Abstract
The ATR-CHEK1 pathway is upregulated and overactivated in Ataxia Telangiectasia (AT) cells, which lack functional ATM protein. Loss of ATM in AT confers radiosensitivity, although ATR-CHEK1 pathway overactivation compensates, leads to prolonged G2 arrest after treatment with ionizing radiation (IR), and partially reverses the radiosensitivity. We observed similar upregulation of the ATR–CHEK1 pathway in a subset of oral squamous cell carcinoma (OSCC) cell lines with ATM loss. In the present study, we report copy number gain, amplification, or translocation of the ATR gene in 8 of 20 OSCC cell lines by FISH; whereas the CHEK1 gene showed copy number loss in 12 of 20 cell lines by FISH. Quantitative PCR showed overexpression of both ATR and CHEK1 in 7 of 11 representative OSCC cell lines. Inhibition of ATR or CHEK1 with their respective siRNAs resulted in increased sensitivity of OSCC cell lines to IR by the colony survival assay. siRNA-mediated ATR or CHEK1 knockdown led to loss of G2 cell cycle accumulation and an increased sub-G0 apoptotic cell population by flow cytometric analysis. In conclusion, the ATR-CHEK1 pathway is upregulated in a subset of OSCC with distal 11q loss and loss of the G1 phase cell cycle checkpoint. The upregulated ATR-CHEK1 pathway appears to protect OSCC cells from mitotic catastrophe by enhancing the G2 checkpoint. Knockdown of ATR and/or CHEK1 increases the sensitivity of OSCC cells to IR. These findings suggest that inhibition of the upregulated ATR–CHEK1 pathway may enhance the efficacy of ionizing radiation treatment of OSCC.
INTRODUCTION
In 2013, there will be an estimated 53,500 new cases of oral squamous cell carcinomas (OSCC) and 11,500 deaths related to OSCC in the United States (Siegel et al., 2013). OSCC is the sixth leading cancer worldwide, with 550,000 new patients diagnosed in 2008 and 250,000 cancer-related deaths in 2008 (Ferlay et al., 2010). In addition to alcohol use and smoking as etiologic factors, the incidence of HPV-related OSCC has increased significantly over the last three decades (Chung and Gillison, 2009). Patients with HPV-associated OSCC have an excellent response to minimally invasive surgery followed by adjuvant radiation with or without chemotherapy (Sinha et al., in press) or definitive chemo-radiation therapy associated with an improved overall survival (Fakhry et al., 2008). Approximately 65% of all OSCC patients present with loco-regionally advanced disease involving the lymph nodes, and an additional 10 to 20% have metastatic disease at initial diagnosis (Siegel et al., 2013). The current standard of care for patients with untreated locally advanced OSCC is minimally invasive surgery followed by adjuvant radiation with or without chemotherapy or concurrent chemoradiation. The use of multimodality treatment paradigms integrating cisplatin-based chemotherapy with radiation or surgery has led to modest improvement in outcomes including a 5-year overall survival of 40 to 60% for patients with locally advanced OSCC (Argiris et al., 2008).
Ionizing radiation exerts its effects by inducing DNA double-strand breaks (DSBs). The phosphatidyl inositol 3 kinase (PI3K)-like protein kinases, ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) are activated in response to DNA damage induced by ionizing radiation, and orchestrate a cascade of events that culminate in cell cycle arrest and DNA repair or apoptosis (Shiloh, 2001). In response to DSBs caused by ionizing radiation (IR), ATM phosphorylates and activates a number of downstream effectors including CHEK2 on serine (SQ) or threonine (TQ) residues (Shiloh, 2003). Similarly, ATR is activated in response to stalled replication forks induced by UV radiation or chemotherapeutic drugs (Helt et al., 2005). The ATR gene, located in chromosomal band 3q24, is frequently gained or amplified in OSCC (Table 1), while ATM at 11q22.3 is lost in a subset of OSCC (Parikh et al., 2007). After DNA damage, ATM undergoes auto-phosphorylation at Ser1981 leading to a conformational change and activation of the ATM protein (Bakkenist and Kastan, 2003). The activation of ATM or ATR initiates a signaling cascade that involves the phosphorylation of substrates and culminates in cell cycle arrest and DNA repair (Shiloh, 2001; Helt et al., 2005). At the cellular level, ATM loss is associated with chromosomal instability, radiosensitivity, as well as a loss of the G1 checkpoint in response to ionizing radiation (Meyn, 1999; Uhrhammer et al., 1999). ATR has been shown to play an important role in regulating chromosomal fragile site stability (Casper et al., 2002). Loss of functional ATM protein in AT cells leads to loss of the G1 phase cell cycle checkpoint in response to ionizing radiation. An ATM–deficient (AT) cell line (AT5BIVA) was demonstrated to have an overactivated ATR-CHEK1 pathway which results in a prolonged G2 arrest after ionizing radiation (Wang et al., 2003). Inhibition of this upregulated ATR-CHEK1 pathway sensitizes the AT cells even further to ionizing radiation (Wang et al., 2003). Thus, the ATR-CHEK1 pathway has overlapping functions and can respond not only to stalled replication forks, but to ionizing radiation in the presence of a deficient ATM-CHEK2 pathway.
TABLE 1.
