Abstract
Background
To detect the expression of lncRNA HOXA11-AS and its biological effect in breast cancer.
Material/Methods
In this study, fluorescent quantitative real-time PCR (qRT-PCR), MTT assay and clone formation assay, flow cytometry, Transwell assay and wound healing assay, immunofluorescence, and Western blot analysis were conducted to detect the expression of lncRNA HOXA11-AS, cell proliferation activity, cell apoptosis rate and cell cycle distribution, the changes of cell invasion and metastasis capacity, and the expressions of molecular markers of epithelial-mesenchymal transition (EMT), respectively. Additionally, a nude mouse metastatic tumor model was established to study the influence of lncRNA HOXA11-AS on invasion and metastasis capacity of breast cancer cells.
Results
The qRT-PCR experiment results showed that HOXA11-AS expression in breast cancer tissue of 50 patients was relatively higher than that in tissue adjacent to cancer. MTT assay suggested that tumor cell proliferation capacity was suppressed followed by the knockdown of lncRNA HOXA11-AS expression in MDA-MB-231 and MCF-7 cells; flow cytometry results demonstrated that interfering in lncRNA HOXA11-AS could induce tumor cell apoptosis and promote cell cycle progression to be arrested in G1/G0 stage; experiments in vivo/vitro manifested that interfering in lncRNA HOXA11-AS could inhibit tumor cell invasion and migration capacity by affecting the expressions of EMT-related molecular markers (E-cadherin, N-cadherin, Vimentin).
Conclusions
High expression of lncRNA HOXA11-AS promotes breast cancer invasion and metastasis by affecting EMT, and interfering in lncRAN HOXA11-AS expression provides a theoretical basis and important molecular target for inhibiting the distant metastasis of breast cancer in clinical practice.
MeSH Keywords: Breast; Epithelial-Mesenchymal Transition; Lymphatic Metastasis; Neoplasm Invasiveness; RNA, Long Noncoding
Background
Breast cancer is one of the most common malignant tumors in women worldwide [1]. Despite advances in operative treatment, chemotherapy, and molecular targeting treatment, the overall prognosis of breast cancer is not satisfactory. Distant metastasis is one of the main causes of treatment failure [2]. Breast cancer invasion and metastasis is a multi-factor and multi-step biological process, in which the activation or silence of many genes and signal pathways are involved, and its detailed molecular mechanism remains unclear [3]. Therefore, the molecular mechanism of breast cancer invasion and metastasis occurrence needs to be explained in detail, which will provide an important theoretical basis for clinical treatment of late-stage breast cancer.
Long non-coding RNA (lncRNA) is a kind of non-coding RNA that is longer than 200bp in length [4]. According to the site of lncRNA in the genome, it can be classified into 5 types: sense lncRNA, antisense lncRNA, bidirectional lncRNA, intronic lncRNA, and intergenic lncRNA [5]. The main functions of lncRNA include: (1) transcribing and interfering, (2) participating in chromosome rearrangement and histone modification, (3) modifying alternative splicing sequence, and (4) stabilizing mRNA [6]. lncRNA can regulate gene expression at the transcriptional level, post-transcriptional level, and epigenetic level, thus participating in the occurrence and development of tumors [7]. Recent studies have shown that LncRNAs serve as carcinogenic factors or tumor suppressors in breast cancer with abnormal expression [8,9]. However, this research group first found that lncRNA HOXA11-AS expression was relatively high in breast cancer and promoted breast cancer invasion and metastasis.
LncRNA HOXA11-AS is the member of the homeobox gene (HOX) family, located at section 7p15.2 in the chromosome, and its total length is 3885bp [10]. There have been few reports about the biological function and molecular mechanism of lncRNA HOXA11-AS in tumors. High expression of lncRNA HOXA11-AS can bind to epigenetic complexes PRC2 and DNMT1 in stomach tissue and cells, and mediate methylation in the downstream target gene promoter region, and then promote the occurrence and development of stomach cancer [11]. Tong et al. found that the relative expression quantity of lncRNA HOXA11-AS was negatively correlated with tumor node metastasis (TNM) staging, tumor size, and carcinoembryonic antigen (CEA) level in colon cancer [12]. Recent studies have revealed that HOXA11-AS could serve as an important biological marker and therapeutic target for other types of tumors [13–15]. However, there is no report about the role of lncRNA HOXA11-AS in breast cancer.
EMT is a kind of biological phenomenon in which epithelial cells lose epithelial characteristics and then obtains interstitial cell phenotype, which is closely related to embryogenesis and distant metastasis of tumor tissue [16,17]. EMT easily occurs in tumor cells and the tumor cells tends to metastasize; this biological process is mainly manifested as down-regulated expression of E-cadherin, substrate degradation, and up-regulated expressions of cadherin and Vimentin [18,19]. In recent years, many studies have reported that lncRNA is involved in tumor invasion and metastasis by regulating EMT. lncRNA UCA1 can promote tumor cell invasion and metastasis in breast cancer, and its underlying molecular mechanism is by activating the Wnt/β-catenin signal pathway and promoting EMT [20]. Hao et al. [21] found that, as an important regulatory factor, lincRNA 01186 was involved in lung cancer cell invasion and metastasis by regulating EMT. Additionally, our results revealed interference in lncRNA HOXA11-AS could result in changes in the expressions of EMT molecular markers such as E-cadherin, N-cadherin, and Vimentin.
