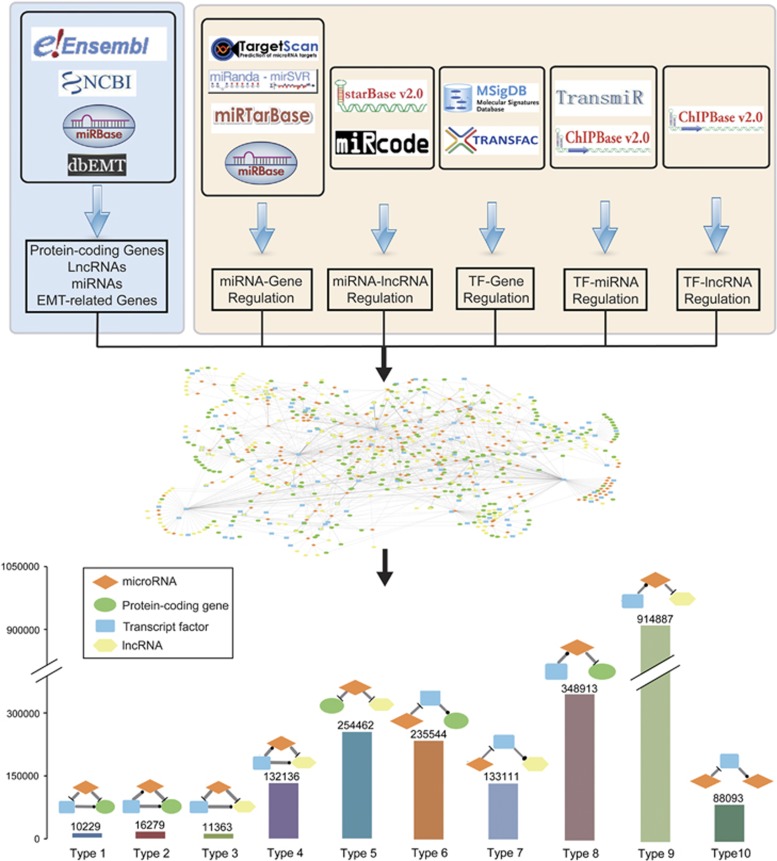
The Epithelial-mesenchymal transition (EMT) is a fundamental cellular process that governs biological processes, such as embryonic development, tumor cell metastasis, organ fibrosis and tissue regeneration.1 EMT allows polarized, immotile epithelial cells to transform into motile mesenchymal cells.2 This switch is mediated by regulatory factors such as transcription factor (TF), microRNA (miRNA) and long non-coding RNA (lncRNA) at transcriptional and post-transcriptional levels.3, 4 These regulatory factors play co-regulational roles on the EMT process and the regulatory loops act as motifs, which are the basic units in the regulatory network.5 However, a database with comprehensive EMT regulatory relationships is still lacking. EMT-Regulome comprehensively collected the interactions between regulators and targets, including TF-target, miRNA-target, miRNA-lncRNA, TF-lncRNA and TF-miRNA and identified 10 types of co-regulation motifs for EMT process (Figure 1). EMT-Regulome provides the option for searching EMT-related regulatory interactions, motifs and networks, which facilitates the investigations of the mechanisms of EMT involved in diseases, such as tumor cell invasion. The database is freely available at http://www.medsysbio.org/EMTRegulome.
Figure 1.
The flow chart of the EMT-Regulome database
In current version, EMT-Regulome contains 1740 miRNAs, 8783 lncRNAs and 13 025 coding genes, which includes 370 TF coding genes and 350 EMT-related genes (Supplementary Figure 1A). We collected regulatory relationships, including TF-targets, miRNA-targets, miRNA-lncRNA, TF-lncRNA and TF-miRNA, from public databases (Figure 1). Then, we integrated the regulation into a comprehensive network and searched 10 types of motifs that are the basic units in the regulatory network (Figure 1). Especially, for the fifth motif type, known as competing endogenous RNA (ceRNA), we used hypergeometric test to measure whether two RNA components significantly shared miRNAs. The motifs containing at least one EMT-related gene from dbEMT,6 which collected experimentally verified EMT-related genes from literatures, were defined as EMT-related motifs in the EMT-Regulome database. The statistics of motifs are shown in Figure 1. The EMT-Regulome database provides a user-friendly interface that allows users to conveniently search, browse and download data (Supplementary Figure 2). In the ‘Search’ page, the query function can be performed using Symbol or ID of coding gene/miRNA/lncRNA with specified motif types. The search results are composed of three parts:(i) information box of gene, miRNA or lncRNA, which is offered as a search condition, including brief functional description, location in chromosome and external link to NCBI, miRBase or Ensembl for more detail; (ii) motifs are grouped by types, and source databases of their regulation are provided. (iii) returned motifs could be visualized as a network and are allowed to be exported as PNG files in various layouts by the Cytoscape web tool (http://js.cytoscape.org).
As a case analysis, the EMT-Regulome database was applied to identify potential activated EMT motifs in the mesenchymal subtype of ovarian cancer by employing expression profiles from The Cancer Genome Atlas (TCGA). Yang et al.7 have classified the TCGA ovarian cancer samples into integrated epithelial (iE) subtype and integrated mesenchymal (iM) subtype labels. Here, the activated motifs were defined as the nodes in motifs that were differentially expressed in iM compared with the iE ovarian cancer samples (T-test) and the edges in motifs were differentially co-expressed in iM type (Pearson correlation test). All the activated EMT-related motifs in ovarian cancer are available in the download page or by searching specific nodes with statistical control in search page.
In conclusion, EMT-Regulome provides a comprehensive landscape of EMT-related regulation motifs to facilitate the investigation of the mechanism of EMT involved in the pathological and physiological processes. The features of EMT-Regulome include, (i) comprehensively integrated different EMT-related regulatory relationships from multiple databases; (ii) systematically provided EMT-related motifs, such as feed-forward loop and ceRNA with statistical control; (iii) all the EMT-related regulatory relationships can be visualized as network and be downloaded.
Acknowledgments
The authors acknowledge the efforts of all of researchers who have contributed the data to the public databases of TargetScan, miRanda, miRBase, miRTarBase, StarBase, miRcode, MsigDB, TRANSFAC, Transmir, ChIPBase, dbEMT, NCBI, Gencode, TCGA and TANRIC. This work was supported by the National Natural Science Foundation of China [grant numbers 61673143, 31671187, 81572935 and 81372213] and Postdoctoral Scientific Research Developmental Fund [grant number: LBH-Q16166].
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
The authors declare no conflict of interest.
Supplementary Material
References
- Nieto MA et al Cell 2016; 166: 21–45. [DOI] [PubMed]
- Yang J, Weinberg RA Dev Cell 2008; 14: 818–829. [DOI] [PubMed]
- Wu CY et al. Trends Genet. TIG 2012; 28: 454–463. [DOI] [PubMed] [Google Scholar]
- Richards EJ et al J Biol Chem 2015; 290: 6857–6867. [DOI] [PMC free article] [PubMed]
- Zhang HM et al Brief Bioinformatics 2015; 16: 45–58. [DOI] [PubMed]
- Zhao M et al Sci Rep 2015; 5: 11459. [DOI] [PMC free article] [PubMed]
- Yang D et al Cancer Cell 2013; 23: 186–199. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.