ATR and CHEK1 Copy Number Changes in 20 Oral Cancer Cell Lines UPCI:SCC003–182 and Two Ovarian Cancer Cell Lines, SKOV3 and OVCAR-3
| Cell Line | 11q13 Amp | ATM loss | ATR | ATR:CEP3 ratio | Idic 3q | CHEK1 | CHEK1:CEP11 ratio | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| % Cells with gain | % Cells with loss | % Cells with no change | % Cells with gain | % Cells with loss | % Cells with no change | ||||||
| Normal | – | – | 100 | 1.0 | – | – | – | 100 | 1.0 | ||
| UPCI: | |||||||||||
| SCC003 | + | + | 53 | 1 | 46 | 1.6 | − | 1 | 93 | 6 | 0.71 |
| SCC029B | + | + | 89 | 2 | 9 | 1.4 | − | – | 99 | 1 | 0.61 |
| SCC032 | + | + | 57 | – | 43 | 1.3 | − | 1 | 99 | – | 0.67 |
| SCC040 | + | + | 53 | – | 47 | 1.6 | − | 5 | 94 | 1 | 0.56 |
| SCC066 | − | − | 2 | 3 | 95 | 1.0 | − | 1 | 1 | 98 | 1.01 |
| SCC070 | + | + | 89 | 1 | 10 | 2.1 | + | – | 98 | 2 | 0.57 |
| SCC077 | + | + | 85 | 1 | 14 | 2.2 | + | 2 | 31 | 67 | 0.67 |
| SCC078 | + | + | 9 | 3 | 88 | 1.0 | − | – | 94 | 6 | 1.05 |
| SCC084 | + | + | 7 | – | 93 | 1.0 | − | 2 | 98 | – | 0.52 |
| SCC099 | − | − | 3 | 6 | 91 | 1.0 | − | – | 100 | – | 1.0 |
| SCC103 | + | + | 8 | 1 | 91 | 1.1 | − | 2 | 94 | 4 | 0.74 |
| SCC104 | − | + | 98 | – | 2 | 2.2 | + | 1 | 11 | 88 | 0.97 |
| SCC105 | − | + | 2.5 | 0.5 | 97 | 1.1 | − | 12 | 32 | 56 | 0.85 |
| SCC116 | − | − | 3 | 2 | 95 | 1.0 | − | 3 | 10 | 87 | 0.94 |
| SCC122 | − | + | 1 | 9 | 90 | 1.0 | − | 1 | 82 | 17 | 0.71 |
| SCC131 | + | + | 92.5 | – | 7.5 | 2.1 | + | – | 98 | 2 | 0.44 |
| SCC136 | + | + | 78.5 | – | 21.5 | 1.3 | − | – | 96 | 4 | 0.53 |
| SCC142 | − | + | 98.5 | – | 1.5 | 2.1 | + | – | 91 | 9 | 0.67 |
| SCC172 | + | + | 3 | 5 | 92 | 1.1 | − | 5 | 90 | 5 | 0.71 |
| SCC182 | − | − | 1 | 1 | 98 | 1.0 | − | 3 | – | 97 | 1.01 |
| SKOV3 | − | − | 4 | 3 | 93 | 1.1 | − | 1 | – | 99 | 1.0 |
| OVCAR-3 | − | + | 99 | – | 1 | 9 | − | 8 | – | 92 | 1.1 |
One hallmark of most solid tumors, including OSCC is chromosomal instability (Hanahan and Weinberg, 2000, 2011). During their development, OSCC acquire genetic alterations, which promote proliferation, spread, and invasion into surrounding tissues as well as therapeutic resistance. In a large prospective multicenter study of 560 OSCC patients, disruptive TP53 mutations were associated with an inferior overall survival (Poeta et al., 2007). Despite the high degree of cytogenetic heterogeneity, specific chromosomal changes occur regularly and are involved in OSCC development and progression, including 11q13 amplification (including the cyclin D1 gene), gains of 3q21–3q29 involving ATR, 5p, 8q, 18q and 22q and losses involving 3p, 8p, 9p, distal 11q, 13q and 21q (Gollin, 2001; Jin et al., 2002). In a study of 113 primary OSCC, gain or amplification of 3q21–29 and 11q13, and loss of 8p21–22 were associated with a poor progression-free interval and a statistically significant reduction in disease-specific overall survival by Kaplan-Meier analysis (Bockmuhl et al., 2000). Along with cyclin D1 (CCND1), a number of genes located in the 11q13 amplicon are amplified and overexpressed (Huang et al., 2002). Distal 11q loss precedes 11q13 amplification during OSCC development and contributes to chromosomal instability and an impaired DNA damage response to ionizing radiation (Parikh et al., 2007; Reshmi et al., 2007). In our study, we test the hypotheses that an upregulated ATR-CHEK1 pathway protects OSCC from mitotic catastrophe by enhancing the G2 checkpoint and that inhibition of this checkpoint sensitizes OSCC to ionizing radiation.
MATERIALS AND METHODS
Cell Culture
OSCC were cultured in M10 medium composed of Minimum Essential Medium (Gibco Life Technologies, Grand Island, NY), supplemented with 1% non-essential amino acids, 1% L-glutamine, 0.05 mg/ml gentamicin, and 10% fetal bovine serum (FBS) (Gibco Life Technologies) at 37°C in a 5% CO2 incubator. GM09607 (Coriell Cell Repositories, Camden, NJ), HEK293, MDA-MB-231 and MCF7 were cultured in Dulbecco’s Modified Eagles Medium (DMEM) (Gibco Invitrogen), supplemented with 1% non-essential amino acids, 0.05 mg/ml penicillin-streptomycin with 0.2 mM L-glutamine and 10% FBS. SKOV3 cells were cultured in McCoys 5A medium supplemented with 0.05 mg/ml penicillin-streptomycin with 0.2 mM L-glutamine and 10% FBS. The OVCAR-3 ovarian cancer cell line was cultured in RPMI 1640 supplemented with 0.05 mg/ml penicillin-streptomycin with 0.2 mM L-glutamine and 10% FBS. Normal human oral keratinocytes (NHOK) cells were cultured in serum-free KGM-2 medium (Clonetics, Walkersville, MD) supplemented with bovine pituitary extract as described in the manufacturer’s instructions (Clonetics). The UPCI:SCC cell lines (White et al., 2007) and NHOK were initiated from de-identified tissues acquired from the University of Pittsburgh Head & Neck Cancer Tissue Bank after patient consent according to our Unified Tissue Bank IRB approval.
Clonogenic Survival Assay
Clonogenic survival assays were carried out to determine cell survival in response to treatment. Two thousand cells from a single cell suspension of each cell line were seeded into 60 mm petri dishes. After 24 hr, specific cell cultures were treated with 2.5, 5, and 10 Gy doses of γ-irradiation with a 137Cs source at a dose rate of 2.83 Gy/min (Gammacell 1,000 Elite irradiator, Nordion International, Ottawa, Canada). Mock-treated cells cultured in parallel were used as controls to determine the relative plating efficiency. After 14 days, the cell colonies were fixed with 70% ethanol, stained with 10% Giemsa stain (Sigma-Aldrich, St. Louis, MO) and counted. All experiments were performed in triplicate and results reported with two standard deviations from the mean.
Fluorescence In Situ Hybridization (FISH)
Control centromere enumeration probes, CEP 3 (D3Z1) and CEP 11 (D11Z1) were obtained from Abbott Molecular, Inc. (Des Plaines, IL). Individual BAC probes mapping to ATR (RP11–427D1, RP11–383G6), ATM (RP11–241D13), CHEK1 (RP11–712D22), and CCND1 (RP11–699M19) were obtained from Children’s Hospital of Oakland Research Institute (CHORI, Oakland, CA). After isolating BAC DNA, it was labeled using a nick translation kit from Abbott Molecular, Inc. (Des Plaines, IL) according to the manufacturer’s instructions. OSCC cells were treated with 0.1μg/ml Colcemid™ (Irvine Scientific, Santa Ana, CA) and harvested after 5 hr using standard cytogenetic methods. Slides were prepared, treated with RNase, dehydrated using a graded ethanol series, and denatured with 70% formamide. The slides were hybridized with the appropriate FISH probe(s) for 16 hr at 37°C in a humidified chamber. Post-hybridization washes were performed and the slides were counterstained with 4′,6-dia-midino-2-phenylindole (DAPI) and mounted with antifade. At least 100 nuclei and 20 metaphases were analyzed per specimen. Images were captured using an Olymp us BX61 epifluorescence microscope (Olympus Microscopes, Melville, KY) and the Genus software platform on the Cytovision System (Leica Microsystems, San Jose, CA). ATR gain was defined as >50% cell nuclei with ATR/CEP 3 copy number ratio >1.5. ATR amplification was defined as >50% cell nuclei with ATR/ CEP 3 copy number ratio >5. CHEK1 loss was defined as loss of one or more copies of the CHEK1 gene compared with CEP 11 in >50% of cell nuclei studied.