In this study, we performed experiments in vivo/vitro to explore the biological role of lncRNA HOXA11-AS in breast cancer. Our results indicated that lncRNA HOXA11-AS expression was relatively high in breast cancer tissue and cells, and the knockdown of lncRNA HOXA11-AS could promote cell apoptosis and inhibit proliferation, and suppress cell invasion and metastasis by affecting EMT in breast cancer cells.
Material and Methods
Material
Breast cancer tissue specimens
Patients who were not treated by preoperative chemotherapy, radiotherapy and molecular targeting treatment were selected as research objects. Specimens of breast cancer and tissue adjacent to cancer cut off by operation from 68 patients in our hospital were collected. The specimens cut off by operation were quickly placed in liquid nitrogen at −180°C for preservation. At the same time, clinical pathologic data of patient corresponding to each specimen were collected. Tissue specimen collections were made with full informed consent of all patients following institutional ethics guidelines that were reviewed and approved by Tengzhou Central People’s Hospital.
Breast cancer cell strains
Human breast cancer cells (MDA-MB-231, MDA-MB-468, MDA-MB-435, SKBR3 and MCF-7) and normal human mammary epithelial cell (MCF-10A) were purchased from Shanghai Cell Institute of Chinese Academy of Sciences, and all cells were kept well in laboratory and cell passage could be performed steadily. Total RNA extraction kit and transfection reagent Lipofectamine 2000 were purchased from Invitrogen TM Life Technologies, and reverse transcription kit and fluorescence quantification PCR detection kit were purchased from Takara Company.
Primer synthesis of siRNA and qRT-PCR interfering in HOXA11-AS
Effective interference sequence of HOXA11-AS: 1#5′-CTACCATCCCTGAGCCTTA-3′, 2# 5′-TGACATCCGAGGAGA CTTC-3′, 3# 5′-CGTAATCGCCGGTGTAACT-3′ and control sequence si-NC: 5′-CCTATCTGGTCAACACGTATT-3′. Upstream and downstream primer sequences of HOXA11-AS performed by real-time fluorescence quantification PCR as follow: F, 5′-TGCCAAGTTGTACTTACTACGTC-3′, and R, 5′-GTTGGAGGAGTAGGAGT ATGTA-3′, and internal reference β-actin-F: 5′-ATAGCACAGCCTGGATAGC AACGTAC-3, β-Actin-R: 5′-CACCTTCTACAATGAGCTGCGTGT G-3′. The shRNA sequence of HOXA11-AS: CACCAGGCCAAGTCCGAGTTC CATTTCTTCGAAAAGAAATGGAACTCGGACTTGGCC. The above sequences were synthesized by Invitrogen Limited Company.
Detection of HOXA11-AS expression via real-time fluorescence quantification PCR
Total RNA was extracted from breast cancer and corresponding tissue adjacent to cancer by use of Trizol kits. An ultraviolet spectrophotometer was used to detect RNA concentration, and agarose gel electrophoresis was used to measure RNA quality. According to the directions in the PrimeScriptTM RT Master Mix (Perfect Real-Time) kit, cDNA was synthesized and used for subsequent real-time fluorescence quantification PCR. qRT-PCR reaction system (20 μL): 10 μL SYBR qPCR Mix, 0.8 μL (10 μmol/L) upstream primer and 0.8 μL (10 μmol/L) downstream primer, 2 μL cDNA product, 0.4 μL 50×ROX reference dye, and RNase-free water was added and complemented until 20 μL. Reaction condition: After pre-degeneration at 95°C for 1 min, 95°C for 30 s, 60°C for 40 s, and 40 cycles in total. Three parallel duplicate wells were designed in the experiment, and all specimens were repeatedly detected 3 times. Relative quantification method was utilized, and −ΔCt and 2−ΔΔCt were respectively represented as the relative expression quantity of target gene. All specimens were operated on ice.
Cell culture and the transfection of human breast cancer cell strain
RPMI 1640 culture medium containing 10% fetal calf serum was used to culture human breast cancer cells MDA-MB-231, MDA-MB-468, MDA-MB-435, SKBR3, and MCF-7 and normal human mammary epithelial cell MCF-10A, and the solutions were placed into a cell incubator for culture at 37°C. When cell convergence degree reached about 60%, according to lipofection transfection instructions, HOXA11-AS- siRNA and si-NC were respectively transferred into cells by Lipofectamine2000.
Detection of breast cancer cell strain proliferation via MTT assay and clone formation assay
The cell strains were respectively inoculated into 96-well plates at the cell concentration of 3×103 cells/well, and siRNA was used to transfect cells. We added 20 μL MTT solution (1.55 g/L) into each well at 0 h, 24 h, 48 h, 72 h, and 96 h after transfection. After these cells were cultured for 4 hours at 37°C, 150 μL DMSO was added into each well. Absorbance values were read and cell growth curves were drawn. The cells in the experiment group and treatment group were inoculated into 6-well plates at 1000 cells/well. The culture medium was changed after 4 days, cells were fixed by formalin after 14 days, and crystal violet staining was performed.
