Abstract
We survey the historical development of scientific knowledge surrounding Vitamin B3, and describe the active metabolite forms of Vitamin B3, the pyridine dinucleotides NAD+ and NADP+ which are essential to cellular processes of energy metabolism, cell protection and biosynthesis. The study of NAD+ has become reinvigorated by new understandings that dynamics within NAD+ metabolism trigger major signaling processes coupled to effectors (sirtuins, PARPs, and CD38) that reprogram cellular metabolism using NAD+ as an effector substrate. Cellular adaptations include stimulation of mitochondrial biogenesis, a process fundamental to adjusting cellular and tissue physiology to reduced nutrient availability and/or increased energy demand. Several metabolic pathways converge to NAD+, including tryptophan-derived de novo pathways, nicotinamide salvage pathways, nicotinic acid salvage and nucleoside salvage pathways incorporating nicotinamide riboside and nicotinic acid riboside. Key discoveries highlight a therapeutic potential for targeting NAD+ biosynthetic pathways for treatment of human diseases. A recent emergence of understanding that NAD+ homeostasis is vulnerable to aging and disease processes has stimulated testing to determine if replenishment or augmentation of cellular or tissue NAD+ can have ameliorative effects on aging or disease phenotypes. This experimental approach has provided several proof of concept successes demonstrating that replenishment or augmentation of NAD+ concentrations can provide ameliorative or curative benefits. Thus NAD+ metabolic pathways can provide key biomarkers and parameters for assessing and modulating organism health.
1. Historical background of Vitamin B3 and NAD+
Over two centuries ago, a Spanish doctor Gasper Casal described a disease in poor farmers, whose diet was poor in meat and mainly dependent on Indian corn or maize [1]. This disease was characterized by dermatitis, diarrhea, dementia and death and was later termed pellagra [2]. Pellagra was a peril amongst the malnourished populations in southern Europe for over two hundred years and became an epidemic in the southern states of the U.S. in the early 1900s [3]. By 1912, South Carolina alone had over 30,000 cases of pellagra with 40% eventual mortality [4]. Between 1907 and 1940, 100,000 Americans died from pellagra [4]. In 1914, Joseph Goldberger related pellagra with a nutrient deficiency associated with a corn-based diet [5]. He later suggested that a deficiency in a specific amino acid caused pellagra [6], and identified a water soluble substance as the “pellagra preventive factor” in the 1920s [7]. He also recommended the use of dried yeast as a cheap dietary source to prevent the disease [8]. In 1937, Conrad Elvehjem discovered that nicotinic acid and nicotinamide alternatively, cured “black tongue”, a correlated disease to pellagra in dogs [9]. Nicotinic acid and nicotinamide are now collectively recognized as Vitamin B3. It is now clear that the chronic lack of dietary Vitamin B3 and the amino acid tryptophan, precursors to nicotinamide adenine dinucleotide (NAD+), are the cause of pellagra. Vitamin B3 obtained from NAD+ and NADP+ hydrolysis in meat and tryptophan are markedly deficient in a corn-based diet. Eventually, due to key breakthroughs in knowledge and diet, the epidemic of pellagra was relieved in the U.S. especially with the fortification of Vitamin B3 in bread starting in 1938 [10].
While the clinical investigation of pellagra was underway, the metabolic importance of nicotinamide adenine dinucleotide (NAD+) was also being recognized. In 1906, Harden and Young discovered that yeast extract that was boiled and filtered facilitated a rapid alcoholic fermentation in unboiled yeast extract [11]. They named the responsible component in the extract coferment or cozymase, which was a mixture of components that are essential to carbohydrate utilization. In 1923, von Euler-Chelpin and Myrbäck purified cozymase, from which they identified a nucleoside sugar phosphate [12]. A decade later, Otto Warburg isolated NAD+ from cozymase and showed its role in hydrogen transfer during fermentation [13, 14]. The chemistry of NAD+ and discovery of its key role in human health converged with the discovery of the cure of pellagra. The biosynthetic pathways to produce NAD+ in cells was more fully elucidated by the work of Arthur Kornberg in the 1940s [15] and by the work of Preiss and Handler in the late 1950s [16, 17]. In the meantime, the role of NAD+ in metabolism was being elucidated by scientists such as Krebs [18], and this included fuller descriptions of its integral roles in glycolysis, the TCA cycle and mitochondrial oxidative phosphorylation (See Figure 1). In more recent decades, non-redox roles for NAD+ have been elucidated. NAD+ possess multiple crucial functions in cell signaling pathways including ADP-ribosylation reactions and sirtuin activities. Remarkably, NAD+ homeostasis is not only important for the prevention of pellagra, but is also associated with extended lifespan, increased resistance against infectious and inflammatory diseases [19, 20] and is likely very important in resisting a number of other disease processes [21] such as cardiovascular disease, metabolic syndrome, neurodegenerative diseases and even cancer.
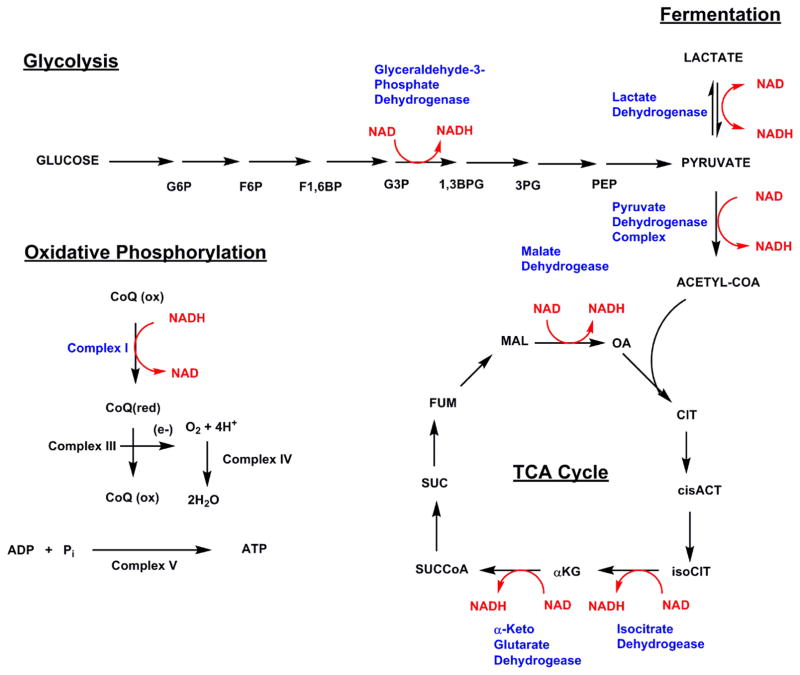
Figure 1. Integration of NAD+ and NADH into cellular energy metabolism.
NAD+ reduction to NADH is featured in glycolysis, pyruvate dehydrogenase, and the tricarboxylic acid cycle (TCA cycle). NADH oxidation to NAD+ occurs in the cytoplasm by action of lactate dehydrogenase and in mitochondria by action of Complex I. These oxidations maintain the high NAD+/NADH ratio maintained by mammalian cells and the majority of mitochondrial production of ATP is directly linked to Complex I activity. Abbreviations: glucose 6-phosphate, G6P; fructose-6-phosphate, F6P; fructose-1,6-bisphosphate, F1,6BP; glyceraldehyde-3-phosphate; 3-phosphoglycerate, 3PG; phosphoenol pyruvate; oxaloacetate, OA; citrate, CIT; cis-aconitate, cisACT; isocitrate, isoCIT; alpha-ketoglutarate, αKG; succinylCoA, SUCCoA; succinate, SUC; fumarate, FUM; malonate, MAL. Coenzyme Q, CoQ.
2. Roles of NAD+ in energy producing metabolic pathways
NAD+ is an important co-enzyme for hydride transfer enzymes essential to multiple metabolic processes including glycolysis, pyruvate dehydrogenase complex, the TCA cycle and oxidative phosphorylation. The enzymes using NAD+ in hydride-transfer are known as dehydrogenases or oxidoreductases, which catalyze reduction of NAD+ into NADH (Examples shown in Figure 1). Mitochondrial NADH is then utilized by the electron transport chain and therein participates as a substrate in mitochondrial ATP production through oxidative phosphorylation (Figure 1).
The first enzyme utilizing NAD+ in glycolysis is glyceraldehyde 3-phosphate dehydrogenase [22]. It is the sixth step in glycolysis, wherein glyceraldehyde 3-phosphate is oxidized to D-glycerate 1,3-bisphosphate in the cytosol (See Figure 1) via transfer of a hydride equivalent to NAD+. As one glucose can be converted into two glyceraldehyde 3-phosphate equivalents, one mole of glucose can generate 2 moles of NADH. Maintenance of high NAD+/NADH in cytoplasm is sustained by two NADH utilization pathways, which combine to maintain a flux of hydride removal from hydride rich carbon substrates inherent to cellular energy metabolism. NADH equivalents generated in the cytoplasm via glycolysis are transferred to the mitochondria by shuttling mechanisms such as the malate-aspartate shuttle, in which NADH in the cytosol is oxidized to NAD+ and NAD+ in mitochondria is reduced to NADH. Alternatively, NADH is oxidized back to NAD+ in the cytoplasm via lactate dehydrogenase, correspondingly producing lactate from pyruvate.
The products of glycolysis are two moles of pyruvate, 2 moles of NADH and two moles of ATP. Pyruvate has multiple possible fates. In lactate dehydrogenase reaction pyruvate is reduced to lactate. (See Figure 1) [23]. For maximal energy yield pyruvate is alternatively acted upon by the pyruvate dehydrogenase complex to form acetylCoA with concomitant NAD+ reduction to NADH [24]. AcetylCoA can then enter the TCA cycle, where NAD+ equivalents are reduced to NADH moieties in several key steps by isocitrate dehydrogenase (IDH), oxoglutarate dehydrogenase (OGD) and malate dehydrogenase. IDH is a key step in the TCA cycle, which oxidizes isocitrate to oxalosuccinate, which is then decarboxylated to form α-ketoglutarate. IDH exists in three isoforms [25], with IDH3 located in mitochondria and used to support the TCA cycle. IDH1 and IDH2 catalyze the same reaction and use NADP+ as cofactor. The next enzyme in the cycle, OGD, catalyzes the reaction from α-ketoglutarate to succinyl CoA, with reduction of NAD+ to NADH. OGD is a key regulatory point in the TCA cycle and is inhibited by its product succinylCoA and NADH. ADP and calcium are allosteric activators of the enzyme [26]. OGD is considered to be a redox sensor in the mitochondria. Increased NADH/NAD+ ratio is associated with increased ROS production and inhibited OGD activity. Once ROS levels are removed, OGD activity can be restored [27]. Malate dehydrogenase completes the TCA cycle and produces the third equivalent of NAD+ reduction to NADH from one mole of acetyl-CoA that enters the cycle.
NADH formed from glycolysis (via the malate-aspartate shuttle) or the TCA cycle can react at Complex I, also known as the NADH/coenzyme Q reductase in the mitochondrial electron transport chain [28]. Each NADH consumed by the mitochondria results in the net production of 3 ATP molecules (Figure 1). The complete oxidation of one glucose molecule generates 2 NADH equivalents in cytosol and 8 NADH molecules in mitochondria, enabling production of 30 ATP equivalents from NADH of the total of 36 ATP equivalents derived from the whole process of catabolizing glucose to CO2 and H2O.
The NAD+/NADH ratio plays a crucial role in regulating the intracellular redox state, especially in the mitochondria and nucleus. Free NAD+/NADH ratio varies between 60 to 700 in eukaryotic cells, although the estimated mitochondrial NAD+/NADH ratios are possibly maintained at closer to 7–10 [29, 30]. Total NAD+ levels in mammalian cells appear to be maintained at 200–500 μM under most conditions. Mitochondrial NAD+ content appears to be relatively more abundant than cytosolic NAD+ in metabolically active cells and tissues, e.g. cardiac myocytes and neurons, probably because of the needs of these tissues for significant energy and ATP production [31]. Interestingly, the mitochondrial NAD+ pool is likely more stable compared to the cytosolic pool, possibly to preserve oxidative phosphorylation and to maintain cell survival even in stressed cells [32]. Under cytoplasmic NAD+ depletion, mitochondrial NAD+ levels can still be maintained for up to 1 day [33]. Remarkably, the mitochondria pool of NAD+ can provide the threshold parameter for the survival of the cell [32].