FISH on Tissue Sections
We also examined copy numbers of ATR and CHEK1 on 5 μm tissue sections of primary OSCC tumors. FISH was carried out as described previously (Parikh et al., 2007). Briefly, the slides were deparaffinized in Histoclear 3 × 5 min, treated with Skip DeWax at 80°C for 10 min (Insitus Biotechnologies, Albuquerque, NM), followed by 0.2 N HCl at 37°C for 20 min. The slides were digested with pepsin in protease buffer 80°C for 20 min, washed with 2× SSC, fixed with 10% formalin for 10 min, dehydrated in a graded ethanol series, and denatured with 70% formamide. The slides were hybridized with the appropriate probe for 16 hr at 37°C in a humidified chamber. Posthybridization washes were performed and the slides were counterstained with DAPI and mounted with antifade. Two hundred nuclei were analyzed for each OSCC primary tumor and normal adjacent tissue, respectively. Copy number gain, amplification or loss was assessed as described above.
Anaphase Bridge Formation Assay
To evaluate the presence of the ATR gene in anaphase bridges, OSCC and AT cell lines were plated in chamber slides and grown until they reached 80% confluence. Colcemid™ (0.1 μg/ml) (Irvine Scientific) was added to each chamber slide and the slides were incubated at 37°C in 5% CO2 for 24 hr. The medium was aspirated and cells fixed with 3:1 methanol:acetic acid for a period of 45 min. FISH using a BAC probe to the ATR gene, and commercial centromere probes to chromosomes 3 and 11 (CEP 3 and CEP 11) was carried out as described above. Fifty anaphase bridges per cell line were evaluated for the presence of ATR, CEP3 and CEP11.
Cell Cycle Analysis by Flow Cytometry
For cell cycle analysis, cells were seeded in 60 mm culture dishes and allowed to attach overnight. 24 hr after treatment (mock or IR), cells were trypsinized and washed with phosphate-buffered saline (PBS), fixed with 70% ethanol, and treated with 50 μg/ml propidium iodide (Molecular Probes, Life Technologies, Carlsbad, CA) for 45 min at 37°C. The stained cells were analyzed using a Coulter Epics XL Flow Cytometer in the UPCI Flow Cytometry Facility.
Quantitiative Reverse Transcriptase PCR (QRT-PCR)
Cell line RNA was extracted using TRIzol® Reagent and Qiagen RNeasy columns. RNA aliquots for the quantitative real-time PCR (QRT-PCR) analysis were DNase I treated. Reverse transcription and QRT-PCR were carried out as described earlier (Huang et al., 2002). Predesigned primer and probe sets for gene expression of ATR and CHEK1 as well as the endogenous control, 18S ribosomal RNA were obtained from Applied Biosystems, Inc. (Foster City, CA). The QRT-PCR was carried out using an Applied Biosystems 7300 Real-Time PCR system.
siRNA Treatment
RNA interference of ATR and CHEK1 was performed using SMARTpool siRNA sequences obtained from Dharmacon (Lafayette, CO). Nonspecific scrambled siRNA was used as a control (Dharmacon). For transfection, the OSCC cell lines were seeded in 60 mm petri dishes and transfected with siRNA duplexes using Lipofectamine 2000 (Life Technologies, Invitrogen) according to the manufacturer’s instructions at a final siRNA concentration of 90 to 100 nM. Transfection efficiency was calculated using a nonspecific siRNA labeled with a fluorescent tag called siGLO (Dharmacon). Forty-eight hours post-transfection, OSCC cell lines (UPCI:SCC066 and 104) were treated with 0.4 μM aphidicolin (Sigma-Aldrich, St. Louis, MO), a DNA polymerase-α inhibitor and harvested as described above 24 hr after aphidicolin treatment. Similarly, UPCI:SCC066 and 104 were treated with 5 Gy ionizing radiation, 48 hr after siRNA transfection and harvested 24 hr post-ionizing radiation.
RESULTS
ATR and CHEK1 are Overexpressed in a Subset of OSCC
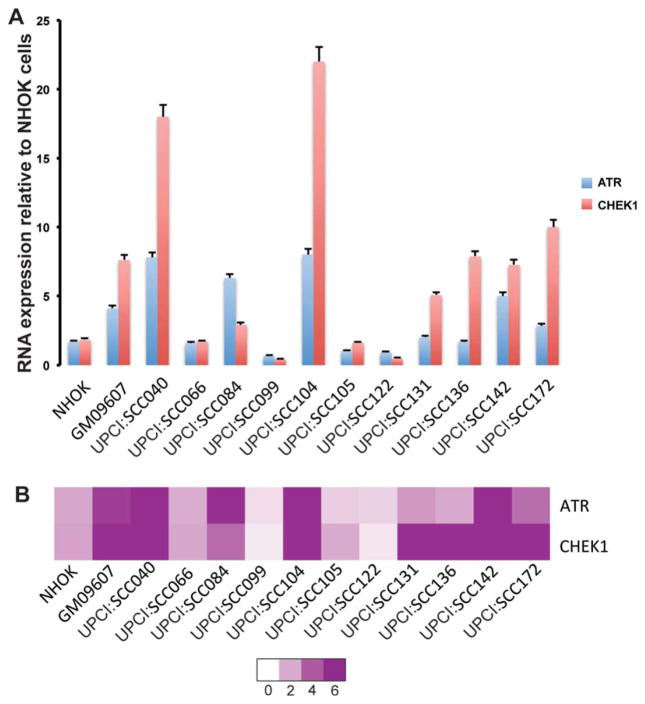
We studied ATR and CHEK1 RNA expression using QRT–PCR in 11 representative OSCC cell lines. NHOK and HEK293 cells served as our negative controls and GM09607 (an AT cell line from the Coriell Institute, Camden, NJ) with ATR and CHEK1 overexpression was our positive control. UPCI:SCC066, 099, 105, and 122 showed ATR and CHEK1 expression equal to or lower than the negative control cell lines (Fig. 1). GM09607, with an upregulated ATR–CHEK1 pathway, demonstrated a four–fold increase in ATR expression and an eightfold increase in CHEK1 when compared with NHOK cells (Fig. 1). Seven of 11 OSCC cell lines, UPCI:SCC040, 084, 104, 131, 136, 142 and 172 demonstrated high ATR and CHEK1 expression compared with our negative controls. It should be noted that all cell lines with ATM loss (UPCI:SCC104, 131, 136, and 142) except for UPCI:SCC122 (Table 1) demonstrated significantly higher ATR and CHEK1 RNA expression than the NHOK cells. Elevated ATR and CHEK1 expression was comparable to or higher in OSCC compared with AT cell line, GM09607 with an upregulated ATR-CHEK1 pathway.