Detection of cell cycle and apoptosis changes via flow cytometry
The concentration of breast cancer cells at logarithmic growth phase was adjusted to 3×105/mL, and the cells were inoculated into 6-well plates at 2 ml/well. At 48 h after transfection, the cells were divided into an experiment group and a control group. After they were cultured continuously for 48 h, cells in the 2 groups were collected. PBS was used to wash cells, and the cells were collected again. Annexin V/PI was used for double staining, and the cells were kept in the dark for 10 min. Flow cytometry was used to determine the cell apoptosis rate of each group; the cells were collected with the same method and under the same condition. Pre-cooled 75% ethyl alcohol was used to resuspend cells, and the solution was fixed at −20°C overnight. Flow cytometry PI staining method was used to detect DNA content in the cells, and each time phase of the cell cycle was classified as G1/G0 stage, S stage, and G2/M stage, and the percentage of each time phase was calculated by special software.
Detection of cell migration and invasion capacity changes via Transwell experiment and wound healing assay
The interference sequence and the sequence in control group were transiently transfected into model cells, and when the transfected cell grew to 80–90% with good status, trypsin was used to digest cells. Serum-free medium was used to resuspend cells into single-cell suspension, the cell count was performed and concentration was adjusted to 3×105 cells/mL; 200 μL above cell suspension was added into the upper chamber of a Transwell chamber (50 mg/L BD Matrigel 1: 8 diluent was used to coat the upper chamber surface of the Transwell chamber bottom film). We added 600 μL of 1640 culture medium containing 20% FBS into the lower chamber, and the solutions were placed into an incubator to culture for 24 h under standard conditions. This was repeated in 3–5 specimens for each group. Formalin was used to fix cells, and crystal violet staining was performed.
Detection of EMT-related molecular marker expression changes via Western blotting and immunofluorescence experiment
Cells in the experiment group and control group were collected, and cell lysis buffer was added. Total protein was extracted and Bradford method was used to quantify protein, followed by SDS-PAGE electrophoresis. The protein separated by electrophoresis was electro-transferred onto PVDF film, and 5% skim milk was used to seal the protein. Then, 1: 1000 rabbit anti-human E-cadherin antibody, rabbit anti-human Vimentin antibody, and rabbit anti-human N-cadherin antibody were added into the protein, and the solution was kept at 4°C overnight. TBST buffer solution was used to wash films 3 times for 10 min/time. Horseradish peroxidase link-coupled anti-mouse or horseradish peroxidase link-coupled anti-rabbit was added (diluent ratio of 1: 2000), and the solution was cultured for 1 h at 37°C. TBST was utilized to wash films 3 times for 10 min/time, then TBS was used to wash films for 10 min. A luminescence tablet was used for color development, and GAPDH served as internal reference. The detailed steps of the immunofluorescence experiment have been described previously [22].
Research on the influence of HOXA11-AS on tumor cell metastasis capacity via experiment in vivo
Cells in the experiment group and control group were injected into BALB/c nude mice via the caudal vein, and the experimental lung metastatic model was established. The nude mice were kept in a constant environment, the status of nude mice was observed and they were weighed every 3 days. After nude mice were treated for 21 days, they were killed uniformly. Their lung tissues and liver tissues were taken out for HE staining, and the number and size of metastatic nodules were observed.
Immunohistochemistry
The expression levels of EMT molecular markers were detected by immunohistochemical method. Brown-yellow or brown-sepia section was regarded as a positive result, and standardization analysis was performed by IMS cell image analysis system and medical image analysis software for the detection results [23].
Statistical analysis
All experiments were repeated 3 times, and statistical software SPSS 15.0 was used for analysis. Data are presented as mean ± standard deviation, and the t test was performed. p<0.05 suggested the differences were statistically significant.
Results
Up-regulated expression of HOXA11-AS in breast cancer tissue and cells
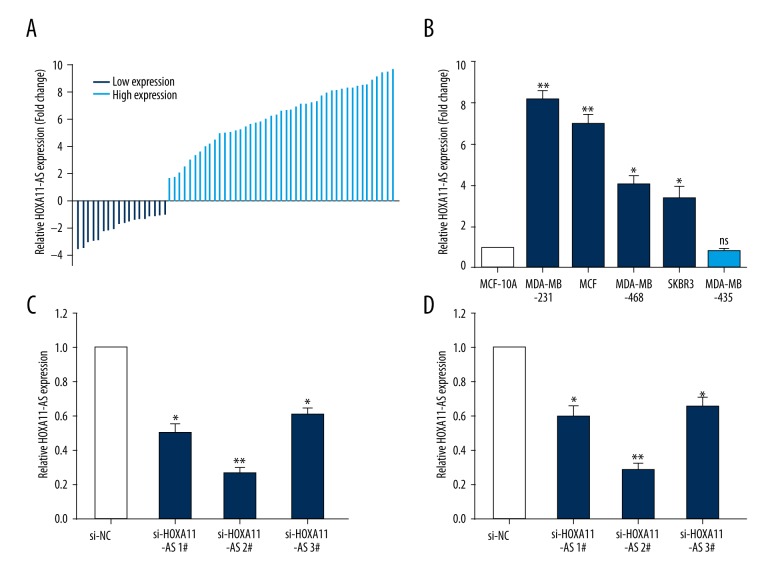
To investigate the influence of HOXA11-AS on the biological function of breast cancer cells, we first used qRT-PCR to detect breast cancer tissue and tissue adjacent to cancer of 68 patients, and the result revealed that HOXA11-AS expression in breast cancer tissue of 50 patients was relatively higher than that in tissue adjacent to cancer (Figure 1A). Next, we used qRT-PCR to detect the relative expression quantities of 5 breast cancer cell lines (MDA-MB-231, MDA-MB-468, MDA-MB-435, SKBR3, and MCF-7) and normal human mammary epithelial cells (MCF-10A). The result indicated that the expression quantity of HOXA11-AS in 4 breast cancer cell lines was up-regulated compared with that in MCF-10A, while the relative expression quantity of HOXA11-AS in MDA-MB-435 was not significantly different (Figure 1B).