Inhibition of mitochondrial electron transport chain activity decreases mitochondrial conversion of NADH to NAD+ and reduces the mitochondrial NAD+/NADH ratio. Complex I/III inhibitors can decrease the intracellular NAD+/NADH ratio by more than 10 fold. This decreased ratio changes the ratio of α-ketoglutarate/citrate ratio and limits acetyl-CoA entry into the TCA cycle [27]. Thus feedback of NADH into metabolism is a key factor determining the rate of catabolism and energy production. Overall oxidative metabolism is decreased when mitochondrial NADH/NAD+ level is elevated. NAD+ within the nucleus also plays significant signaling roles in controlling and regulating metabolic pathways [34, 35]. For example, nuclear NAD+ alters sirtuin 1 (SIRT1) activity [34, 35], which in turn regulates the activity of the downstream transcriptional regulators such as Forkhead box O (FOXOs) that play important role in metabolism, stress resistance and cell death [19, 36]. A decline in nuclear NAD+ level and elevation of NADH results in the accumulation of hypoxia-inducible factor 1 alpha (HIF-1α) stimulating the Warburg effect, a hallmark of cancer cell metabolism called aerobic glycolysis [37]. Aging has been shown to promote the decline of nuclear and mitochondrial NAD+ levels, and the risk of cancer development may be increased by this phenomenon [37], although the role of dinucleotides in cancer risk is currently being vigorously investigated.
3. NADPH/NADP+ roles
A structural analogue to NAD+ is NADP+ which incorporates a 2′-phosphate on the adenosine ribose moiety absent in NAD+. This structural and chemical difference enables a distinctive role for NADP+ and its reduced form NADPH in cells [38]. For example, intracellular levels of NADP+ and NADPH are maintained at significantly lesser amounts than NAD+ and NADH. In addition, the NADP+/NADPH ratio is preferentially maintained to favor the reduced form, very unlike the corresponding NAD+/NADH ratio [38, 39]. NADPH is essential for survival, and is important for detoxification of oxidative stress. For example, NADPH donates a hydride equivalent to generate antioxidant molecules, such as reduced glutathione, reduced thioredoxin and reduced peroxiredoxins [40–42], which help eliminate cellular reactive oxygen species (ROS) [43]. NADPH is also required in the activity of detoxifying enzymes, such cytochrome P450 that function in xenobiotic metabolism [44]. In inflammatory pathways, NADPH acts as a substrate for NADPH oxidase in neutrophils and phagocytes, which use these enzymes to kill pathogens by generating superoxides [45]. NADPH serves as a key electron donor in the synthesis of fatty acid, steroid and DNA molecules [38]. NADP+ is utilized in the pentose phosphate pathway to regenerate NADPH in a pathway which can ultimately produce ribose-5-phosphate for nucleotide synthesis [46]. Interestingly, in a non-redox capacity, NADP+ serves as a precursor for nicotinic acid adenine dinucleotide phosphate (NAADP), a potent calcium mobilizing messenger which regulates calcium homeostasis [47]. Numerous studies indicate that maintenance of NADP+ and NADPH levels are vital to ensure the survival of cells especially in oxidative stress [38].
NADP+ is generated from NAD+ in cells, by action of the enzyme NADK. A well-studied cytosolic NADK preferentially uses NAD+ (Km = 1.07 mM for NAD+) as a substrate over NADH in human cells, and the NADP+ is rapidly converted to NADPH by transdehydrogenase activity [48]. Interestingly, the overexpression or down-regulation of cytoplasmic NADK influences exclusively NADPH level without altering the level of NADP+, NAD+ and NADH [48]. The activity of NADK can be activated by oxidative stress and calcium/calmodulin [49] and inhibited by high NADPH levels [50]. A putative mitochondrial human NADK has been reported [51–53]. The reported Km for NAD+ is 22 μM [53], suggesting it is typically saturated with NAD+ substrate, unlike the cytoplasmic counterpart. This may indicate greater demand for NADP+ maintenance in mitochondria, although the lifetimes of the NADP+/(NADPH) moiety in cells or in subcellular compartments are unknown.
Robust NADP+ reduction to NADPH is key to maintenance of high NADPH/NADP+ ratio [38]. In the cytosol, NADPH is generated from NADP+ by glucose-6-phosphate dehydrogenase, 6-gluconate phosphate dehydrogenase, IDH1, 2 or cytosolic NADP+-dependent malate dehydrogenase enzymes. The NADPH generated in the cytoplasm is believed to be responsible for NADPH oxidase-dependent ROS generation. In mitochondria, NADPH is generated from NADP+ by IDH3, mitochondrial malate dehydrogenase or transhydrogenases. NADP+ and NADPH concentration changes and how they mediate downstream biological effects are of current interest, and these concentration changes may be influenced by NAD+ responsive mediators, such as SIRT3, which upregulates IDH3 activity and thereby affects mitochondrial NADPH/NADP+ ratio [54]. More research in this area is clearly needed, as the cross talks between NAD+ metabolism and NADPH/NADP+ dynamics are still poorly understood. Moreover, how cellular, hormonal or nutritional stimuli affect NADPH/NADP+ dynamics are still generally poorly understood as well.
4. Roles of NAD+ in signaling pathways
The realization that NAD+ could be directly involved in the regulation of cell biological processes through changes in global signaling events was revealed by two sets of studies made in the 1960s. The first of these were pioneered by Chambon, who published two papers in 1963, and 1966, investigating if ATP could become incorporated into nuclear proteins. He initially reported an nicotinamide mononucleotide (NMN) stimulated activity [55], which turned out to be the incorporation of the ADP-ribose moiety of NAD+ into acid-precipitated proteins [56]. Further analysis of the protein adducts by digestion with snake venom phosphodiesterase indicated a polymer, later determined to be poly-ADPribose [56]. This insightful and brilliant investigative work laid the foundation for decades of subsequent biochemical and biological investigations into the roles of ADP-ribosyltransfer in modulating protein activity in mammalian cells.
Interestingly, at nearly the same time, it became apparent that selected bacterial toxins might act via a similar kind of mechanism. In 1964, Collier and Pappenheimer determined that a key protein virulence factor of the organism Corynebacterium Diphtheria (the cause of the human disease Diphtheria) caused inhibition of protein synthesis in mammalian cells in an NAD+-dependent manner [57]. Honjo et al. later determined that Diphtheria toxin ADP-ribosylates the elongation factor Ef-2, which is required to complete protein synthesis within mammalian cells [58]. This covalent modification of a mammalian protein by ADPribosylation clearly altered protein function, and provided the prototype biochemical event upon which to postulate that NAD+ can serve as a protein modifying agent, with a direct consequence to a cellular protein activity.
Stemming from these seminal studies emanating from the 1960s, and with the advent of modern proteomics and bioinformatics approaches, it has become apparent that there are a variety of signaling enzymes that harness NAD+ as a substrate, and transfer the ADPribose unit to proteins by mono-ADP-ribosylation mechanisms, or by poly-ADP-ribosylation mechanisms thereby altering protein functions (For a recent comprehensive review on this topic, see Hassa et al.[59]). For example, the parent poly-ADPRibosyl polymerase (PARP), PARP1, along with other family members (PARP2, 5a and 5b), are responsible for conferring poly-ADP-ribosylpolymerase activity [60, 61], whereas another 11 PARPs are mono-ADP-ribosyltransferases. Surprisingly, although a number of these are likely to have key functions in NAD+ mediated signaling processes, only a subset of functions have been firmly described for family-members in this group.
In addition to PARP-related ADPribosyltransferases, there is another major group of ADP-ribosyltransferases, which are called sirtuins [62, 63]. Technically, sirtuins are ADPribosyltransferases because they use this mechanism chemically to effect deacylation of substrates. The connection of sirtuins to the ADP-ribosylating toxins and PARPs was not immediately apparent, as the sirtuins are distinct in structure, sequence and chemistry from known NAD+-consuming enzymes, responsible for ADPribosyltransfer [63]. However, sirtuins have been shown to react NAD+ with acyl-lysine modified protein substrates, leading to formation of transiently-ADP-ribosylated acyl-groups, causing subsequent deacylation of these substrates [63]. This protein deacylation activity leads to regulated changes in cell behaviors, linked to these deacylation events. Most remarkable of these are at the level of chromatin, where histone and transcription factor deacylations are responsible for regulation of a whole spectrum of gene regulatory changes (as reviewed recently by Kraus and co-workers [35]).
The activities of some ADPribosylating enzymes are regulated partially in a manner independent of the availability of NAD+ substrate. A case in point is the regulation of PARP1 and PARP2 by the formation of DNA strand breaks [59]. However, a key emerging premise in understanding the activities and roles of NAD+ utilizing enzymes, is that these enzymes are regulated dynamically by cellular NAD+ metabolism. Specifically, the idea that nicotinamide and NAD+ concentrations act as inhibitors or drivers, respectively, of ADPribosylating enzymes has become a key concept, supported by biochemical measurements which determine the apparent steady-state Km enzyme parameters for NAD+ (e.g. sirtuins) in the 100–200 μM range [64, 65], squarely within the range of physiological NAD+ concentrations. In fact, recent work examining the effects of supraphysiological NAD+ concentrations, obtainable by genetic modifications (CD38 [66] and PARP1 [67] knockouts for example), confirm that increased NAD+ causes activation of signaling programs linked to and requiring sirtuins, leading to downstream events such as increased mitochondrial biogenesis [67, 68]. Naturally, there are some caveats associated with these knockouts, since they also eliminate key biochemical activities from cells and tissues. However, evidence indicates that SIRT1 activity is enhanced in these high NAD conditions to produce deacetylation of peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC1α), activating this co-transcriptional regulator, to enhance transcription of a broad subset of genes responsible for formation of new mitochondria [67]. Indeed, low calorie diets, fasting [32] and exercise [69] appear to enhance NAD+ synthesis [32], which leads to activation of sirtuins [70] and presumably PARP family members. It is notable that PARP1 deletion leads to significant upregulation of NAD+ levels [67], indicating that baseline PARP activity is a key factor in establishing normal tissue NAD+ homeostasis, and implying that significant crosstalk exists between NAD+ metabolism, sirtuins and PARP enzymes [71].
5. Overall view of mammalian NAD+ metabolism
Consistent with the centrality of NAD+ to cellular bioenergetics, and supportive of the relatively high concentrations of NAD+ metabolites in cells (200–500 μM typically in mammalian cells), several distinct pathways are involved in the biosynthesis of NAD+. In humans this includes de novo pathways from the amino acid precursor tryptophan, and additional pathways, including from different nicotinoyl precursors, such as nicotinic acid, nicotinamide as well as nucleosides nicotinamide riboside (NR) and nicotinic acid riboside (NAR). Broadly, NAD+ metabolism can be viewed in four main categories, 1. De Novo Synthesis, 2. Scavenging Pathways from preformed precursors (nicotinic acid, nicotinamide riboside and nicotinic acid riboside), 3. Core Recycling Pathway through nicotinamide. Finally, 4. ADPR-transfer/NAD hydrolysis, which occurs through a variety of enzymatic pathways, leading to cleavage of the N-glycosidic bond of nicotinamide to the ribose ring, thereby liberating nicotinamide and providing an ADPR-nucleophile product. Thus, three general types of synthesis pathways converge to produce NAD+, while consumption pathways comprised of several types of NAD+ consumers deplete NAD+. Steady-state NAD+ levels are set where the magnitude of the rate of turnover of NAD+ is equalized by the net formation rate contributed by the separate synthetic pathways. Evidence indicates the entire NAD+ pool is consumed and resynthesized in mammals several times a day [72]. Under normal cellular and tissue conditions, synthesis of NAD+ is affected by the availability of possible precursors, so that availabilities of nicotinic acid, nicotinamide riboside, nicotinamide and tryptophan can alter NAD+ synthetic rates, thus affecting NAD+ level. Not each of these precursors is bioequivalent in this respect. Strikingly, nicotinamide is not limiting in many tissues, except possibly in liver, so availability of nicotinamide is not crucially tied to NAD+ formation rate, and even when administered at fairly substantial doses via diet [73]. The identity and not necessarily only quantity of NAD+ precursor is important to the biological NAD+ level. The importance of increased NAD+ levels to signaling pathways emphasizes the relevance of the different NAD biosynthetic pathways, which are examined in greater detail in the following section.