Figure 1.
(A) Increased ATR and CHEK1 RNA expression in OSCC cell lines assayed using QRT-PCR. (B) Heat map showing increased ATR and CHEK1 expression in OSCC by QRT-PCR.
Copy Number and Structural Changes Involving the ATR and CHEK1 Genes
We carried out dual-color FISH using BAC probes mapping to ATR (3q23) compared with CEP 3 and CHEK1 (11q24.2) compared with CEP 11, respectively. Table 1 summarizes our FISH results for copy number gain in relation to the ploidy of each cell line, which was determined from chromosome 3 and/or 11 centromere enumeration (CEP 3, CEP 11) and consensus karyotypes (Martin et al., 2008). We observed copy number gain of the ATR gene in seven of 20 OSCC cell lines and ATR translocation in one of 20 OSCC cell lines. In addition, we observed that ATR copy number gain was associated with isochromosome 3q formation in five of 20 OSCC cell lines (Fig. 2, Table 1). In UPCI:SCC084, we did not observe copy number gain of the ATR gene, but by metaphase FISH, we observed a translocation of one copy of the ATR gene to a derivative chromosome 11 characterized by 11q13 amplification. In addition to OSCC, we observed amplification (8–10 copies per cell) in an ovarian tumor cell line, OVCAR–3. Interestingly we observed that CHEK1 underwent copy number loss in all OSCC cell lines with 11q13 amplification. Loss of CHEK1 was also seen in UPCI:SCC122 and 142, which did not show 11q13 amplification. The exact mechanism of CHEK1 overexpression despite partial copy number loss is unclear at this time. Of the 20 OSCC cell lines studied, we observed ATR gain or amplification in eight and CHEK1 loss in 13, suggesting that both of these events occur frequently in OSCC.
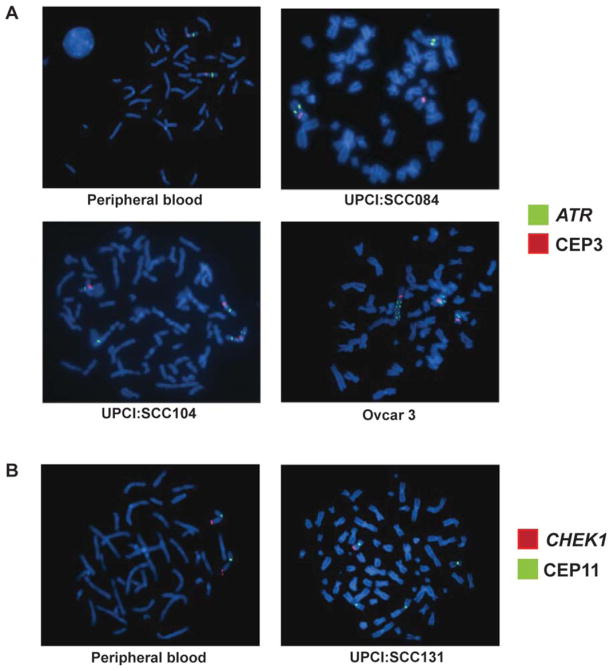
Figure 2.
(A) Two copies of the ATR gene (green) and two CEP3 (red) signals in a normal lymphocyte metaphase cell. (B) We observed a translocation of the ATR gene (green) in UPCI:SCC084. (C) In UPCI:SCC104, the ATR gene (green) is gained compared with CEP3 as a result of isodicentric chromosome 3 formation. (D) In the ovarian tumor cell line, OVCAR-3, we observe amplification of the ATR gene (green) compared with CEP3 (red). (E) A normal peripheral lymphocyte metaphase spread with two CHEK1 (red) and two CEP11 (green) signals. (F) UPCI:SCC131 OSCC cells with partial copy number loss of the CHEK1 gene (red) compared with CEP11 (green).
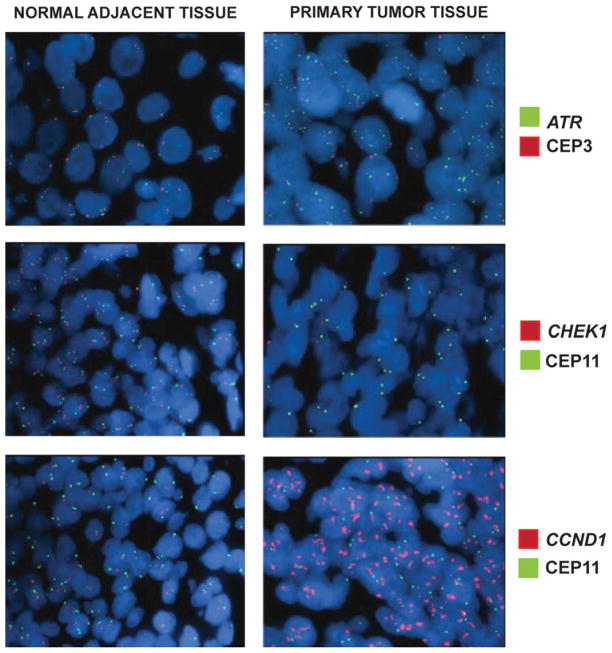
To confirm whether our findings in OSCC cell lines are representative of OSCC tumors, we evaluated five primary OSCC and matched surrounding normal oral mucosa for ATR and CHEK1 copy number alterations (Fig. 3, Table 2.) by FISH. As expected, the primary tumors were heterogeneous. We observed ATR gene copy number gain in 32 to 58% of cells in all five tumors, while the surrounding normal tissue exhibited normal disomic copy number for the ATR gene and CEP 3 by dual-color FISH. Similarly, CHEK1 showed copy number loss in 35 to 65% of cells in the tumors, while the adjacent normal mucosa showed no CHEK1 loss using dual-color FISH. We observed high-level (more than four copies) CCND1 amplification in 65 to 100% of the tumor cells from all five tumors, confirming our previous observation that 11q13 amplification is a relatively frequent early change during tumor development (Martin et al., 2008). These results show that ATR gain and CHEK1 loss are present not only in OSCC cell lines, but also in primary oral/head and neck tumors, and therefore, do not represent artifacts of cell culture.
Figure 3.
Representative images of OSCC paraffin sections demonstrating (A) gain of ATR gene (green) copy number compared with CEP3 (red), (B) loss of the CHEK1 gene (red) compared with CEP11 (green), and (C) amplification of the CCND1 gene (red) compared with CEP11 (green) in tumor tissue, but not in the adjacent normal tissue.
TABLE 2.