Figure 1.
The expression quantity of lncRNA HOXA11-AS in breast cancer tissues and cells. (A) The relative expression quantities of lncRNA HOXA11-AS in 68 breast cancer tissues and cells were detected by qRT-PCR experiment; there were 50 cases with up-regulated expression of lncRNA HOXA11-AS and 18 cases of down-regulated expression of lncRNA HOXA11-AS. (B) The relative expression quantity of lncRNA HOXA11-AS in breast cancer cells compared with that in normal mammary epithelial cells was detected by qRT-PCR. (C, D) Were designed and synthesized into specific interference sequences of HOXA11-AS, which were transiently transfected into breast cancer cells, and qRT-PCR was used to detect the efficiency of interference. (** p<0.01; * p<0.05, ns – no statistical significance).
Research on the biological function of HOXA11-AS in breast cancer cell via experiment in vitro
Our previous research result showed that HOXA11-AS expression was relatively high in breast cancer tissue and cell. We selected 2 kinds of cell lines – MCF-7 and MDA-MB-231 – which had the highest relative expression quantities as model cells, and designed and synthesized specific interference sequence of HOXA11-AS, and used lip2000 to transiently transfect it into MCF-7 and MDA-MB-231 cells. After 48 h, cell RNA was extracted from the experimental group and control group to detect the interference efficiency of si-HOXA11-AS (Figure 1C, 1D), and sequences 2# were selected for subsequent experimentation.
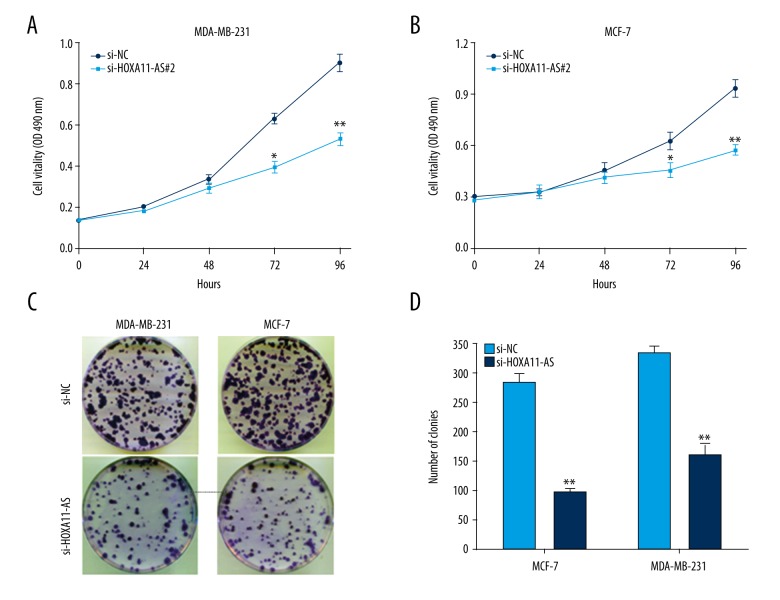
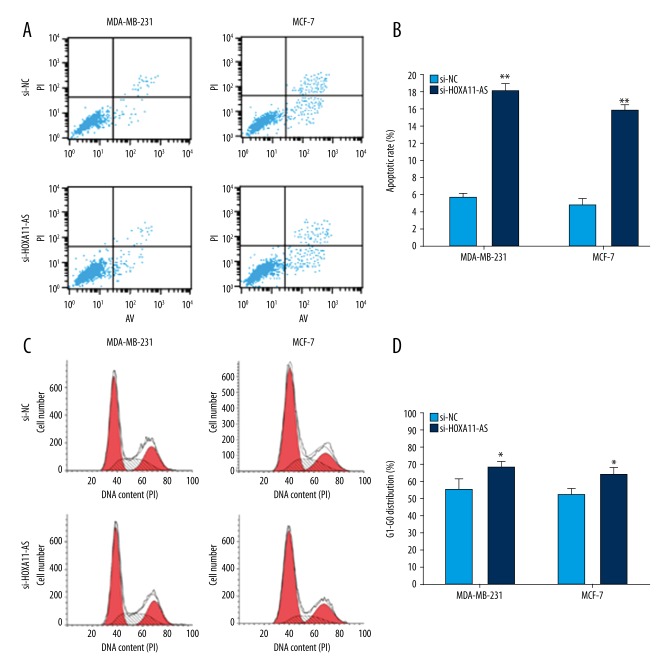
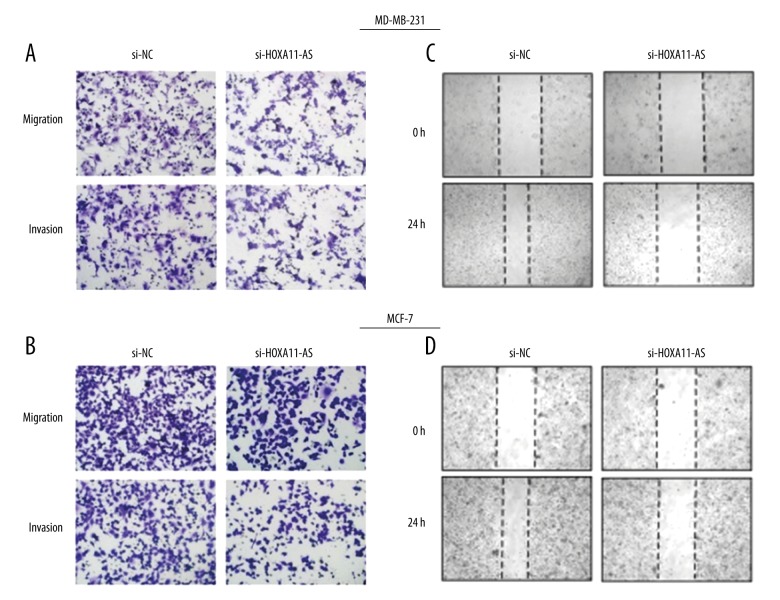
Next, we used MTT assay to investigate the influence of interfering in HOXA11-AS on the proliferation activity of MCF-7 and MDA-MB-231 cells, and the result demonstrated that the proliferation capacity in the experimental group was obviously decreased compared with that in the control group (Figure 2A, 2B), and the result obtained from clone formation assay was consistent with MTT assay (Figure 2C, 2D). To investigate the influence of HOXA11-AS on breast cancer cell apoptosis and cycle distribution, we used lip2000 to transiently transfect si-HOXA11-AS into breast cancer cell lines. After 48 h, cells were collected and the changes in cell cycle distribution and apoptosis were detected by flow cytometry. The results showed that the cell cycle progression in the si-HOXA11-AS group was arrested in G1-G0 stage and cell apoptosis rate was increased compared with that in the si-NC group (Figure 3). Next, we used wound healing assay to investigate whether interfering in HOXA11-AS affected breast cancer cell invasion and metastasis capacity, and the results showed that cell migration and invasion capacity in the si-HOXA11-AS group was markedly decreased compared with that in the control group (Figure 4B). C cells of the si-HOXA11-AS group and si-NC group were obtained by the same method, and they were planted in Transwell chambers. After 48 h, cells crossing the chamber were counted, and the results were consistent with those of the wound healing assay (Figure 4A).