6. Pathway and enzymes to make NAD+ from tryptophan
Tryptophan (Trp) is a substrate for the de novo synthesis of NAD+ in humans, much of which is believed to take place in the liver [74]. Trp is one of the essential amino acids required for protein synthesis and metabolic functions, with primary producers including bacteria, fungi and plants [75]. After proteins are hydrolyzed into amino acids in the GI tract, Trp is available for protein synthesis, and the majority of Trp catabolism occurs through the kynurenine (KYN) pathway [76, 77]. In the brain, in particular, Trp is the building block for several essential molecules, for instance the neurotransmitter serotonin and the sleeping hormone melatonin [78]. These metabolites are synthesized in a pathway independent of the kynurenine pathway (Figure 2). Trp is also considered as a nutritionally relevant precursor to NAD+. In a niacin deficient diets, Trp can be a key source for NAD+ and NADP+ synthesis. However, due to other metabolic fates of this amino acid, it has been estimated that 60 mg of Trp converts to only 1 mg niacin [79].
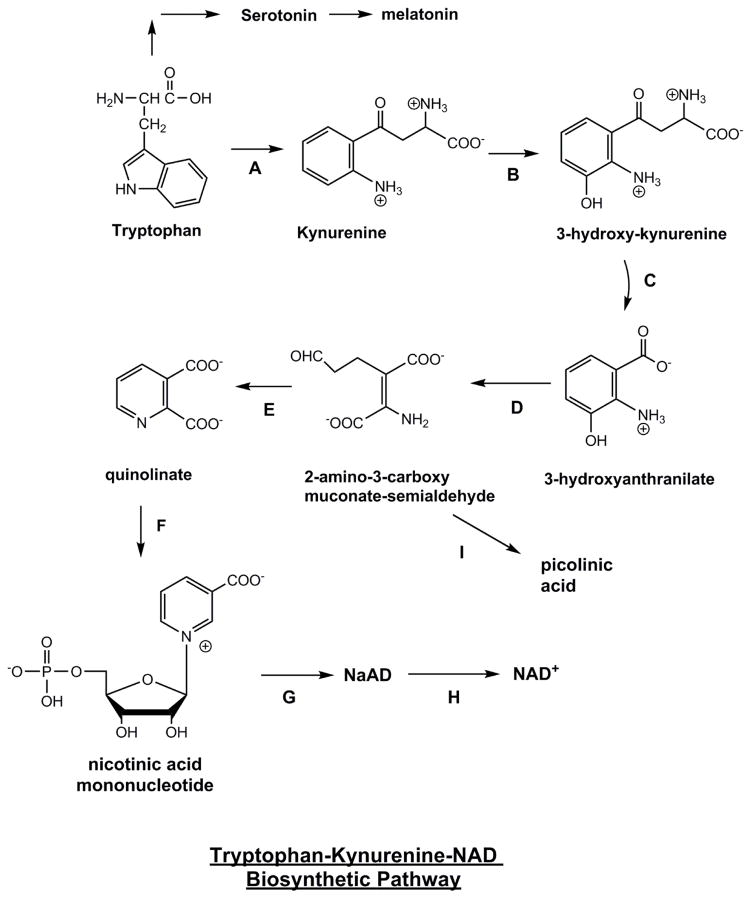
Figure 2. Tryptophan-Derived Biosynthesis of NAD+ via Kynurenine Pathway.
Enzymes are A) tryptophan 2,3-dioxygenase B) kynurenine mono-oxygenase C) kynureninase D) 3-hydroxyanthranilate 3,4,-dioxygenase E) no-enzyme F) Quinolinate phosphoribosyltransferase G) nicotinic acid/nicotinamide mononucleotide adenylyltransferase (Nmnat 1, 2, 3) H) NAD synthetase. I) 2-amino-3-carboxy-muconate-semialdehyde decarboxylase (ACMSD) which acts upon 2-amino-3-carboxy-muconate-semialdehyde to form picolinic acid, as an alternative pathway fed by the kynurenine catabolic pathway, as discussed in the text.
Trp can be converted to NAD+ through an eight-step biosynthesis pathway. The first and rate-limiting step in this pathway is the conversion of Trp to N-formylkynurenine by indoleamine 2,3-dioxygenase (IDO) or tryptophan 2,3-dioxygenase (TDO) [80]. TDO exists primarily in liver [81] and can be activated by Trp [76] or corticosteroids [82], whereas IDO is found in numerous cell types, such as microglia, astrocytes, neurons [83] and macrophages [84] that reside in extra-hepatic tissues and is activated by pro-inflammatory signals [85, 86]. The first stable intermediate in the pathway, KYN, can be metabolized through two distinct pathways to form kynurenic acid or NAD+ [86]. With the enzymatic reaction of kynurenine mono-oxygenase, and kynureninase KYN is metabolized into 3-hydroxyanthranillic acid, which is then converted by 3-hydroxyanthranilate-3,4-dioxygenase into 2-amino-3-carboxymuconate semialdehyde. This metabolic intermediate can be acted upon by 2-amino-3-carboxymuconate semialdehyde decarboxylase (ACMSD) (formally termed picolinic carboxylase) to provide picolinic acid, or non-enzymatically converted into quinolinic acid (QA) [87]. QA is metabolized by quinolinate phosphoribosyltransferase (QPRT) to form nicotinic acid mononucleotide (NAMN) and enter the salvage pathway for NAD+ synthesis in liver [88]. Trp will only be diverted to NAD+ synthesis when the substrate supply far exceeds the enzymatic capacity of ACMSD [89], which helps explain the weak niacin equivalence of 60 mg trp per mg niacin [79].
The daily recommendation of tryptophan intake is 4 mg/kg body weight for adults and 8.5-6 mg/kg body weight for infants to adolescents [90]. Being an alternative but weak source for NAD+, it is an interesting question why the Trp pathway to NAD is evolutionarily well-retained in humans (Cats do not use Trp efficiently to make NAD+ [89]). Studies have shown that the de novo biosynthesis rate of NAD+ from Trp is unchanged by the absence or presence of nicotinamide in the diet, as the urinary excretion of intermediates in the Trp pathway are not altered with 0 or up to 68.6 mg/day nicotinamide [91]. Upregulated Trp derived NAD+ biosynthesis pathway does not appear to compensate for a deficiency of other NAD+ precursors. Oral intake of Trp as high as 15 g/day renders low acute toxicity, with side effects such as drowsiness and headache [92]. Overconsumption of tryptophan causes toxicity in certain animal species. The LD50 value is 1.6 g/kg body weight in rats and 2 g/kg body weight in mice and rabbits with i.p. or i.v. administration. Oral LD50 is around 3 times that of i.p. dose [90]. Increases in Trp catabolism may result in adverse effects in the human body. Several intermediates and products in the KYN pathway, including QA, 3-hydroxyl-L-kynurenine and kynurenic acid, are neurotransmitters and display key roles in central nervous system [87]. High levels of QA in brain have been associated with neurodegenerative conditions such as Huntington’s disease [93] or seizure [94]. QA and KYN have also been shown to induce anxiety in mice [95, 96]. Therefore, it may not be ideal to use Trp as a primary dietary or pharmacologic source to enrich NAD+ level.
7. Pathway and enzymes to make NAD+ from nicotinic acid
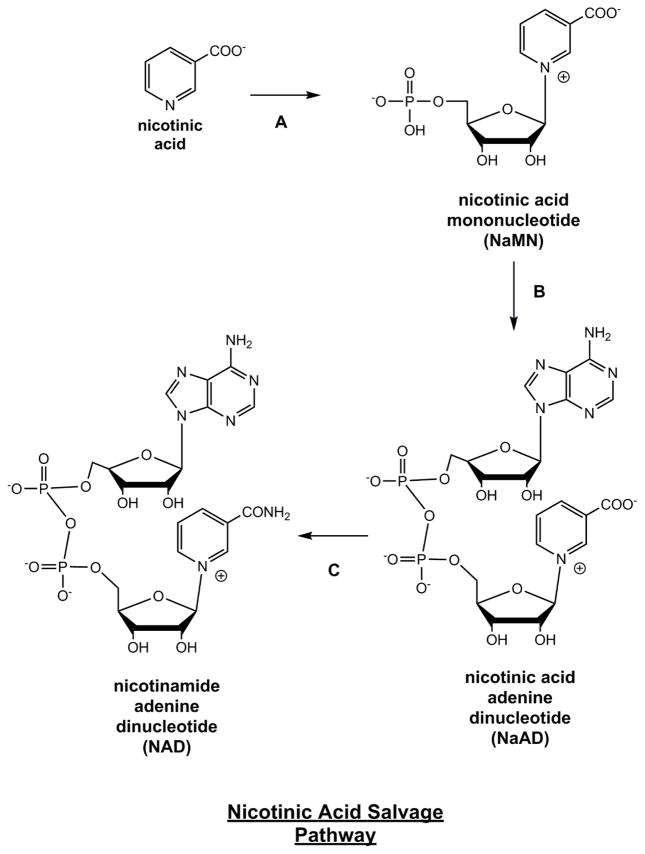
In humans and mammals, nicotinamide and nicotinic acid are routed in non-overlapping pathways to NAD+. These separate paths stand in contrast to the NAD+ metabolism observed in flies, worms, yeast and many bacteria. In flies, yeast, worms and many bacteria the breakdown product of NAD+, nicotinamide, is directly converted to nicotinic acid by a highly conserved enzyme called nicotinamidase [97]. This key transformation connects the pathways of nicotinamide and nicotinic acid derived NAD+ biosynthesis in these organisms, as they represent consecutive metabolites in NAD+ production. On the other hand, humans and mammals lack nicotinamidase activity [19]. Consequently, nicotinamide is directly metabolized to NAD+ independent of nicotinic acid. Nicotinic acid is converted to the intermediate nicotinic acid phosphoribosyltransferase (NaMN) by action of the enzyme nicotinate phosphoribosyltransferase (Npt) (Figure 3). NaMN is common to the nicotinic acid salvage pathway and the tryptophan quinolinate pathway. Adenylation of NaMN can be accomplished by NaMN adenylyltransferase (nmnat). This enzyme has 3 isoforms nmnat-1, nmnat-2 and nmnat-3. Compartmentalization of these in cells are known, with the nmnat-1 nuclear, nmnat-2 golgi associated, and nmnat-3 mitochondrial. These enzymes can accept either NaMN or NMN as nucleotide substrates, with the NaMN being formed to nicotinic acid adenine dinucleotide (NaAD+). The terminal enzyme in this pathway is NAD+ synthetase. It converts the NaAD+ to NAD+ in an ammonia and ATP-dependent process. NAD+ synthetase in humans also combines a glutaminase activity which provides a source of the ammonia to complete the reaction. This basic set of transformations is found in nearly all organisms that can recycle nicotinic acid, and has become famously called the Preiss-Handler pathway, after Preiss and Handler who first described it over 60 years ago [16, 17, 98].
Figure 3. Nicotinic Acid Salvage Pathway (Preiss-Handler Pathway).
Enzymes are A) nicotinate phosphoribosyltransferase B) nicotinic acid/nicotinamide mononucleotide adenylyltransferase (Nmnat 1, 2, 3) C) NAD+ synthetase.