ATR, CHEK1, and CCND1 (Cyclin D1) Copy Number Changes by FISH on Paraffin-Embedded Tissue Sections from Primary Oral Cancer (T) and Adjacent Normal Mucosa (N)
| ATR | CHEK1 | CCND1 | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Amp/gain (%) | Normal (%) | Loss (%) | Amp/gain (%) | Normal (%) | Loss (%) | Amp/gain (%) | Normal (%) | Loss (%) | |
| 0402584 | |||||||||
| N | 2 | 98 | – | – | 99 | 1 | 8 | 98 | – |
| T | 54 | 46 | – | – | 35 | 65 | 100 | – | – |
| 3L621340 | |||||||||
| N | 1 | 99 | – | – | 98 | 2 | 1 | 99 | – |
| T | 58 | 42 | – | – | 65 | 65 | 99 | 1 | – |
| 1H620781 | |||||||||
| N | 2 | 98 | – | – | 98 | 2 | 3 | 94 | 3 |
| T | 32 | 66 | 2 | – | 51 | 49 | 65 | 34 | 1 |
| 3G040281 | |||||||||
| N | 1 | 98 | 1 | 1 | 98 | 1 | – | 100 | – |
| T | 42 | 58 | – | – | 44 | 56 | 100 | – | – |
| 1F620600 | |||||||||
| N | - | 98 | 2 | – | 99 | 1 | 3 | 95 | 2 |
| T | 42 | 57 | 1 | – | 37 | 63 | 68 | 32 | – |
Mechanism of ATR Gain and ATR Translocations
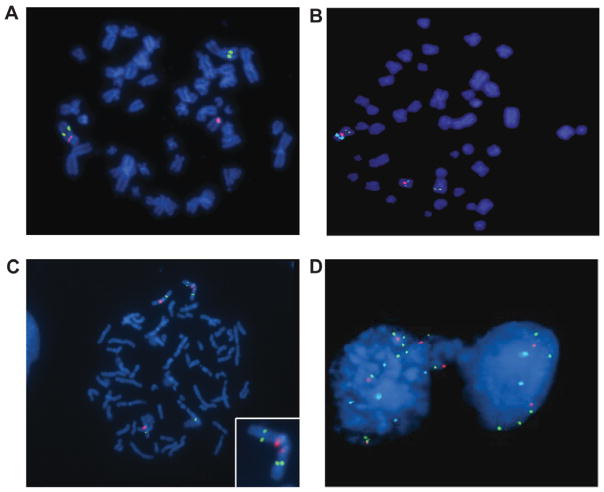
It has been reported previously in OSCC that translocations of chromosome 3 are commonly associated with 11q13 amplification, and that frequently, chromosome 3 segments cap the amplified chromosome 11 (Jin et al., 2002). We observed that three of 20 OSCC cell lines studied (UPCI:SCC078, 084 and 172) had a translocation between the derivative chromosome 11 der(11) with 11q13 amplification and segments of 3q including the ATR gene (Figs. 4A and 4B) by three-color FISH. These results confirm that this combination of rearrangements is common in head and neck tumors (Jin et al., 2002). Conventional karyotyping using Giemsa staining revealed t(3;11) in all 20 metaphases studied from each of the three OSCC cell lines, UPCI:SCC078, 084 and 172 (Martin et al., 2008).
Figure 4.
(A) A metaphase spread from UPCI:SCC084 shows translocation of one copy of the ATR gene (green) from chromosome 3 marked by CEP3 labeled red to another chromosome, which (B) further analysis reveals is a derivative chromosome 11 (red) with amplified CCND1 (aqua). (C) A representative image of a chromosome spread from UPCI:SCC104 revealing an isodicentric chromosome 3 (CEP3 in red) with ATR copy number gain (green). (D) Image showing the ATR gene in an anaphase bridge in the UPCI:SCC104 cell line, which may have resulted from the isodicentric chromosome 3.
We observed isodicentric chromosome 3q in five of 20 OSCC cell lines examined (UPCI:SCC070, 077, 104, 131 and 142). Isodicentric chromosomes are prone to being pulled to opposite poles during cell division, resulting in an anaphase bridge between the two daughter cells. We determined the frequency of the ATR gene and CEP 3 in anaphase bridges and compared it to the presence of CEP 11 in anaphase bridges in a subset of four OSCC cell lines and GM09607 the (AT cell line) (Table 3). The ATR gene mapped at a higher frequency to anaphase bridges in UPCI:SCC104 and 131, both of which have gain of chromosome 3, compared with UPCI:SCC066 and 105, both of which have normal copy numbers of the ATR gene (Table 3). Thus, the higher frequency of ATR and CEP 3 probes in anaphase bridges suggests ongoing selection for 3q gain.
TABLE 3.
Increased Frequency of ATR in Anaphase Bridges in OSCC and GM09607
| Cell line | 11q13 Amp | ATR Gain | Frequency of ATR in anaphase bridges (%) | Frequency of CEP3 in anaphase bridges (%) | Frequency of CEP11 in anaphase bridges (%) |
|---|---|---|---|---|---|
| UPCI:SCC066 | − | − | 4 | 4 | 6 |
| UPCI:SCC104 | − | + | 26 | 26 | 14 |
| UPCI:SCC105 | − | − | 6 | 6 | 22 |
| UPCI:SCC131 | + | + | 28 | 28 | 24 |
| GM09607 | − | − | 16 | 16 | 10 |
OSCC Cell Lines with ATR-CHEK1 Upregulation Accumulate in G2 Phase in Response to Ionizing Radiation and are Radioresistant
We treated eight representative OSCC cell lines, NHOK cells, and GM09607 with 5 Gy of ionizing radiation and evaluated their cell cycle profiles. Five of the eight OSCC cell lines with an upregulated ATR–CHEK1 pathway (UPCI:SCC084, 104, 131, 136 and 142) demonstrated loss of the G1 cell cycle checkpoint with predominant accumulation of cells in the G2 phase after treatment with 5 Gy IR (Table 4). All five cell lines with loss of the G1 checkpoint and ATR-CHEK1 upregulation also demonstrated increased resistance to ionizing radiation (Parikh et al., 2007). Since loss of p53 signaling was also observed in these cell lines, the resistance to ionizing radiation could be a result of both loss of p53-mediated apoptotic pathways and an upregulated ATR–CHEK1 pathway which promotes G2 accumulation and homologous recombination repair after DNA damage.
TABLE 4.