Figure 2.
The influence of HOXA11-AS on the proliferation capacity of breast cancer cells. (A) MTT assay detected that cell proliferation vitality was inhibited in breast cancer cells after the interference in HOXA11-AS. (B) The result of clone formation assay showed that cell proliferation capacity was reduced after the interference in HOXA11-AS expression. (** p<0.01; * p<0.05).
Figure 3.
The influence of HOXA11-AS on the biological function of breast cancer cells. Flow cytometry detected and found that the apoptosis rate of breast cancer cell was increased after interference of A and B in HOXA11-AS. Flow cytometry detected and found that breast cancer cell cycle progression was arrested at G1-G0 stage after C and D interfered in HOXA11-AS. (** p<0.01; * p<0.05).
Figure 4.
The influence of HOXA11-AS on cell migration and invasion capacity. Transwell assay was used to detect the changes of breast cell migration and invasion capacity after interference of A and B in HOXA11-AS. Wound healing assay was used to detect the changes of cell migration capacity after interference of C and D in HOXA11-AS.
Research on the influence of HOXA11-AS on breast cancer cell invasion and metastasis via in vivo experiment
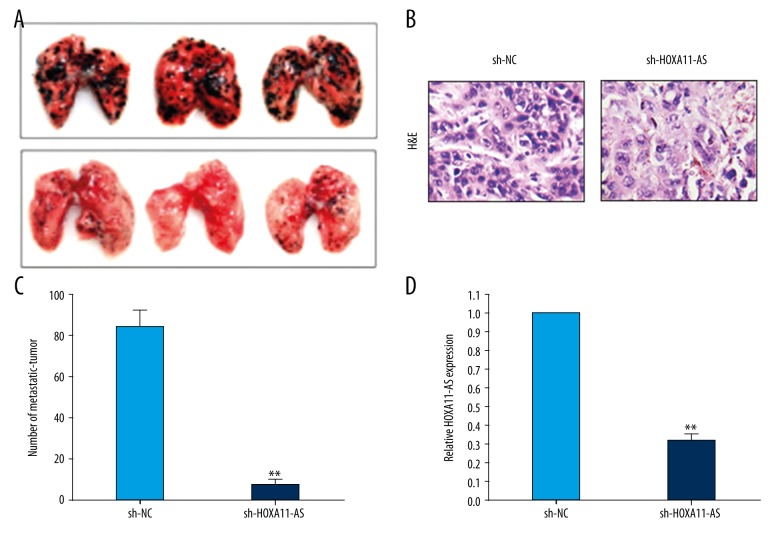
Next, we established a nude mouse metastatic tumor model to study the influence of interference in HOXA11-AS on breast cancer cell invasion and metastasis capacity via experiment in vivo. Firstly, the MDA-MB-231 cell line that had the highest transfection efficiency was screened out as the model cell, and cells in the sh-HOXA11-AS group and empty plasmid group were injected into nude mice via the caudal vein. Nude mouse growth was observed and weighed every 3 days. At 21 days after cells were injected into the caudal vein, the nude mice were killed uniformly, and lungs were removed and photographed (Figure 5A). As shown in Figure 5C, the number of metastatic tumors in lungs in the sh-HOXA11-AS group was clearly decreased compared with that in the control group. Secondly, we removed the metastatic tumors in lungs and the total RNA was extracted from tumor tissue. The relative expression quantity of HOXA11-AS in metastatic tumors was detected by qRT-PCR method, and the result showed that HOXA11-AS expression quantity in the metastatic tumors transfected from sh-HOXA11-AS cells was significantly decreased compared with that in the control group (Figure 5D). Thirdly, the metastatic tumor was fixed and sectioned, and immunohistochemical analysis was performed. The nude mouse metastatic tumor establishment was demonstrated to be successful by HE staining (Figure 5B).