8. Core recycling pathway from nicotinamide
For mammalian cells the central challenge in NAD+ homeostasis is successful recycling of nicotinamide, released from NAD+ consuming processes, back to NAD+. Published data for NAD+ turnover in vivo indicate half lives of as little as 4–10 hours [72]. For resynthesis that equates to a minimal need to recycle 200–600 umol/kg per day of tissue in rats [73]. Assuming comparable numbers for a 75 kg human, 3 g of nicotinamide is required to be resynthesized to NAD+ up to several times per day (assuming 300 μmoles NAD/kg wet tissue)[99]. These levels are far below the amounts available from food intakes (1 lb tuna provides 100 mg vitamin B3, and 1 lb beef provides 30 mg B3, whereas 4 cups of broccoli provide only 4 mg of B3). These facts implicate efficient nicotinamide recycling as the basis for effective NAD+ maintenance in humans, and a consistent lack of pellagra in the developed world.
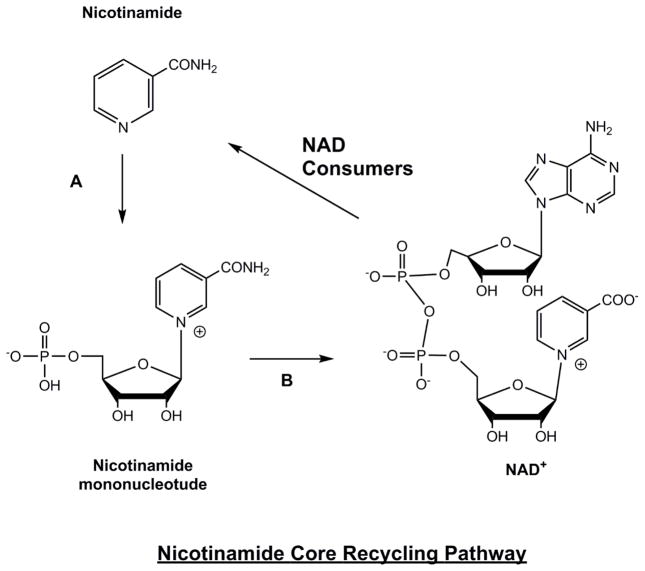
The nicotinamide recycling reaction is catalyzed by an enzyme called nicotinamide phosphoribosyltransferase (nampt). Nampt couples nicotinamide with PRPP to form nicotinamide mononucleotide (NMN) (Figure 4). Kinetic studies indicate that ATPase activity is also coupled to this process, which drives the equilibrium toward NMN formation [100, 101]. A valuable pharmacologic tool has been developed to inhibit this enzyme, called FK866. This inhibitor, which has a binding constant of 0.3 nM [102] can be applied to mammalian cells and leads to rapid depletion of cellular NAD+, causing levels to reach 30% within 4–8 hours. In vivo, this inhibitor has been evaluated as a possible anti-cancer agent, since it limits the abilities of tissues to recycle nicotinamide caused by anti-cancer genotoxins. Recently Nuncioni and coworkers showed that NAD+ contents can be significantly reduced in several tissues, including blood, spleen and heart with systemic administration of FK866 alone to mice [103].
Figure 4. Nicotinamide Core Recycling Pathway.
Enzymes are A) nicotinamide phosphoribosyltransferase B) nicotinic acid/nicotinamide mononucleotide adenylyltransferase (Nmnat 1, 2, 3). NAD+ is constantly consumed in cells by action of NAD+ consumers, including sirtuins, PARP enzymes, CD38 etc., creating a constant supply of nicotinamide, which must be recycled to maintain NAD+ homeostasis. The importance and efficiency of this process is discussed in the text.
Experiments to assess the role of Nampt in setting the NAD+ level in cells confirms that the level of the enzyme, and not nicotinamide concentrations themselves, have the largest effect on setting NAD+ level. For instance, Sinclair and Sauve and co-workers [32] used overexpression of Nampt or knockdown of Nampt in mammalian cultured cells to establish that Nampt levels regulate how much cellular NAD+ is available in cells. As expected, knockdown of Nampt caused reduced NAD+ levels in cells and also within the mitochondrial compartment. Conversely, overexpression caused increased cellular NAD contents and increased mitochondrial NAD+ levels. An interesting effect of overexpression of Nampt was noted on cell resistance to genotoxic stress, in that Nampt overexpressors proved resistant to apoptosis [32]. These protection effects required mitochondrial sirtuin activities, notably SIRT3 and possibly SIRT4 [32]. Interestingly, Nampt levels appear to be upregulated by dietary intake and exercise. Increased levels in liver were observed for fasted rats [32], and exercised rats [69]. Nampt levels also increase in humans in exercised legs as compared with matched unexercised legs [104].
Although comprehensive data on Nampt activity in humans is unclear, due to lack of published data, the efficiency of this nicotinamide recycling appears to be very high. For example, there is a relatively low Vitamin B3 requirement published by the Food and Nutrition Board at the Institute of Medicine which states that only 16 mg day Vitamin B3 is required for male adults above 16 and 14 mg day for females above 14 [105]. This implies a net loss of not greater than 0.5% total NAD+ per day, to maintain niacin homeostasis, if no additional input is available from outside sources. This is remarkable, if one considers the possibility that the entire NAD+ pool is being replaced 2–4 times per day, suggesting that only 0.1–0.2 % nicotinamide is lost per turnover cycle. This result implicates a highly efficient NAD+ resynthesis capacity in humans, made more impressive if one considers that nicotinamide is neutral, small (MW = 122) and a polar hydrophobic (LogP = −0.4), likely to passively diffuse through cell membranes, a phenomenon demonstrated in experimental studies [106]. The existence of nicotinamide methyltransferases and downstream catabolic enzymes that degrade nicotinamide to pyridone-related catabolites are a likely source for lost nicotinamide, and the amount of nicotinamide degradation products have been noted in some neurodegenerative diseases, suggesting enhanced NAD+ breakdown [21].
9. Pathways and enzymes to make NAD+ from nucleosides
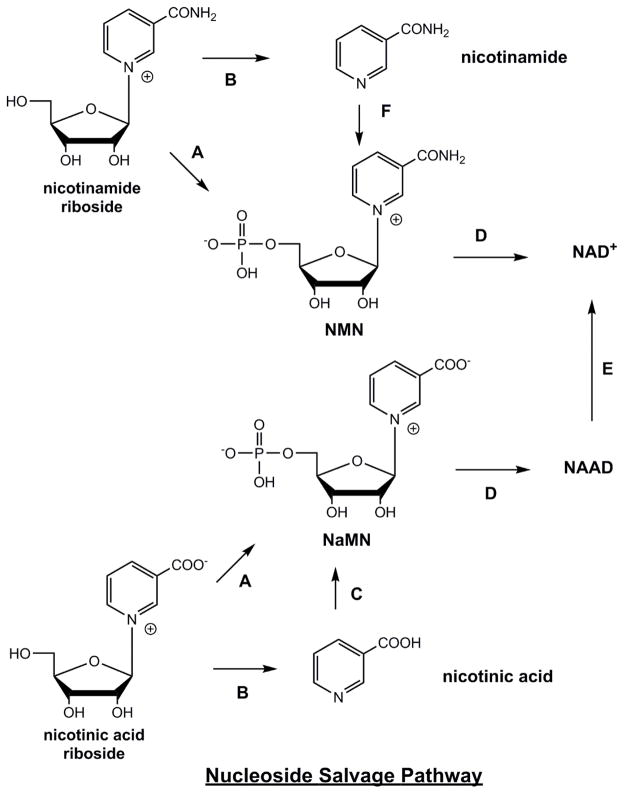
Additional relevant pathways to NAD+ are from nucleosides, including nicotinamide riboside (NR) and nicotinic acid riboside (NaR)(Figure 5). These pathways are facilitated by action of nicotinamide riboside kinases (Nrk1 and Nrk2) [107]. These enzymes are encoded by the human genome, and by genomes of other mammalian organisms and have been shown to enzymatically convert NR and NaR to NMN and NaMN respectively [108, 109]. X-ray crystallographic evidence, as well as biochemical data, show that these enzymes bind NR and NaR into an active site that discriminates against purines and pyrimidine nucleosides, thus making these enzymes preferentially specific for NR derivatives, as well as able to accommodate the antimetabolite drugs tiazofurin (TZ) and benzamide riboside (BR) [108]. NR and NaR raise NAD+ levels dose-dependently and up to 2.7 fold in mammalian cells [110] in a manner unlike the behavior of nicotinamide or nicotinic acid at the same concentrations. It is proposed that a transporter can convey NR and NaR into mammalian cells, and an NR transporter (Nrt1) in yeast has been identified [111]. Evidence for such a transporter is provided by studies of transport of BR and TZ [112]. Human concentrative nucleoside transporter 3 was most efficacious as a nucleoside transporter of both BR and TZ in transfected xenopus oocytes [112]. Recent data suggests that both NR and NaR could be produced in mammalian cells and released extracellularly, and could thereby be produced in mammals as NAD+ precursors [113], suggesting possible intercellular metabolic networks involving NR and NaR creation, release and transport into other cells. In addition, an NR degrading and possibly NaR degradative pathway may also help explain some of the effects of NR and NaR on cells and tissues, as first described by Kornberg [114]. The potency of NR in increasing cellular NAD+ has led to investigations to determine if it can treat diseases such as neurodegenerative disorders, or metabolic syndromes, based on the idea that reduced NAD+ level might be a risk factor in these conditions [32, 68, 70].
Figure 5. Nucleoside Salvage Pathway.
Enzymes are A) nicotinamide riboside kinase 1 and nicotinamide riboside kinase 2 (Nrk1, Nrk2) B) purine nucleoside phosphorylase C) nicotinic acid phosphoribosyltransferase D) nicotinic acid/nicotinamide mononucleotide adenylyltransferase (Nmnat 1, 2, 3) E) NAD+ synthetase F) nicotinamide phosphoribosyltransferase.
10. NAD+ metabolism as a target of therapy
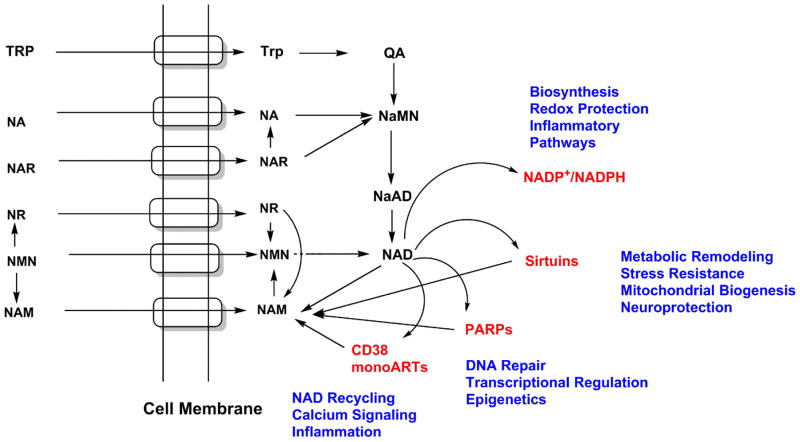
Figure 6 provides a comprehensive view of NAD+ metabolic pathways as found in humans and how these pathways intersect and converge on the central metabolite NAD+. This figure also conveys the fact that different NAD+ precursors can enter NAD+ biosynthetic pathways and can converge to NAD+ via independent and also overlapping pathways. This distinctiveness in how different NAD+ precursors are metabolized is now clearly recognized with the understanding that these distinct NAD+ precursors have different properties and have distinct biological effects.
Figure 6. Intracellular Import of NAD Precursors and Intracellular Conversions to NAD+.
Depiction of how different NAD precursors are transported into cells via identified or putative transport systems in cell membranes (embedded cylindrical domains). As can be visualized by the figure, networks of biosynthetic pathways converge to two main NAD biosynthetic intermediates, namely NaMN and NMN. NAD itself provides an action point for several other downstream processes, including biosynthesis of NADP/NADPH and for signaling processes, which regenerates nicotinamide for the core nicotinamide recycling pathway.