Results of Flow Cytometry Analysis of Untreated and 5 Gy IR Treated Normal Human Keratinocytes (NHOK), AT Cell Line (GM09607), and UPCI:SCC Cell Lines
| UPCI cell line | ATR-CHEK1 upregulation | Untreated | 5 Gy ionizing radiation | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Sub-G0 (%) | G1 (%) | S (%) | G2M (%) | Sub-G0 (%) | G1 (%) | S (%) | G2M (%) | ||
| NHOK | − | 0.1 | 71 | 13 | 15.9 | 3 | 75 | 2 | 20 |
| GM09607 | + | 2 | 52.2 | 6.8 | 39 | 6 | 34 | 8.9 | 51.1 |
| SCC066 | − | 1.2 | 70 | 13.3 | 15.5 | 1.9 | 50.3 | 16.1 | 31.7 |
| SCC084 | + | 0.7 | 60 | 10 | 29.3 | 2.4 | 32 | 12 | 53.6 |
| SCC104 | + | 0.9 | 53 | 16 | 30.1 | 2.8 | 22 | 18 | 57.2 |
| SCC105 | − | 0.7 | 67 | 13 | 19.3 | 4 | 51 | 15 | 30 |
| SCC116 | − | 1.1 | 61.9 | 15 | 22 | 4 | 49 | 15 | 32 |
| SCC131 | + | 1 | 66.5 | 15.5 | 15 | 2.5 | 32 | 11 | 55.5 |
| SCC136 | + | 1 | 59.9 | 10 | 29.1 | 1.9 | 31.6 | 25.5 | 41 |
| SCC142 | + | 1.3 | 62.2 | 14.2 | 22.3 | 2.6 | 31 | 16 | 40.4 |
ATR and CHEK1 siRNA Sensitize a Subset of OSCC to Ionizing Radiation-Induced DNA Damage
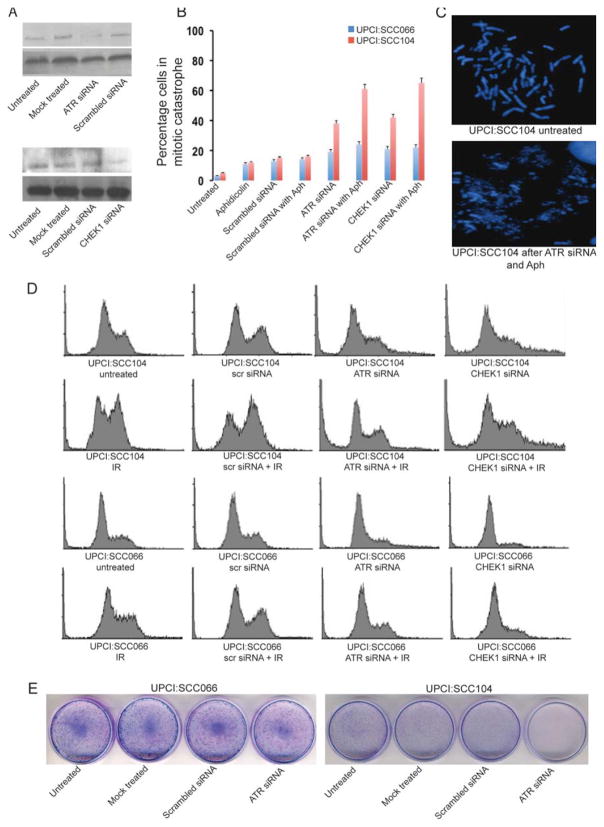
We used siRNA to reduce expression of ATR and CHEK1 in two representative OSCC cell lines, UPCI:SCC066 and 104. We confirmed a reduction in ATR and CHEK1 protein levels by immunoblotting at the end of 72 hr. We observed a high level of ATR and CHEK1 knockdown by their specific siRNAs, while scrambled, nonspecific siRNA did not alter ATR and CHEK1 protein expression (Fig. 5A). We analyzed the cell cycle profiles of UPCI:SCC066 and 104 after ATR and CHEK1 knockdown using the respective siRNAs (Fig. 5D). We observed 18% of untreated UPCI:SCC104 cells in S phase and 29% of cells in G2 phase. Twenty-four hours after treatment with 5 Gy IR, we observed a significantly higher percentage of UPCI:SCC104 cells (nearly 55%) in G2 phase compared with 31% of UPCI:SCC066 cells suggestive of a loss of the G1 checkpoint in SCC104. Pretreatment with ATR si RNA resulted in a significant reduction in the G2 population of irradiated UPCI:SCC104 cells from 55% to 18%, and an increase in the sub-G0 population (dead cells) from 1% to nearly 18%. Even in un-irradiatedUPCI:SCC104 cells, ATR siRNA reduced G2 accumulation and increased the sub-G0 population (15%). UPCI:SCC066 cells demonstrated a modest reduction in G2 accumulation following treatment with ATR siRNA regardless of radiation treatment, but showed no corresponding increase in the dead (sub-G0) cell population. We observed similar radiosensitization following treatment with CHEK1 siRNA of UPCI:SCC104, but not UPCI:SCC066. Thus, UPCI:SCC104 was radiosensitized robustly following inhibition of the upregulated ATR–CHEK1 pathway with the respective siRNAs. Our results suggest that UPCI:SCC104, an OSCC cell line with ATR and CHEK1 overexpression becomes substantially more sensitive to IR following knockdown of the ATR-CHEK1 pathway using ATR or CHEK1 siRNA. Clonogenic cell survival assays for UPCI:SCC104 confirmed a complete loss of cell survival in response to ATR and CHEK1 siRNA in untreated cells (Fig. 5E) and cells treated with 5 Gy IR.
Figure 5.
(A) ATR and CHEK1 knockdown using siRNA in UPCI:SCC104. (B) Comparison of the frequency of mitotic catastrophe in UPCI:SCC066 (blue) and UPCI:SCC104 (red) in response to ATR and CHEK1 siRNA treatment with or without aphidicolin (Aph). (C) Representative images of mitotic catastrophe in UPCI:SCC104 after treatment with ATR siRNA and aphidicolin (D) Flow cytometric analysis following treatment with ATR and CHEK1 siRNA. Cell cycle profiles of UPCI:SCC104 and UPCI:SCC066 following mock treatment with scrambled siRNA, ATR siRNA and CHEK1 siRNA in unirradiated cells or cells irradiated with 5 Gy of IR. In comparison with UPCI:SCC066, UPCI:SCC104 shows increased accumulation of irradiated cells in the G2M phase. On specific knockout of ATR or CHEK1 with specific siRNA, we observed elimination of the G2M accumulation of irradiated cells and an increase in the sub-G0 dead cell population.
DISCUSSION
Gain or amplification of 11q13 (harboring the cyclin D1 gene (CCND1)) and loss of distal 11q (containing MRE11A (11q21), ATM (11q22.3), H2AFX (11q23.3), and CHEK1 (11q24.2)) occur in ~46% of OSCC according to the Broad Institute The Cancer Genome Atlas (TCGA) copy number portal, analysis version 2013-02-21 stddata__2013_02_03 (Beroukhim et al., 2010). We showed previously in OSCC that loss of distal 11q results in a qualitative and quantitative reduction in γ-H2AX focus formation, increased chromosomal instability in the form of chromosomal breaks and anaphase bridges, and radioresistance (Parikh et al., 2007). In the present study, we examined the role of the ATR-CHEK1 pathway in radioresistance in OSCC cells with and without distal 11q loss.