Figure 5.
Study on the influence of HOXA11-AS on the migration capacity of breast cancer cells via experiment in vitro. (A) MDA-MB-231 cells and (sh-NC/MDA-MB-231) cells in control group which were used to interfere in HOXA11-AS were injected into nude mice through the caudal vein to establish the nude mouse metastatic tumor model, and then the mice were killed uniformly and their lung tissues were taken to photograph. (B) HE staining proved that the establishment of the nude mouse metastatic tumor model was successful. (C) The number of metastatic tumors in the lung was counted and statistically analyzed. (D) The relative expression quantities of HOXA11-AS in the experiment group and control group were detected by qRT-PCR. (** p<0.01; * p<0.05, ns – no statistical significance)
HOXA11-AS promotes breast cancer cell invasion and metastasis by regulating EMT
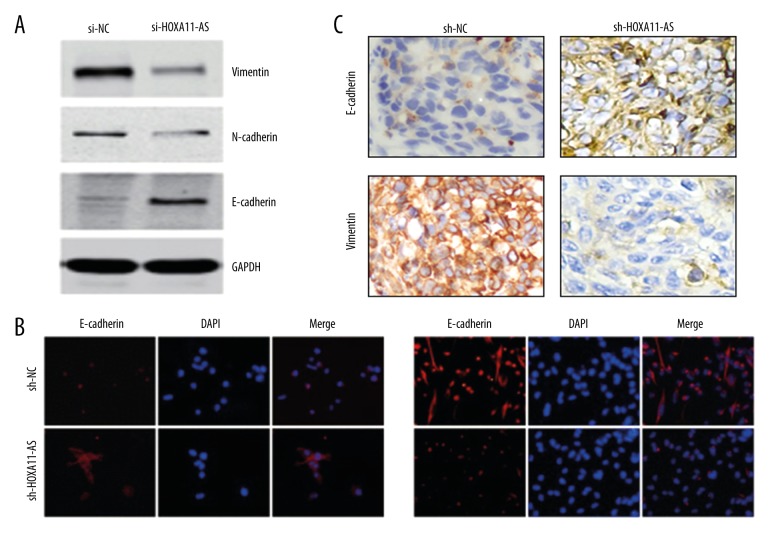
The above results showed that interfering in HOXA11-AS expression could significantly inhibit cell invasion and metastasis capacity in breast cancer cell. However, there is no report about how HOXA11-AS regulates the downstream molecular pathway of tumor cell migration and invasion. Recent research has reported that epithelial-mesenchymal transition is closely related to tumor invasion and metastasis. Chen et al. [24] found that lncRNA HOTTIP could promote esophageal cancer invasion and metastasis by inducing EMT. We conducted Western blot analysis and found that after HOXA11-AS was interfered, EMT-related molecular markers and E-cadherin expression were up-regulated, and the expressions of N-cadherin and Vimentin were down-regulated compared with those in the control group (Figure 6A). We verified the above conclusions by immunofluorescence (Figure 6B). Then, the expressions of E-cadherin and Vimentin in the above metastatic tumors were detected by immunohistochemistry, and the result was consistent with Western blot and immunofluorescence analysis (Figure 6C). Based on the results of the above experiments in vivo/vitro, we conclude that the knockdown of HOXA11-AS can inhibit the occurrence of breast cancer cell invasion and metastasis by regulating EMT.
Figure 6.
The influence of HOXA11-AS on EMT. (A) The changes in the expressions of EMT molecular markers were detected by Western blot analysis after the interference in HOXA11-AS. (B) The changes in the expressions of EMT molecular markers were detected by immunofluorescence experiment after the interference in HOXA11-AS. (C) The changes in the expressions of EMT molecular markers were detected by immunohistochemical experiment for the obtained nude mouse metastatic tumor model.