The unique properties of NAD+ precursors was first appreciated by the determination that high doses of nicotinic acid could cause lowering of serum lipids, including free fatty acids and low density lipoprotein (LDL)- cholesterol, and could increase the “good” cholesterol form called high density lipoprotein (HDL)- cholesterol [115]. Kirkland and co-workers explored effects of high doses of nicotinamide or nicotinic acid on tissue NAD+ levels in rats, and concluded that these precursors had different abilities to raise NAD+ levels in different tissues [73]. Nicotinamide had ability to increase NAD+ level in liver (47%), but was weaker in kidney (2%), heart (20%), blood (43%) or lungs (17%). Nicotinic acid raised NAD+ in liver (47%), and impressively raised kidney (88%), heart (62%), blood (43%) and lungs (11%) [73] [both nicotinamide and nicotinic acid were administered at 1000 mg/kg diet].
The reinvestigation of less-studied Vitamin B3 forms such as nicotinamide riboside and NMN for their possible NAD+ enhancing benefits became important with the recognition that NAD+ may act as a key signaling molecule in cellular physiology. The Brenner laboratory showed that NR raises NAD+ levels substantially in yeast [116]. Our laboratory then explored the ability of NR to increase NAD+ in mammalian cell lines and determined that cellular NAD+ levels increased as much as 270% above controls in several different cell lines [110]. This potency of NR to increase cellular NAD+ levels implied a novel mechanism of metabolism for this compound, possibly through action of the newly characterized Nrk enzymes. Our laboratory similarly showed that the NR relative, NaR was also able to increase NAD+ level in mammalian cells by as much as 1.9 fold [110], possibly through direct synthesis of NaMN.
11. NAD+ increases as a novel modality to treat diseases
The hypothesis that augmentation of NAD+ level could stimulate adaptive changes in cellular bioenergetic and survival adaptation has been experimentally examined. This hypothesis is anchored by biochemical demonstrations that human NAD+ consuming enzymes, such as sirtuins (and other mammalian sirtuins), have kinetic parameters (Km) which make them intrinsically sensitive to changes in NAD+ concentrations in the physiologic range found in cells. Key additional support for this idea is that NAD+ is an intrinsic regulator of cell bioenergetics as revealed by studies showing that the Nampt level is upregulated by dietary stress or by exercise. This suggests that some of the health beneficial effects of diet and exercise could derive in part from upregulated NAD+ production [69, 117, 118]. If so, this raises additional interest in the therapeutic prospects of raising NAD+ levels as a possible intervention to effect beneficial changes in human physiology.
Metabolic Syndrome
The NAD+ enhancing effects of the compound NR were explored for potential therapeutic effects in a mouse model of metabolic syndrome. The Auwerx and Sauve laboratories found that NR enhanced NAD+ contents in several mammalian tissues, and induced mitochondrial biogenesis as determined by increased cristae content and increased expression of mitochondrial proteins, such as Complex V [119]. This data provided a pharmacologic mirroring of effects found in at least two genetic models where NAD+ levels were increased, where animals were protected from weight gain caused by high fat diets. For example, PARP1−/− animals displayed overexpression of mitochondrial proteins in skeletal muscle [67]. CD38−/− animals were protected from weight gain also showed impressive mitochondrial biogenesis in skeletal muscle [66].
NAD+ Signaling by activation of SIRT1 and PGC1α to promote mitochondrial biogenesis
The mechanisms by which NAD+ increases can lead to mitochondrial biogenesis are still being examined, but one fundamental mechanism of action is through activation of SIRT1, and stimulated activity of the co-transcriptional activator PGC1α [20, 120]. SIRT1 deacetylation of PGC1α leads to activation and possibly stabilization of this protein, whereby it can coordinate with nuclear transcription factors that control mitochondrial biogenesis genes [121]. Thus, increases in NAD+ levels caused by genetic modifications (CD38−/− or PARP1−/−) or pharmacologic interventions such as NR administration lead to increased PGC1α deacetylation, increased transcription of genes in the mitochondrial biogenesis pathway, and increased oxidative activity as determined by assays of mitochondrial activity [119]. Similar consequences are associated with activation of SIRT1 and PGC1α signaling applying the putative SIRT1 activator resveratrol [122, 123]. Resveratrol has since been appreciated to have complex effects, including activation of AMPK [124] as well as potentiation of cAMP signaling [125]. Since these additional pathways are known to stimulate mitochondrial biogenesis it remains to be determined if and how these pathways contribute to the observed effects of NAD+ enhancement on mitochondrial biogenesis.
Mitochondrial Disorders
Proof of concept studies establishing that increased NAD+ level stimulates mitochondrial biogenesis, place NAD+ squarely in the center of key signaling pathways with major impact for bioenergetic and survival physiology. Translationally directed followups to these provocative studies addressed the possibility that enhanced NAD+ production could provide a stimulative and ameliorative benefit in mitochondrial disorders. Three independent studies in animal models of mitochondrial disease largely supported the initial findings and showed that increased NAD+ production achieved by one of the following: 1) by administration of NAD+ precursors (NR), 2) by PARP inhibition or 3) by PARP genetic knockout could improve mitochondrial function [67], improve exercise intolerance [68], and could improve mitochondrial protein levels [68, 126, 127]. In addition, sirtuins have been shown to be protect mitochondria from stress. SIRT3, known to be influenced by NAD+ level, can reduce oxidative stresses through SOD activation [128, 129] and increased SIRT7 activity can alleviate mitochondrial protein folding stress [130]. It can be proposed that combinations of NAD+ concentration increases and SIRT7 induction can suppress mitochondrial stress and promote mitochondrial integrity. Collectively, these results have stimulated interest in the potential benefits of enhancing tissue NAD+ as a means to treat mitochondrial diseases [131].
DNA Repair Syndromes
Cockayne’s Syndrome is an accelerated aging disease involving mutations in either Cockayne syndrome group A (CSA) or CSB proteins, involved in DNA repair, leading to progressive neurodegeneration [132, 133]. Bohr and coworkers verified that one feature of deficiency in CSB mutant animals is activation of PARP1, and increased PAR levels in CSBm/m cells [134]. Accompanying this increase in NAD+ turnover is a severe metabolic disruption including defects in weight gain due to a hypermetabolic phenotype and increased levels of lactate in brain tissues, such as the cerebellum [134]. Application of PARP inhibitors as a means to reverse this metabolic effect proved successful, in that the inhibitor PJ34 could increase oxygen consumption rate/extracellular acidification (OCR/ECAR) ratio, a measure of improvement in the normalization of catabolism [134]. Administration of NR for one week as an NAD+ repletion agent enabled improvements in NAD+ level in cerebellum of WT and CSBm/m animals. Moreover, ATP homeostasis was also substantially improved in this tissue. NR treated CSBm/m mice had cerebellum mitochondria that had corrected defects in membrane potential and ROS production [134].
Alzheimer’s Disease
The ability of NR to penetrate into brain was recently verified, where it was then shown to provide improvements in Alzheimer’s Disease (AD) neuropathology in the Tg2576 mouse model of this disease [135]. Previous work had shown that the effects of calorie-restriction (CR) in animal models of AD provided reduced neuropathology, as determined by plaque burden, and improvements in behavioral scores measuring memory and cognition [135]. This was shown to be accompanied by increased NAD+ levels and increased NAD+/nicotinamide ratios in CR treated animals as compared to ad libitum fed Tg2576 transgenic mice [70]. These results suggested the possibility that increased NAD+ levels could provide a component in the protective mechanisms that are produced by CR. Administration of NR was shown to increase NAD+ levels in brain and caused reduced production of Aβ1-42. NR was shown to promote PGC1α levels in transgenic animals. In parallel, the authors showed that PGC1α−/− animals exhibited markedly worse neuropathology in the Tg2576 background [135]. Taken together, NR promotes NAD+ levels, increases PGC1α and improves behavioral and molecular markers indicative of resistance to AD progression.
Fatty Liver Disease
Recent investigations into the ability of NAD+ to potentiate oxidative metabolism and to improve mitochondrial function and density led several collaborating groups, including ourselves, to investigate the effects of NR administration in models of liver disease, such as fatty liver disease [136]. These studies provided evidence that NAD+ elevation is protective of this disease in at least two disease models, such as high fat combined with high sucrose (HFHS), as well as in apolipoprotein E (ApoE)−/− animals fed a high fat high cholesterol (HFHC) diet. NR administered at 500 mg/kg protected from fatty liver induced by HFHS as determined by Oil-Red O staining, as well as fibrosis and lipogenesis markers [136]. In an anti-inflammatory effect, plasma TNFα was reduced to control levels in NR fed animals but was elevated 2 fold in HFHC fed animals. As hypothesized, NR administration led to increased oxidative metabolism as measured in isolated liver tissue; for example O2 consumption was elevated, as was citrate synthase activity. Activation of the mitochondrial unfolded protein response was also observed, suggesting that activation of mitochondrial protein stress may be a key to the benefits observed with NR, and therefore NAD+ increase [136]. Similar findings were obtained in ApoE−/− animals challenged with HFHC [136]. At this stage, more mechanistic studies are needed to understand how NR-induced NAD+ increases exert these protective effects. For one possible link to nuclear signaling, overexpression of SIRT7 can prevent the spontaneous development of fatty liver disease [137, 138] and SIRT7 suppresses mitochondrial protein folding stress by repressing NRF1 activity [130]. The full set of molecular players that participate to produce the protective effects of NR in prevention of fatty liver disease may belong to several signaling pathways.
Hepatic Carcinoma
An illuminating study in the area of cancer biology in liver examined effects of NAD+ and unconventional prefoldin RPB5 interactor (URI) driven dysregulation of hepatocellular mTOR/S6K1 signaling pathways on development of hepatocellular carcinoma (HCC). Using overexpression of an oncogenic protein called URI in liver, Tummala et al.[139] found that livers became fibrotic and developed progressive hepatocellular dysplasia, resembling transformative cell phenotypes common to early stage human hepatocarcinoma. Diethylnitrosoamine induced accelerated hepatocellular carcinoma in heterozygous URI+/− and homozygous URI+/+ transgenic animals, with increased gene dosage of URI providing shortened induction to HCC in liver. Interestingly, NAD+ levels in URI+/+ homozygous livers were found to be substantially lower (35–50% of control) at 3 weeks of life. To investigate if NAD+ repletion could mitigate the adverse effects of URI overexpression on liver phenotype, the authors administered NR via diet, and determined that this increased NAD+ levels back to near normal levels, and completely prevented the development of hepatocellular dysplasia observed in untreated controls. Treatment of 12 week old mice with fully developed hepatocellular carcinoma with NR for 48 weeks produced regression of HCC tumors [139].
Inflammatory Conditions
The NLRP3 inflammasome complex, a component of innate immune surveillance, is known to be activated in a number of disease associated conditions, and is known to be overactivated in the physiology of obesity and is involved in disease states such as insulin resistance [140]. Overactivation of innate immunity can be triggered by pathogen-associated molecular patterns or damage associated molecular patterns (DAMPs), such as those derived from mitochondrial dysfunction [141]. DAMPs can stimulate assembly of the inflammasome, and lead to production of cytokines such as pro IL-1β or pro IL-18, which can amplify inflammation [140]. Sack and coworkers [142] determined that nutritional intakes increase activation of the inflammasome, whereas fasting attenuates these activations. They found that NR treatment of human derived macrophages had attenuated inflammasome activation in a SIRT3 dependent manner, suggesting that NAD+ signaling can be a potential means to inhibit excessive inflammation triggered by nutritional inputs [142].
Cardiomyopathy
Andrews and coworkers recently demonstrated that deletion of transferrin receptor 1 (Tfr1) leads to a profound defect in iron loading in the heart, causing early lethality in Tfr1−/− mice due to the iron deficit [143]. This impairment in iron loading causes mitochondrial defects, and leads to cardiodilation and early death at post-natal day 10. Remarkably, administration at birth of NR causes up to 50% improvements in survival of mice (day 15) [143]. Although mechanistically the effect of NR was not fully elucidated, the presumptive effect may be through improvement in mitochondrial function and maintenance, as this effect of NR has been noted in other situations as discussed in this review. Notably, NR decreased accumulation of p62, suggesting improvements in mitochondrial mitophagy [143]. The effect of improving NAD+ status on improving survival outcomes in a model of heart disease is notable, given the broad impact of this disease in human populations, and its substantial mortality.