In AT cells, which lack functional ATM protein, the ATR-CHEK1 pathway is upregulated and overactivated compared with normal cells (Wang et al., 2003). The prolonged G2 arrest in AT cells in response to ionizing radiation is abrogated by CHEK1 inhibition using siRNA. We demonstrate a similar G2 arrest caused by overactivation of the ATR-CHEK1 pathway in OSCC with distal 11q (ATM) loss. This overactivation of the ATR-CHEK1 pathway in response to ionizing radiation promotes G2 accumulation of cells, protecting them, since entering mitosis with DNA damage would lead to mitotic catastrophe, a p53-independent form of cell death. Here, we demonstrate that depletion of ATR or CHEK1 using siRNA leads to loss of G2 accumulation of cells in response to ionizing radiation, increased mitotic catastrophe, and radiosensitization of OSCC cell lines. In the current era of targeted therapy and personalized genomic medicine, our biomarker comprised of distal 11q loss and/or upregulation of the ATR-CHEK1 pathway may be valuable in identifying a subgroup of OSCC patients whose tumors would be expected to be resistant to radiation therapy but would be expected to respond to combined radiotherapy with ATR-CHEK1 pathway inhibition.
Ionizing radiation used to treat OSCC generates reactive oxygen species in the tumor microenvironment. Hypoxia induces chromosomal breaks and promotes chromosomal rearrangements including deletions, translocations and chromosomal amplification (Coquelle et al., 1998). Hypoxia also leads to dedifferentiation of tumor cells and results in tumor heterogeneity (Axelson et al., 2005) and can select for tumor cells with a high level of chromosomal instability that lack p53-mediated apoptotic pathways in response to DNA damaging agents (Graeber et al., 1996). This eventually leads to development of an aggressive subclone of tumor cells with poor response to treatment. In response to hypoxia, a rapid localization and activation of ATR with focus formation is observed at the sites of hypoxia-induced stalled replication forks (Hammond et al., 2002, 2003). The activated ATR–CHEK1 pathway protects the cells from hypoxia-induced DNA damage. Inhibition of ATR leads to sensitization of cells to hypoxia and reoxygenation induced DNA damage (Hammond et al., 2004). These studies assume significant importance in light of our findings in OSCC, wherein the ATR–CHEK1 pathway is highly upregulated. The overexpressed ATR and CHEK1 proteins in OSCC may serve to protect these cells from hypoxia-induced cell death and provide survival benefit in response to ionizing radiation-induced DNA damage. Cyclin D1 (CCND1) interacts with RAD51 and promotes homologous recombination during the S and G2 phases of the cell cycle (Bartek and Lukas, 2011; Jirawatnotai et al., 2011). After exposure to hydroxyurea, Thr309 phosphorylation of RAD51 and homologous recombination repair by RAD51 occurs in a CHEK1-dependent manner (Sorensen et al., 2005). Thus ATR/CHEK1 upregulation and CCND1 amplification in OSCC represent two synergistic events that enable tumor cells to repair DNA damage after treatment with chemotherapy or radiation therapy and escape cell death.
Our finding of an upregulated ATR-CHEK1 pathway in OSCC has substantial translational value since a number of ATR and CHEK1 inhibitors are currently under development. UCN-01, 7-hydroxystaurosporine, is an example of a firstgeneration CHEK1 inhibitor that causes a complete disruption of the G2 checkpoint in response to ionizing radiation (Busby et al., 2000). UCN-01 was abandoned due to an unfavorable toxicity profile, however it has spurred the development of a number of other first- and second-generation CHEK1 inhibitors. The first generation CHEK1 inhibitor, PF00477736 is effective as a single agent in Philadelphia chromosome-positive acute lymphoblastic leukemia (Iacobucci et al., 2011). Another first generation CHEK1 inhibitor, LY2606318 is currently being evaluated in phase I and II clinical trials combined with gemcitabine or pemetrexed for non-small cell lung cancer (NCT01139775). The CHEK1 inhibitor, AZD7762 was shown to be of benefit in triple negative breast cancer (Ma et al., 2012) and enhances radiosensitization of TP53-mutated pancreatic cancer cells (Vance et al., 2011). In pancreatic cancer cell lines, the use of the novel selective ATR inhibitor, VE-821 led to enhanced sensitivity of these cells to IR and gemcitabine-induced DNA damage under both normoxic and hypoxic conditions (Prevo et al., 2012). In breast and ovarian cancer cell lines, a potent new ATR inhibitor, NU6027 reduced G2 arrest and was synthetically lethal in combination with either poly (ADP-ribose) polymerase (PARP) inhibition or XRCC1 deficiency (Peasland et al., 2011). The Merck CHEK1 inhibitor (MK8776/SCH900776) has been shown to be effective in combination with cytarabine in refractory AML (Karp et al., 2012; Schenk et al., 2012). The second-generation CHEK1 inhibitor, LY2606368 appears to function effectively as a stand-alone or monotherapy in preclinical ovarian cancer cell lines and pancreatic cancer xenografts (McNeely et al., 2011; Wu et al., 2011). Additional pre-clinical and early phase studies with ATR and CHEK1 small molecule inhibitors are ongoing in multiple laboratories, including our own.
In conclusion, we have demonstrated that the presence of an upregulated ATR-CHEK1 pathway in OSCC is associated with resistance to ionizing radiation. Inhibition of this pathway using siRNA to ATR or CHEK1 sensitizes OSCC cell lines to ionizing radiation. Therefore, it is likely that inhibition of the ATR-CHEK1 pathway will sensitize not only OSCC, but also other tumors with an overactivated ATR-CHEK1 pathway to radiation therapy and or chemotherapy.
Acknowledgments
Supported by NIH/NIDCR/NCI, Grant numbers: R01DE014729, P30CA047904, and P50CA097190; University of Pittsburgh Cancer Institute; Pittsburgh Foundation Learmonth Fund; the Teresa and H. John Heinz III Charitable Fund; Joan Gaines Cancer Research Fund at the University of Pittsburgh.
The authors thank Dr. Andrew Remes from the University of Pittsburgh Office of Enterprise Development and Dr. Maria Vanegas of the University of Pittsburgh Office of Technology Management for helpful discussions. The authors are grateful for Ms. Jennifer Ridge Hetrick for serving as Honest Broker in this study. Molecular cytogenetic studies were carried out in the University of Pittsburgh Cell Culture and Cytogenetics Facility.