Discussion
If non-metastatic and primary breast cancer is found early and surgically treated in time, patient 5-year and 10-year survival rates are higher [25]. However, for metastatic, secondary, and recurrent breast cancer, surgery alone cannot inhibit cancer cell spread and tumor regeneration because of the metastasis of tumor cells, and even though the operation is combined with chemotherapy, radiotherapy, and molecular targeting treatment to treat metastatic breast cancer, its efficacy is also limited [26]. Tumor cell metastasis is one of the main causes of death for breast cancer patients. Therefore, screening out the biomarkers which can be used to early predict breast cancer cell metastasis and looking for the treatment means or molecular targets which can inhibit breast cancer cell metastasis are of clinical value and social significance [27].
With the rapid development and application of second-generation sequencing technology, accumulating evidence indicates that lncRNA is expressed abnormally in tumor tissues [28–30]. The IncRNA molecule can specifically regulate the occurrence and development of tumors, which has potential use as a novel tumor therapeutic target and biomarker. Because it lacks an open reading frame and sequence conservation, lncRNA was initially considered as the “dark matter” of gene transcription, even as “rubbish of transcription” without a biological function [31]. In recent years, research has revealed that lncRNA is involved in multiple cell processes, such as chromosome silence, genome imprinting, histone modification, transcriptional activation, and interference and intranuclear protein translocation, and it has a very important biological function [32,33]. For instance, lncRNA BLCAT1 promotes intestinal cancer cell proliferation by epigenetically silencing P15, thus participating in the occurrence and development of tumors [34]. As a cancer suppressor gene, lncRNA0086 promotes tumor apoptosis by targeting and regulating miRNA-214 in nasopharyngeal cancer [35], but there has been no report about the biological effect of HOXA11-AS on breast cancer.
Recent research has showed that the abnormal expression of HOXA11-AS is closely related to tumor malignant phenotype formation. Low expression of HOXA11-AS in tumors found by high-throughput chip technology in epithelial ovarian cancer can function as an independent factor to predict prognosis. The same low expression of HOXA11-AS is found in colon cancer, and overexpression of HOXA11-AS can inhibit tumor cell migration, invasion, and proliferation [33,34]. The relatively high expression of HOXA11-AS in breast cancer tissue and cells was found for the first time in this research, and we found that the knockdown of HOXA11-AS expression can inhibit cell proliferation, invasion, and metastasis, and promote apoptosis via experiments in vivo/vitro. The above experiments demonstrated that as an important regulatory factor, HOXA11-AS was closely related to the occurrence and development of breast cancer. Moreover, research on the specific inhibitor of HOXA11-AS also provides a new direction for inhibiting breast cancer metastasis.
Although HOXA11-AS is involved in the distant metastasis of tumor in colon cancer and ovarian cancer, as an important gene regulatory factor, its underlying molecular mechanism affecting tumor cell invasion and metastasis remains unclear. The occurrence mechanism of tumor cell invasion and metastasis includes the interpretation of extra cellular matrix (ECM) and enhancement of tumor cell motility and EMT [36–40]. EMT plays an important role in the biological behavior of tumor migration and metastasis, and epithelial cells lose cell polarity via EMT and lose epithelial phenotype such as the connection with basement membrane, and then obtains higher migration and invasion ability. Recent research has reported that EMT mediated by lncRNA exists in diverse tumors [21,24]. In combination with the latest achievements in scientific research, we conducted Western blot and immunofluorescence analysis and found that the knockdown of HOXA11-AS expression in breast cancer cell increased the expression of E-cadherin (the EMT molecular marker) and inhibited the expressions of N-cadherin and Vimentin.
Conclusions
In conclusion, this research first demonstrated: HOXA11-AS expression was relatively higher in breast cancer tissue and cells. Inhibiting HOXA11-AS expression could suppress tumor cell proliferation, invasion, and metastasis and promote tumor cell apoptosis. In addition, further experiments in vivo/vitro demonstrated that HOXA11-AS was involved in breast cancer cell invasion and metastasis by regulating the EMT process. The above results indicate that HOXA11-AS can act as a biological marker for the distant metastasis in breast cancer patients and to predict prognosis. Blocking the interaction between HOXA11-AS and EMT also provides an important theoretical basis for controlling breast cancer recurrence and metastasis.