Noise Induced Hearing Loss
Hearing loss affects 100s of millions of people worldwide, from a variety of causes. Nevertheless, new treatments to prevent hearing loss are largely unavailable. In a recent study Brown and co-workers [144] showed that high intensity wide spectrum sound could cause loss of hearing in a mouse model of hearing loss. Overexpression of the mitochondrial sirtuin SIRT3 or the WLDs mouse which encodes a triplicate repeat of the NAD+ biosynthetic gene NMNAT1, could largely prevent the loss of hearing. To investigate if increased NAD+ content could also prevent the noise induced hearing loss, these investigators administered NR before, before and after, or only after noise exposure and showed that all three treatments fully protected hearing in all frequencies [144]. This finding strongly hinted that approaches which can augment NAD+ levels in the neurons and tissues of the ear could provide protection from trauma induced hearing loss and could also be meaningful in progressive hearing loss syndromes. These ideas will require additional study to determine if this approach can be broadly applied in hearing loss treatment.
Aging and the Diseases of Aging
Although there is not a firm understanding of all the factors that cause biological aging to occur, there are a number of factors that appear to be common to aging, particularly in mammals. Among these are loss of regenerative potential, defects in DNA repair and mitochondrial decline. Although there is not a clear consensus on the pathways and causes that lead to these effects, the findings that enhancements in cellular NAD+ can improve outcomes in DNA repair syndromes (i.e. Cockaynes Syndrome) and in mouse models featuring mitochondrial deficiency (mito-disorders and Tfr1−/−) suggests that NAD+ stimulated pathways as modify aging phenotypes toward more youthful conditions (See current review on this by Verdin [145]). Mechanistically NAD can elevate the activity level of sirtuins, such as SIRT3 and SIRT7, as SIRT3 and SIRT7 have been shown to be depleted in aged stem cells and reintroduction of SIRT3 or SIRT7 reverses stem cell aging by reducing mitochondrial stresses [130, 146].
This idea that NAD+ could be central to aging has been directly evaluated in a fascinating and timely study authored by Sinclair and co-workers [37]. Leaping off from the idea that mitochondrial decline and increased ROS could be a central cause of aging, as advocated by Harman [147] these workers proposed that the decline is linked to imbalances in mitochondrial proteinse encoded by nuclear and mitochondrial genomes. Imbalanced mitochondrial and nuclear gene transcription for mitochondrial proteins impairs mitochondrial activity in aged animals. Mitochondrial encoded proteins, in particular, were found to be adversely affected by age. A key cellular factor in maintaining a correct balance is the sirtuin SIRT1, as well as the SIRT1 substrate NAD+. Intriguingly, NAD+ homeostasis was found to be significantly impaired in older animals (22 months), causing NAD+ levels to drop to 40% of the levels observed in young animals (6 months)[37]. This suggested that replenishing NAD+ might rebalance mitochondrial-derived and nuclear-derived mitochondrial protein production. Thus, aged mice were fed the compound NMN for one week, and this treatment was able to restore NAD+ levels in 22 month animals to amounts at observed at 6 months. Several key mitochondrial fitness parameters improved, including increased mitochondrial-encoded transcripts and increased ATP levels. This reversal in key molecular phenotypes, by a relatively straightforward NAD+ enhancement strategy provided new insights into the role NAD+ might play in human aging, and in mitochondrial decline, and suggested new ways to intervene to mitigate these effects.
12. Conclusions
The centrality of energy metabolism in organisms, and the integration of key metabolic components into signaling pathways that modulate organism health and physiology, make it clear that some of the more abundant and central factors, such as NAD+ can have unexpected roles in maintaining healthy physiology and could be important in the development of pathology. As this review attempts to illuminate, current knowledge of NAD+ metabolic pathways and knowledge of the ways in which NAD+ regulates key processes in cells and tissues is undergoing a current reblossoming of interest. This has been bolstered by identification and investigation of “newer” Vitamin B3 forms, such as NMN and NR, which provide new opportunities to pharmacologically modulate NAD+ metabolism and to possibly alleviate disease conditions. These newer NAD+ precursors have shown impressive effects in a number of proof of concept studies that favorably mitigate, prevent or cure animal models of disease including AD, cardiovascular disease, metabolic syndromes, mitochondrial disorders, cancer and even aging. These developments set the stage for deeper investigative understanding into the mechanisms by which some diseases are sensitive to NAD+ status, and may pinpoint details of how deterioration of NAD+ homeostasis increases the susceptibility to human disease conditions.
Acknowledgments
Acknowledgements and Conflicts of Interest
AAS acknowledges support from the NIH for this work. AAS also acknowledges he owns intellectual property related to nicotinamide riboside and novel derivatives thereof. AAS and Cornell University receive royalties on commercial sales of NR from Chromadex Inc. AAS is also a consultant and co-founder of Metro MidAtlantic Biotech LLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rajakumar K. Pellagra in the United States: A Historical Perspective. Southern Medical Journal. 2000;93 [PubMed] [Google Scholar]
- 2.Hegyi J, Schwartz RA, Hegyi V. Pellagra: dermatitis, dementia, and diarrhea. International journal of dermatology. 2004;43:1–5. doi: 10.1111/j.1365-4632.2004.01959.x. [DOI] [PubMed] [Google Scholar]
- 3.Sydenstricker VP. The history of pellagra, its recognition as a disorder of nutrition and its conquest. The American journal of clinical nutrition. 1958;6:409–414. doi: 10.1093/ajcn/6.4.409. [DOI] [PubMed] [Google Scholar]
- 4.Morabia A. Joseph Goldberger’s research on the prevention of pellagra. Journal of the Royal Society of Medicine. 2008;101:566–568. doi: 10.1258/jrsm.2008.08k010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberger J. The Etiology of Pellagra: The Significance of Certain Epidemiological Observations with Respect Thereto. Public Health Reports (1896–1970) 1914;29:1683–1686. [Google Scholar]
- 6.Goldberger J, Tanner WF. Amino-Acid Deficiency Probably the Primary Etiological Factor in Pellagra. Public Health Reports (1896–1970) 1922;37:462–486. doi: 10.1111/j.1753-4887.1987.tb06349.x. [DOI] [PubMed] [Google Scholar]
- 7.Goldberger J, Wheeler GA, Lillie RD, Rogers LM. A Further Study of Butter, Fresh Beef, and Yeast as Pellagra Preventives, with Consideration of the Relation of Factor P-P of Pellagra (And Black Tongue of Dogs) to Vitamin B. Public Health Reports (1896–1970) 1926;41:297–318. [Google Scholar]
- 8.Goldberger JTW. A Study of the Pellagra-preventive Action of Dried Beans, Casein, Dried Milk, and Brewers’ Yeast, with a Consideration of the Essential Preventive Factors Involved. The American Journal of the Medical Sciences. 1925;169:623. [Google Scholar]
- 9.Elvehjem CA, Madden RJ, Strong FM, Woolley DW. RELATION OF NICOTINIC ACID AND NICOTINIC ACID AMIDE TO CANINE BLACK TONGUE. Journal of the American Chemical Society. 1937;59:1767–1768. [Google Scholar]
- 10.Bollet AJ. Politics and pellagra: the epidemic of pellagra in the U.S. in the early twentieth century. The Yale journal of biology and medicine. 1992;65:211–221. [PMC free article] [PubMed] [Google Scholar]
- 11.Harden A, Young WJ. The Alcoholic Ferment of Yeast-Juice. Part II.--The Conferment of Yeast-Juice. Proceedings of the Royal Society of London B: Biological Sciences. 1906;78:369–375. [Google Scholar]
- 12.von Euler-Chelpin H. Nobel Lecture. The Nobel Foundation; 1929. Fermentation of Sugars and Fermentative Enzymes. [Google Scholar]
- 13.Warburg O, Chrisitan W, Griese A. Hydrogen-trnasferring co-enzyme, its composition and mode of functioning. Biochem Z. 1935;280 [Google Scholar]
- 14.Warburg O, Christian W. Pyridine, the hydrogen transfusing component of fermentative enzymes. Helv Chim Acta. 1936;19:79–88. [Google Scholar]
- 15.Kornberg A. The participation of inorganic pyrophosphate in the reversible enzymatic synthesis of diphosphopyridine nucleotide. The Journal of biological chemistry. 1948;176:1475. [PubMed] [Google Scholar]
- 16.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. The Journal of biological chemistry. 1958;233:488–492. [PubMed] [Google Scholar]
- 17.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. The Journal of biological chemistry. 1958;233:493–500. [PubMed] [Google Scholar]
- 18.Krebs HA, Veech RL. Equilibrium relations between pyridine nucleotides and adenine nucleotides and their roles in the regulation of metabolic processes. Advances in enzyme regulation. 1969;7:397–413. doi: 10.1016/0065-2571(69)90030-2. [DOI] [PubMed] [Google Scholar]
- 19.Sauve AA. NAD+ and vitamin B3: from metabolism to therapies. The Journal of pharmacology and experimental therapeutics. 2008;324:883–893. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- 20.Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell metabolism. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu P, Sauve AA. Vitamin B3, the nicotinamide adenine dinucleotides and aging. Mechanisms of ageing and development. 2010;131:287–298. doi: 10.1016/j.mad.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Berg TJJM, Stryer L. Biochemistry. Section 16.1. W H Freeman; New York: 2001. Glycolysis Is an Energy-Conversion Pathway in Many Organisms. [Google Scholar]
- 23.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. The Journal of clinical investigation. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akram M. Citric acid cycle and role of its intermediates in metabolism. Cell biochemistry and biophysics. 2014;68:475–478. doi: 10.1007/s12013-013-9750-1. [DOI] [PubMed] [Google Scholar]
- 25.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tretter L, Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360:2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLain AL, Szweda PA, Szweda LI. alpha-Ketoglutarate dehydrogenase: a mitochondrial redox sensor. Free radical research. 2011;45:29–36. doi: 10.3109/10715762.2010.534163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sazanov LA. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nature reviews. Molecular cell biology. 2015;16:375–388. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 29.Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. The Biochemical journal. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veech RL, Guynn R, Veloso D. The time-course of the effects of ethanol on the redox and phosphorylation states of rat liver. The Biochemical journal. 1972;127:387–397. doi: 10.1042/bj1270387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying W, Alano CC, Garnier P, Swanson RA. NAD+ as a metabolic link between DNA damage and cell death. Journal of neuroscience research. 2005;79:216–223. doi: 10.1002/jnr.20289. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A. Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. The Journal of biological chemistry. 2010;285:34106–34114. doi: 10.1074/jbc.M110.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. The Journal of biological chemistry. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochimica et biophysica acta. 2010;1804:1666–1675. doi: 10.1016/j.bbapap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacological reviews. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxidants & redox signaling. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 39.Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides--small molecules with a multitude of functions. The Biochemical journal. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. European journal of biochemistry/FEBS. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 41.Cejudo FJ, Ferrandez J, Cano B, Puerto-Galan L, Guinea M. The function of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system in plastid redox regulation and signalling. FEBS letters. 2012;586:2974–2980. doi: 10.1016/j.febslet.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Tribble DL, Jones DP. Oxygen dependence of oxidative stress. Rate of NADPH supply for maintaining the GSH pool during hypoxia. Biochemical pharmacology. 1990;39:729–736. doi: 10.1016/0006-2952(90)90152-b. [DOI] [PubMed] [Google Scholar]