References
- Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelson H, Fredlund E, Ovenberger M, Landberg G, Pahlman S. Hypoxia-induced dedifferentiation of tumor cells–a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin Cell Dev Biol. 2005;16:554–563. doi: 10.1016/j.semcdb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA repair: Cyclin D1 multitasks. Nature. 2011;474:171–172. doi: 10.1038/474171a. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl U, Schluns K, Kuchler I, Petersen S, Petersen I. Genetic imbalances with impact on survival in head and neck cancer patients. Am J Pathol. 2000;157:369–375. doi: 10.1016/S0002-9440(10)64549-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby EC, Leistritz DF, Abraham RT, Karnitz LM, Sarkaria JN. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 2000;60:2108–2112. [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res. 2009;15:6758–6762. doi: 10.1158/1078-0432.CCR-09-0784. [DOI] [PubMed] [Google Scholar]
- Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol Cell. 1998;2:259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLO-BOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Gollin SM. Chromosomal alterations in squamous cell carcinomas of the head and neck: window to the biology of disease. Head Neck. 2001;23:238–253. doi: 10.1002/1097-0347(200103)23:3<238::aid-hed1025>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–1843. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem. 2003;278:12207–12213. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Dorie MJ, Giaccia AJ. Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer Res. 2004;64:6556–6562. doi: 10.1158/0008-5472.CAN-04-1520. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Helt CE, Cliby WA, Keng PC, Bambara RA, O’Reilly MA. Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. J Biol Chem. 2005;280:1186–1192. doi: 10.1074/jbc.M410873200. [DOI] [PubMed] [Google Scholar]
- Huang X, Gollin SM, Raja S, Godfrey TE. High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc Natl Acad Sci USA. 2002;99:11369–11374. doi: 10.1073/pnas.172285799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I, Cattina F, Pomella S, ILonetti A, Derenzini E, Brighenti E, Ferrari A, Papayannidis C, Falzacappa MVV, Guadagnuolo V, Aluigi M, Ottaviani E, Formica S, Abbenante MC, Soverini S, Russo D, Pane F, Pellicci PG, Baccarani M, Martinelli G. The novel small molecule Chk1/Chk2 inhibitor PF-00477736 (Pfizer) is highly active as single agent in Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL). Blood (ASH Annual Meeting Abstracts); 53rd ASH Annual Meeting and Exposition; Nov 2011; 2011. p. 76. [Google Scholar]
- Jin Y, Jin C, Wennerberg J, Hoglund M, Mertens F. Cyclin D1 amplification in chromosomal band 11q13 is associated with overrepresentation of 3q21–q29 in head and neck carcinomas. Int J Cancer. 2002;98:475–479. doi: 10.1002/ijc.10225. [DOI] [PubMed] [Google Scholar]
- Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, Kunkel TA, van Harn T, Xia B, Correll M, Quackenbush J, Livingston DM, Gygi SP, Sicinski P. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp JE, Thomas BM, Greer JM, Sorge C, Gore SD, Pratz KW, Smith BD, Flatten KS, Peterson K, Schneider P, Mackey K, Freshwater T, Levis MJ, McDevitt MA, Carraway HE, Gladstone DE, Showel MM, Loechner S, Parry DA, Horowitz JA, Isaacs R, Kaufmann SH. Phase I and Pharmacologic Trial of Cytosine Arabinoside with the Selective Checkpoint 1 Inhibitor Sch 900776 in Refractory Acute Leukemias. Clin Cancer Res. 2012;18:6723–6731. doi: 10.1158/1078-0432.CCR-12-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, Lin L, Hoog J, Goiffon RJ, Prat A, Aft RL, Ellis MJ, Piwnica-Worms H. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122:1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CL, Reshmi SC, Ried T, Gottberg W, Wilson JW, Reddy JK, Khanna P, Johnson JT, Myers EN, Gollin SM. Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature. Oral Oncol. 2008;44:369–382. doi: 10.1016/j.oraloncology.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely SC, Burke TF, DurlandBusbice S, Barnard DS, Marshall MS, Bence AK, Beckmann RP. LY2606368, a second generation Chk1 inhibitor, inhbiits growth of ovarian carcinoma xenografts either as monotherapy or in combination with standard-of-care agents. Proceedings of the AACR-NCI-EORTC International Conference: Molecular Targets and Cancer Therapeutics; San Francisco, CA: AACR; 2011. [Google Scholar]
- Meyn MS. Ataxia-telangiectasia, cancer and the pathobiology of the ATM gene. Clin Genet. 1999;55:289–304. doi: 10.1034/j.1399-0004.1999.550501.x. [DOI] [PubMed] [Google Scholar]
- Parikh RA, White JS, Huang X, Schoppy DW, Baysal BE, Baskaran R, Bakkenist CJ, Saunders WS, Hsu LC, Romkes M, Gollin SM. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2007;46:761–775. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, Cliby WA, Sarkaria J, Beale G, Edmondson RJ, Curtin NJ. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105:372–381. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J, Westra W, Sidransky D, Koch WM. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, Muschel RJ, Brunner TB. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13:1072–1081. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshmi SC, Huang X, Schoppy DW, Black RC, Saunders WS, Smith DI, Gollin SM. Relationship between FRA11F and 11q13 gene amplification in oral cancer. Genes Chromosomes Cancer. 2007;46:143–154. doi: 10.1002/gcc.20394. [DOI] [PubMed] [Google Scholar]
- Schenk EL, Koh BD, Flatten KS, Peterson KL, Parry D, Hess AD, Smith BD, Karp JE, Karnitz LM, Kaufmann SH. Effects of selective checkpoint kinase 1 inhibition on cytarabine cytotoxicity in acute myelogenous leukemia cells in vitro. Clin Cancer Res. 2012;18:5364–5373. doi: 10.1158/1078-0432.CCR-12-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Sinha P, Hackman T, Nussenbaum B, Wu N, Lewis JS, Jr, Haughey BH. Transoral laser microsurgery for oral squamous cell carcinoma: oncologic outcomes and prognostic factors. Head Neck. 2013 Jun 1; doi: 10.1002/hed.23293. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- Uhrhammer N, Fritz E, Boyden L, Meyn MS. Human fibroblasts transfected with an ATM antisense vector respond abnormally to ionizing radiation. Int J Mol Med. 1999;4:43–47. doi: 10.3892/ijmm.4.1.43. [DOI] [PubMed] [Google Scholar]
- Vance S, Liu E, Zhao L, Parsels JD, Parsels LA, Brown JL, Maybaum J, Lawrence TS, Morgan MA. Selective radiosensitization of p53 mutant pancreatic cancer cells by combined inhibition of Chk1 and PARP1. Cell Cycle. 2011;10:4321–4329. doi: 10.4161/cc.10.24.18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Khadpe J, Hu B, Iliakis G, Wang Y. An overactivated ATR/CHK1 pathway is responsible for the prolonged G2 accumulation in irradiated AT cells. J Biol Chem. 2003;278:30869–30874. doi: 10.1074/jbc.M301876200. [DOI] [PubMed] [Google Scholar]
- White JS, Weissfeld JL, Ragin CC, Rossie KM, Martin CL, Shuster M, Ishwad CS, Law JC, Myers EN, Johnson JT, Gollin SM. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007;43:701–712. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Bi C, Bence AK, Um SL, Yan B, Starling JJ, Marshall MS, Beckmann RP. Antitumor activity of Chk1 inhibitor LY2606368 as a single agent in SW1990 human pancreas orthotopic tumor model. Cancer Res. 2011;72 Abstract nr 1776. [Google Scholar]