Footnotes
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Lemler DJ, Lynch ML, Tesfay L, et al. DCYTB is a predictor of outcome in breast cancer that functions via iron-independent mechanisms. Breast Cancer Res. 2017;19:25. doi: 10.1186/s13058-017-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golse N, Adam R. Liver metastases from breast cancer: What role for surgery? Indications and results. Clin Breast Cancer. 2017 doi: 10.1016/j.clbc.2016.12.012. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Shi J, Dong B, Cao J, et al. Long non-coding RNA in glioma: Signaling pathways. Oncotarget. 2017;8(16):27582–92. doi: 10.18632/oncotarget.15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–88. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 6.Idogawa M, Ohashi T, Sasaki Y, et al. Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int J Cancer. 2017;140(12):2785–91. doi: 10.1002/ijc.30689. [DOI] [PubMed] [Google Scholar]
- 7.Wei W, Liu Y, Lu Y, et al. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017 doi: 10.1002/jcb.25988. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Malih S, Saidijam M, Malih N. A brief review on long noncoding RNAs: A new paradigm in breast cancer pathogenesis, diagnosis and therapy. Tumour Biol. 2016;37:1479–85. doi: 10.1007/s13277-015-4572-y. [DOI] [PubMed] [Google Scholar]
- 9.Ye N, Wang B, Quan ZF, et al. Functional roles of long non-coding RNA in human breast cancer. Asian Pac J Cancer Prev. 2014;15:5993–97. doi: 10.7314/apjcp.2014.15.15.5993. [DOI] [PubMed] [Google Scholar]
- 10.Richards EJ, Permuth-Wey J, Li Y, et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745–57. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 12.Neri S, Miyashita T, Hashimoto H, et al. Fibroblast-led cancer cell invasion is activated by epithelial-mesenchymal transition through platelet-derived growth factor BB secretion of lung adenocarcinoma. Cancer Lett. 2017;395:20–30. doi: 10.1016/j.canlet.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, He RQ, Dang YW, et al. Comprehensive analysis of the long noncoding RNA HOXA11-AS gene interaction regulatory network in NSCLC cells. Cancer Cell Int. 2016;16:89. doi: 10.1186/s12935-016-0366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Xu C, Cai B, et al. Expression and clinicopathological significance of the lncRNA HOXA11-AS in colorectal cancer. Oncol Lett. 2016;12:4155–60. doi: 10.3892/ol.2016.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Zhang J, Liu Y, et al. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016;373:251–59. doi: 10.1016/j.canlet.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 16.Cai HK, Chen X, Tang YH, Deng YC. MicroRNA-194 modulates epithelial-mesenchymal transition in human colorectal cancer metastasis. Onco Targets Ther. 2017;10:1269–78. doi: 10.2147/OTT.S125172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017 doi: 10.1038/nrclinonc.2017.44. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang LT, Chiou SS, Chai CY, et al. Transcription factor SPZ1 promotes TWIST-mediated epithelial-mesenchymal transition and oncogenesis in human liver cancer. Oncogene. 2017 doi: 10.1038/onc.2017.69. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bong AH, Monteith GR. Breast cancer cells: Focus on the consequences of epithelial-to-mesenchymal transition. Int J Biochem Cell Biol. 2017;87:23–26. doi: 10.1016/j.biocel.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20:2819–24. [PubMed] [Google Scholar]
- 21.Hao Y, Yang X, Zhang D, et al. Long noncoding RNA LINC01186, regulated by TGF-beta/SMAD3, inhibits migration and invasion through Epithelial-Mesenchymal-Transition in lung cancer. Gene. 2017;608:1–12. doi: 10.1016/j.gene.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Han H, Li Y, et al. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget. 2016;7:84480–85. doi: 10.18632/oncotarget.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HJ, Lee CH, Kim JW, et al. Use of hepatobiliary phase images in Gd-EOB-DTPA-enhanced MRI of breast cancer hepatic metastasis to predict response to chemotherapy. Clin Imaging. 2017;43:127–31. doi: 10.1016/j.clinimag.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Addison CL, Simos D, Wang Z, et al. A phase 2 trial exploring the clinical and correlative effects of combining doxycycline with bone-targeted therapy in patients with metastatic breast cancer. J Bone Oncol. 2016;5:173–79. doi: 10.1016/j.jbo.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manica GC, Ribeiro CF, Oliveira MA, et al. Down regulation of ADAM33 as a predictive biomarker of aggressive breast cancer. Sci Rep. 2017;7:44414. doi: 10.1038/srep44414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan L, Sun M, Liu GJ, et al. Long Noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15:1082–94. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Xiong Y, Xia R, et al. The pseudogene-derived long noncoding RNA SFTA1P is down-regulated and suppresses cell migration and invasion in lung adenocarcinoma. Tumour Biol. 2017;39:1393398246. doi: 10.1177/1010428317691418. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Li H, Zhang L, et al. Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J Biol Chem. 2017;292:5801–13. doi: 10.1074/jbc.M116.758508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry RB, Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143:3882–94. doi: 10.1242/dev.140962. [DOI] [PubMed] [Google Scholar]
- 30.Ma G, Tang M, Wu Y, et al. LncRNAs and miRNAs: Potential biomarkers and therapeutic targets for prostate cancer. Am J Transl Res. 2016;8:5141–50. [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Li Z, Zheng H, et al. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2) doi: 10.1111/cpr.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su J, Zhang E, Han L, et al. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis. 2017;8:e2665. doi: 10.1038/cddis.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo J, Ma J, Zhao G, et al. Long non-coding RNA LINC0086 functions as a tumor suppressor in nasopharyngeal carcinoma by targeting miR-214. Oncol Res. 2017 doi: 10.3727/096504017X14865126670075. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards EJ, Permuth-Wey J, Li Y, et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745–57. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Xu C, Cai B, et al. Expression and clinicopathological significance of the lncRNA HOXA11-AS in colorectal cancer. Oncol Lett. 2016;12:4155–60. doi: 10.3892/ol.2016.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erdogan B, Webb DJ. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem Soc Trans. 2017;45:229–36. doi: 10.1042/BST20160387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam SW, Clair T, Kim YS, et al. Autotaxin (NPP-2), a metastasis-enhancing motogen, is an angiogenic factor. Cancer Res. 2001;61:6938–44. [PubMed] [Google Scholar]
- 38.Tsutsumi S, Saeki H, Nakashima Y, et al. PD-L1 expression at tumor invasive front is associated with EMT and poor prognosis in esophageal squamous cell carcinoma. Cancer Sci. 2017 doi: 10.1111/cas.13237. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Li X, Prow TW, et al. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]