- 43.Agledal L, Niere M, Ziegler M. The phosphate makes a difference: cellular functions of NADP. Redox report: communications in free radical research. 2010;15:2–10. doi: 10.1179/174329210X12650506623122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandey AV, Fluck CE. NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacology & therapeutics. 2013;138:229–254. doi: 10.1016/j.pharmthera.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Segal AW. The function of the NADPH oxidase of phagocytes and its relationship to other NOXs in plants, invertebrates, and mammals. The International Journal of Biochemistry & Cell Biology. 2008;40:604–618. doi: 10.1016/j.biocel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kruger NJ, von Schaewen A. The oxidative pentose phosphate pathway: structure and organisation. Current Opinion in Plant Biology. 2003;6:236–246. doi: 10.1016/s1369-5266(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 47.Guse AH, Lee HC. NAADP: a universal Ca2+ trigger. Science signaling. 2008;1:re10. doi: 10.1126/scisignal.144re10. [DOI] [PubMed] [Google Scholar]
- 48.Pollak N, Niere M, Ziegler M. NAD kinase levels control the NADPH concentration in human cells. The Journal of biological chemistry. 2007;282:33562–33571. doi: 10.1074/jbc.M704442200. [DOI] [PubMed] [Google Scholar]
- 49.Williams MB, Jones HP. Calmodulin-dependent NAD kinase of human neutrophils. Archives of biochemistry and biophysics. 1985;237:80–87. doi: 10.1016/0003-9861(85)90256-5. [DOI] [PubMed] [Google Scholar]
- 50.Apps DK. Kinetic studies of pigeon liver NAD kinase. European journal of biochemistry/FEBS. 1968;5:444–450. doi: 10.1111/j.1432-1033.1968.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang R. MNADK, a novel liver-enriched mitochondrion-localized NAD kinase. Biology open. 2013;2:432–438. doi: 10.1242/bio.20134259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang R. MNADK, a Long-Awaited Human Mitochondrion-Localized NAD Kinase. Journal of cellular physiology. 2015;230:1697–1701. doi: 10.1002/jcp.24926. [DOI] [PubMed] [Google Scholar]
- 53.Ohashi K, Kawai S, Murata K. Identification and characterization of a human mitochondrial NAD kinase. Nature communications. 2012;3:1248. doi: 10.1038/ncomms2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochemical and biophysical research communications. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 56.Chambon P, Weill JD, Doly J, Strosser MT, Mandel P. On Formation of a Novel Adenylic Compound by Enzymatic Extracts of Liver Nuclei. Biochemical and biophysical research communications. 1966;25:638-&. [Google Scholar]
- 57.Collier RJ, Pappenheimer AM., Jr Studies on the Mode of Action of Diphtheria Toxin. Ii. Effect of Toxin on Amino Acid Incorporation in Cell-Free Systems. The Journal of experimental medicine. 1964;120:1019–1039. doi: 10.1084/jem.120.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Honjo T, Nishizuka Y, Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. The Journal of biological chemistry. 1968;243:3553–3555. [PubMed] [Google Scholar]
- 59.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiology and molecular biology reviews: MMBR. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5 doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annual review of biochemistry. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 63.Yang T, Sauve AA. NAD metabolism and sirtuins: Metabolic regulation of protein deacetylation in stress and toxicity. The AAPS Journal. 2006;8:E632–E643. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sauve AA. Sirtuin chemical mechanisms. Biochimica et biophysica acta. 2010;1804:1591–1603. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cen Y, Youn DY, Sauve AA. Advances in characterization of human sirtuin isoforms: chemistries, targets and therapeutic applications. Current medicinal chemistry. 2011;18:1919–1935. doi: 10.2174/092986711795590084. [DOI] [PubMed] [Google Scholar]
- 66.Barbosa MT, Soares SM, Novak CM, Sinclair D, Levine JA, Aksoy P, Chini EN. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:3629–3639. doi: 10.1096/fj.07-8290com. [DOI] [PubMed] [Google Scholar]
- 67.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell metabolism. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cerutti R, Pirinen E, Lamperti C, Marchet S, Sauve AA, Li W, Leoni V, Schon EA, Dantzer F, Auwerx J, Viscomi C, Zeviani M. NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell metabolism. 2014;19:1042–1049. doi: 10.1016/j.cmet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koltai E, Szabo Z, Atalay M, Boldogh I, Naito H, Goto S, Nyakas C, Radak Z. Exercise alters SIRT1, SIRT6, NAD and NAMPT levels in skeletal muscle of aged rats. Mechanisms of ageing and development. 2010;131:21–28. doi: 10.1016/j.mad.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. The Journal of biological chemistry. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 71.Canto C, Sauve AA, Bai P. Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Molecular aspects of medicine. 2013;34:1168–1201. doi: 10.1016/j.mam.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ijichi H, Ichiyama A, Hayaishi O. Studies on the biosynthesis of nicotinamide adenine dinucleotide. 3. Comparative in vivo studies on nicotinic acid, nicotinamide, and quinolinic acid as precursors of nicotinamide adenine dinucleotide. The Journal of biological chemistry. 1966;241:3701–3707. [PubMed] [Google Scholar]
- 73.Jackson TM, Rawling JM, Roebuck BD, Kirkland JB. Large supplements of nicotinic acid and nicotinamide increase tissue NAD+ and poly(ADP-ribose) levels but do not affect diethylnitrosamine-induced altered hepatic foci in Fischer-344 rats. The Journal of nutrition. 1995;125:1455–1461. doi: 10.1093/jn/125.6.1455. [DOI] [PubMed] [Google Scholar]
- 74.Terakata M, Fukuwatari T, Kadota E, Sano M, Kanai M, Nakamura T, Funakoshi H, Shibata K. The niacin required for optimum growth can be synthesized from L-tryptophan in growing mice lacking tryptophan-2,3-dioxygenase. The Journal of nutrition. 2013;143:1046–1051. doi: 10.3945/jn.113.176875. [DOI] [PubMed] [Google Scholar]
- 75.Moffett JR, Namboodiri MA. Tryptophan and the immune response. Immunology and cell biology. 2003;81:247–265. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x. [DOI] [PubMed] [Google Scholar]
- 76.Gal EM, Sherman AD. L-kynurenine: its synthesis and possible regulatory function in brain. Neurochemical research. 1980;5:223–239. doi: 10.1007/BF00964611. [DOI] [PubMed] [Google Scholar]
- 77.Leklem JE. Quantitative aspects of tryptophan metabolism in humans and other species: a review. The American journal of clinical nutrition. 1971;24:659–672. doi: 10.1093/ajcn/24.6.659. [DOI] [PubMed] [Google Scholar]
- 78.Oxenkrug GF. Tryptophan kynurenine metabolism as a common mediator of genetic and environmental impacts in major depressive disorder: the serotonin hypothesis revisited 40 years later. The Israel journal of psychiatry and related sciences. 2010;47:56–63. [PMC free article] [PubMed] [Google Scholar]
- 79.O.B.V. Institute of Medicine (US) Standing Committee on the Scientific Evaluation of Dietary Reference Intakes and its Panel on Folate, and Choline. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline. National Academies Press (US); Washington (DC): 1998. [PubMed] [Google Scholar]
- 80.Salter M, Beams RM, Critchley MA, Hodson HF, Iyer R, Knowles RG, Madge DJ, Pogson CI. Effects of tryptophan 2,3-dioxygenase inhibitors in the rat. Advances in experimental medicine and biology. 1991;294:281–288. doi: 10.1007/978-1-4684-5952-4_25. [DOI] [PubMed] [Google Scholar]
- 81.Salter M, Pogson CI. The role of tryptophan 2,3-dioxygenase in the hormonal control of tryptophan metabolism in isolated rat liver cells. Effects of glucocorticoids and experimental diabetes. The Biochemical journal. 1985;229:499–504. doi: 10.1042/bj2290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rubin RT. Adrenal cortical activity changes in manic-depressive illness. Influence on intermediary metabolism of tryptophan. Archives of general psychiatry. 1967;17:671–679. doi: 10.1001/archpsyc.1967.01730300031006. [DOI] [PubMed] [Google Scholar]
- 83.Guillemin GJ, Smythe G, Takikawa O, Brew BJ. Expression of indoleamine 2,3-dioxygenase and production of quinolinic acid by human microglia, astrocytes, and neurons. Glia. 2005;49:15–23. doi: 10.1002/glia.20090. [DOI] [PubMed] [Google Scholar]
- 84.Guillemin GJ, Smith DG, Smythe GA, Armati PJ, Brew BJ. Expression of the kynurenine pathway enzymes in human microglia and macrophages. Advances in experimental medicine and biology. 2003;527:105–112. doi: 10.1007/978-1-4615-0135-0_12. [DOI] [PubMed] [Google Scholar]
- 85.Fujigaki S, Saito K, Sekikawa K, Tone S, Takikawa O, Fujii H, Wada H, Noma A, Seishima M. Lipopolysaccharide induction of indoleamine 2,3-dioxygenase is mediated dominantly by an IFN-gamma-independent mechanism. European journal of immunology. 2001;31:2313–2318. doi: 10.1002/1521-4141(200108)31:8<2313::aid-immu2313>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 86.Chen Y, Guillemin GJ. Kynurenine pathway metabolites in humans: disease and healthy States. International journal of tryptophan research: IJTR. 2009;2:1–19. doi: 10.4137/ijtr.s2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vecsei L, Szalardy L, Fulop F, Toldi J. Kynurenines in the CNS: recent advances and new questions. Nature reviews. Drug discovery. 2013;12:64–82. doi: 10.1038/nrd3793. [DOI] [PubMed] [Google Scholar]
- 88.Okuno E, Schwarcz R. Purification of quinolinic acid phosphoribosyltransferase from rat liver and brain. Biochimica et biophysica acta. 1985;841:112–119. doi: 10.1016/0304-4165(85)90280-6. [DOI] [PubMed] [Google Scholar]
- 89.Ikeda M, Tsuji H, Nakamura S, Ichiyama A, Nishizuka Y, Hayaishi O. Studies on the Biosynthesis of Nicotinamide Adenine Dinucleotide. Ii. A Role of Picolinic Carboxylase in the Biosynthesis of Nicotinamide Adenine Dinucleotide from Tryptophan in Mammals. The Journal of biological chemistry. 1965;240:1395–1401. [PubMed] [Google Scholar]
- 90.Moehn S, Pencharz PB, Ball RO. Lessons learned regarding symptoms of tryptophan deficiency and excess from animal requirement studies. The Journal of nutrition. 2012;142:2231s–2235s. doi: 10.3945/jn.112.159061. [DOI] [PubMed] [Google Scholar]
- 91.Fukuwatari T, Shibata K. Effect of nicotinamide administration on the tryptophan-nicotinamide pathway in humans. International journal for vitamin and nutrition research. Internationale Zeitschrift fur Vitamin- und Ernahrungsforschung. Journal international de vitaminologie et de nutrition. 2007;77:255–262. doi: 10.1024/0300-9831.77.4.255. [DOI] [PubMed] [Google Scholar]
- 92.Kimura T, Bier DM, Taylor CL. Summary of workshop discussions on establishing upper limits for amino acids with specific attention to available data for the essential amino acids leucine and tryptophan. The Journal of nutrition. 2012;142:2245s–2248s. doi: 10.3945/jn.112.160846. [DOI] [PubMed] [Google Scholar]
- 93.Schwarcz R, Okuno E, White RJ, Bird ED, Whetsell WO., Jr 3-Hydroxyanthranilate oxygenase activity is increased in the brains of Huntington disease victims. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:4079–4081. doi: 10.1073/pnas.85.11.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwarcz R, Speciale C, Okuno E, French ED, Kohler C. Quinolinic acid: a pathogen in seizure disorders? Advances in experimental medicine and biology. 1986;203:697–707. doi: 10.1007/978-1-4684-7971-3_53. [DOI] [PubMed] [Google Scholar]
- 95.Lapin IP, Mutovkina LG, Ryzov IV, Mirzaev S. Anxiogenic activity of quinolinic acid and kynurenine in the social interaction test in mice. Journal of psychopharmacology (Oxford, England) 1996;10:246–249. doi: 10.1177/026988119601000312. [DOI] [PubMed] [Google Scholar]
- 96.Lapin IP. Neurokynurenines (NEKY) as common neurochemical links of stress and anxiety. Advances in experimental medicine and biology. 2003;527:121–125. doi: 10.1007/978-1-4615-0135-0_14. [DOI] [PubMed] [Google Scholar]
- 97.French JB, Cen Y, Vrablik TL, Xu P, Allen E, Hanna-Rose W, Sauve AA. Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism. Biochemistry. 2010;49:10421–10439. doi: 10.1021/bi1012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Preiss J, Handler P. Enzymatic synthesis of nicotinamide mononucleotide. The Journal of biological chemistry. 1957;225:759–770. [PubMed] [Google Scholar]
- 99.Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2876–2881. doi: 10.1073/pnas.1417921112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burgos ES, Schramm VL. Weak coupling of ATP hydrolysis to the chemical equilibrium of human nicotinamide phosphoribosyltransferase. Biochemistry. 2008;47:11086–11096. doi: 10.1021/bi801198m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Burgos ES, Vetticatt MJ, Schramm VL. Recycling nicotinamide. The transition-state structure of human nicotinamide phosphoribosyltransferase. Journal of the American Chemical Society. 2013;135:3485–3493. doi: 10.1021/ja310180c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer research. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 103.Montecucco F, Bauer I, Braunersreuther V, Bruzzone S, Akhmedov A, Luscher TF, Speer T, Poggi A, Mannino E, Pelli G, Galan K, Bertolotto M, Lenglet S, Garuti A, Montessuit C, Lerch R, Pellieux C, Vuilleumier N, Dallegri F, Mage J, Sebastian C, Mostoslavsky R, Gayet-Ageron A, Patrone F, Mach F, Nencioni A. Inhibition of nicotinamide phosphoribosyltransferase reduces neutrophil-mediated injury in myocardial infarction. Antioxidants & redox signaling. 2013;18:630–641. doi: 10.1089/ars.2011.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brandauer J, Vienberg SG, Andersen MA, Ringholm S, Risis S, Larsen PS, Kristensen JM, Frosig C, Leick L, Fentz J, Jorgensen S, Kiens B, Wojtaszewski JF, Richter EA, Zierath JR, Goodyear LJ, Pilegaard H, Treebak JT. AMP-activated protein kinase regulates nicotinamide phosphoribosyl transferase expression in skeletal muscle. The Journal of physiology. 2013;591:5207–5220. doi: 10.1113/jphysiol.2013.259515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bender DA. Tryptophan and niacin nutrition--is there a problem? Advances in experimental medicine and biology. 1996;398:565–569. [PubMed] [Google Scholar]
- 106.van Roermund CW, Elgersma Y, Singh N, Wanders RJ, Tabak HF. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. The EMBO journal. 1995;14:3480–3486. doi: 10.1002/j.1460-2075.1995.tb07354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 108.Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedyalkova L, Yang T, Sauve AA, Park HW, Brenner C. Nicotinamide riboside kinase structures reveal new pathways to NAD+ PLoS biology. 2007;5:e263. doi: 10.1371/journal.pbio.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dolle C, Ziegler M. Application of a coupled enzyme assay to characterize nicotinamide riboside kinases. Analytical biochemistry. 2009;385:377–379. doi: 10.1016/j.ab.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 110.Yang T, Chan NY, Sauve AA. Syntheses of nicotinamide riboside and derivatives: effective agents for increasing nicotinamide adenine dinucleotide concentrations in mammalian cells. J Med Chem. 2007;50:6458–6461. doi: 10.1021/jm701001c. [DOI] [PubMed] [Google Scholar]
- 111.Belenky PA, Moga TG, Brenner C. Saccharomyces cerevisiae YOR071C encodes the high affinity nicotinamide riboside transporter Nrt1. The Journal of biological chemistry. 2008;283:8075–8079. doi: 10.1074/jbc.C800021200. [DOI] [PubMed] [Google Scholar]
- 112.Damaraju VL, Visser F, Zhang J, Mowles D, Ng AM, Young JD, Jayaram HN, Cass CE. Role of human nucleoside transporters in the cellular uptake of two inhibitors of IMP dehydrogenase, tiazofurin and benzamide riboside. Molecular pharmacology. 2005;67:273–279. doi: 10.1124/mol.104.004408. [DOI] [PubMed] [Google Scholar]
- 113.Kulikova V, Shabalin K, Nerinovski K, Dolle C, Niere M, Yakimov A, Redpath P, Khodorkovskiy M, Migaud ME, Ziegler M, Nikiforov A. Generation, Release, and Uptake of the NAD Precursor Nicotinic Acid Riboside by Human Cells. The Journal of biological chemistry. 2015;290:27124–27137. doi: 10.1074/jbc.M115.664458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rowen JW, Kornberg A. The phosphorolysis of nicotinamide riboside. The Journal of biological chemistry. 1951;193:497–507. [PubMed] [Google Scholar]
- 115.Boden WE, Sidhu MS, Toth PP. The therapeutic role of niacin in dyslipidemia management. Journal of cardiovascular pharmacology and therapeutics. 2014;19:141–158. doi: 10.1177/1074248413514481. [DOI] [PubMed] [Google Scholar]
- 116.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+ Cell. 2007;129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 117.Song J, Ke SF, Zhou CC, Zhang SL, Guan YF, Xu TY, Sheng CQ, Wang P, Miao CY. Nicotinamide phosphoribosyltransferase is required for the calorie restriction-mediated improvements in oxidative stress, mitochondrial biogenesis, and metabolic adaptation. The journals of gerontology. Series A, Biological sciences and medical sciences. 2014;69:44–57. doi: 10.1093/gerona/glt122. [DOI] [PubMed] [Google Scholar]
- 118.Rappou E, Jukarainen S, Rinnankoski-Tuikka R, Kaye S, Heinonen S, Hakkarainen A, Lundbom J, Lundbom N, Saunavaara V, Rissanen A, Virtanen KA, Pirinen E, Pietilainen KH. Weight loss is associated with increased NAD/SIRT1 expression but reduced PARP activity in white adipose tissue. The Journal of clinical endocrinology and metabolism. 2016:jc20153054. doi: 10.1210/jc.2015-3054. [DOI] [PubMed] [Google Scholar]
- 119.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell metabolism. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Houtkooper RH, Auwerx J. Exploring the therapeutic space around NAD+ The Journal of cell biology. 2012;199:205–209. doi: 10.1083/jcb.201207019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jeninga EH, Schoonjans K, Auwerx J. Reversible acetylation of PGC-1: connecting energy sensors and effectors to guarantee metabolic flexibility. Oncogene. 2010;29:4617–4624. doi: 10.1038/onc.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 123.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, Chung JH. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pirinen E, Canto C, Jo YS, Morato L, Zhang H, Menzies KJ, Williams EG, Mouchiroud L, Moullan N, Hagberg C, Li W, Timmers S, Imhof R, Verbeek J, Pujol A, van Loon B, Viscomi C, Zeviani M, Schrauwen P, Sauve AA, Schoonjans K, Auwerx J. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell metabolism. 2014;19:1034–1041. doi: 10.1016/j.cmet.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Khan NA, Auranen M, Paetau I, Pirinen E, Euro L, Forsstrom S, Pasila L, Velagapudi V, Carroll CJ, Auwerx J, Suomalainen A. Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO molecular medicine. 2014;6:721–731. doi: 10.1002/emmm.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metabolism. 12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 129.Tao R, Coleman MC, Pennington D, Ozden O, Park S-H, Jiang H, Kim H-S, Flynn CR, Hill S, McDonald WH, Olivier AK, Spitz DR, Gius D. Sirt3-Mediated Deacetylation of Evolutionarily Conserved Lysine 122 Regulates MnSOD Activity in Response to Stress. Molecular Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. STEM CELL AGING. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science (New York, NY) 2015;347:1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lightowlers RN, Chrzanowska-Lightowlers ZM. Salvaging hope: Is increasing NAD(+) a key to treating mitochondrial myopathy? EMBO molecular medicine. 2014;6:705–707. doi: 10.15252/emmm.201404179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gitiaux C, Blin-Rochemaure N, Hully M, Echaniz-Laguna A, Calmels N, Bahi-Buisson N, Desguerre I, Dabaj I, Wehbi S, Quijano-Roy S, Laugel V. Progressive demyelinating neuropathy correlates with clinical severity in Cockayne syndrome. Clinical neurophysiology: official journal of the International Federation of Clinical Neurophysiology. 2015;126:1435–1439. doi: 10.1016/j.clinph.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 133.Moriel-Carretero M, Herrera-Moyano E, Aguilera A. A unified model for the molecular basis of Xeroderma pigmentosum-Cockayne Syndrome. Rare diseases. 2015;3:e1079362. doi: 10.1080/21675511.2015.1079362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, Wang J, Dunn CA, Singh N, Veith S, Hasan-Olive MM, Mangerich A, Wilson MA, Mattson MP, Bergersen LH, Cogger VC, Warren A, Le Couteur DG, Moaddel R, Wilson DM, 3rd, Croteau DL, de Cabo R, Bohr VA. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell metabolism. 2014;20:840–855. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gong B, Pan Y, Vempati P, Zhao W, Knable L, Ho L, Wang J, Sastre M, Ono K, Sauve AA, Pasinetti GM. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiology of aging. 2013;34:1581–1588. doi: 10.1016/j.neurobiolaging.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gariani K, Menzies KJ, Ryu D, Wegner CJ, Wang X, Ropelle ER, Moullan N, Zhang H, Perino A, Lemos V, Kim B, Park YK, Piersigilli A, Pham TX, Yang Y, Ku CS, Koo SI, Fomitchova A, Canto C, Schoonjans K, Sauve AA, Lee JY, Auwerx J. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology. 2015 doi: 10.1002/hep.28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, Li P, Cheng H-L, Murphy Andrew J, Valenzuela David M, Luo H, Kapahi P, Krauss R, Mostoslavsky R, Yancopoulos George D, Alt Frederick W, Chua Katrin F, Chen D. SIRT7 Represses Myc Activity to Suppress ER Stress and Prevent Fatty Liver Disease. Cell Reports. 5:1479. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ryu D, Jo Young S, Lo Sasso G, Stein S, Zhang H, Perino A, Lee Jung U, Zeviani M, Romand R, Hottiger Michael O, Schoonjans K, Auwerx J. A SIRT7-Dependent Acetylation Switch of GABPβ1 Controls Mitochondrial Function. Cell Metabolism. 20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 139.Tummala KS, Gomes AL, Yilmaz M, Grana O, Bakiri L, Ruppen I, Ximenez-Embun P, Sheshappanavar V, Rodriguez-Justo M, Pisano DG, Wagner EF, Djouder N. Inhibition of de novo NAD(+) synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer cell. 2014;26:826–839. doi: 10.1016/j.ccell.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 140.Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nature medicine. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Anders HJ, Schaefer L. Beyond tissue injury-damage-associated molecular patterns, toll-like receptors, and inflammasomes also drive regeneration and fibrosis. Journal of the American Society of Nephrology: JASN. 2014;25:1387–1400. doi: 10.1681/ASN.2014010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Traba J, Kwarteng-Siaw M, Okoli TC, Li J, Huffstutler RD, Bray A, Waclawiw MA, Han K, Pelletier M, Sauve AA, Siegel RM, Sack MN. Fasting and refeeding differentially regulate NLRP3 inflammasome activation in human subjects. The Journal of clinical investigation. 2015;125:4592–4600. doi: 10.1172/JCI83260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu W, Barrientos T, Mao L, Rockman HA, Sauve AA, Andrews NC. Lethal Cardiomyopathy in Mice Lacking Transferrin Receptor in the Heart. Cell reports. 2015;13:533–545. doi: 10.1016/j.celrep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brown KD, Maqsood S, Huang JY, Pan Y, Harkcom W, Li W, Sauve A, Verdin E, Jaffrey SR. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell metabolism. 2014;20:1059–1068. doi: 10.1016/j.cmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 146.Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, Zhang D, Scadden DT, Chen D. SIRT3 Reverses Aging-associated Degeneration. Cell reports. 2013;3:319–327. doi: 10.1016/j.celrep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harman D. Origin and evolution of the free radical theory of aging: a brief personal history, 1954–2009. Biogerontology. 2009;10:773–781. doi: 10.1007/s10522-009-9234-2. [DOI] [PubMed] [Google Scholar]