Abstract
Background
Recent WHO guidelines identify virologic monitoring for diagnosing and confirming ART failure. In view of this, validation and scale up of point of care viral load technologies is essential in resource limited settings.
Methods
A systematic validation of the GeneXpert® HIV-1 Quant assay (a point-of-care technology) in view of scaling up HIV-1 viral load in India to monitor the success of national ART programme was carried out. Two hundred nineteen plasma specimens falling in nine viral load ranges (<40 to >5 L copies/ml) were tested by the Abbott m2000rt Real Time and GeneXpert HIV-1 Quant assays. Additionally, 20 seronegative; 16 stored specimens and 10 spiked controls were also tested. Statistical analysis was done using Stata/IC and sensitivity, specificity, PPV, NPV and %misclassification rates were calculated as per DHSs/AISs, WHO, NACO cut-offs for virological failure.
Results
The GeneXpert assay compared well with the Abbott assay with a higher sensitivity (97%), specificity (97-100%) and concordance (91.32%). The correlation between two assays (r = 0.886) was statistically significant (p < 0.01), the linear regression showed a moderate fit (R2 = 0.784) and differences were within limits of agreement. Reproducibility showed an average variation of 4.15 and 3.52% while Lower limit of detection (LLD) and Upper limit of detection (ULD) were 42 and 1,740,000 copies/ml respectively. The misclassification rates for three viral load cut offs were not statistically different (p = 0.736). All seronegative samples were negative and viral loads of the stored samples showed a good fit (R2 = 0.896 to 0.982).
Conclusion
The viral load results of GeneXpert HIV-1 Quant assay compared well with Abbott HIV-1 m2000 Real Time PCR; suggesting its use as a Point of care assay for viral load estimation in resource limited settings. Its ease of performance and rapidity will aid in timely diagnosis of ART failures, integrated HIV-TB management and will facilitate the UNAIDS 90-90-90 target.
Keywords: GeneXpert HIV-1 quant, Abbott RealTime PCR, HIV-1, Viral load, Point of care technologies, ART
Background
Success of the ambitious “Treatment 2015” initiative by UNAIDS [1] has accelerated the pace of antiretroviral therapy (ART) scale-up globally. The achievement of this initiative is attributed to the international solidarity and responsibilities shared by diverse disciplines. Upto 2014, an estimated 14.9 million people in developing/low- and middle-income countries received ART [2]. The National AIDS Control Programme in India has successfully reduced the incidence of new HIV-1 infections by 57% and the free roll out of ART has covered nearly 0.8 million HIV infected individuals with nearly 67% coverage of those in need of ART [3]. Furthermore, the wider access to ART has resulted in declining the death rate due to AIDS related co-morbidities [4].
Presently, the free ART programme monitors patients on ART clinically and immunologically with six monthly CD4 count and follows a “Targeted Viral Load” approach for treatment failure. However, several studies have demonstrated poor predictive value of the immunological criteria and reported accumulation of HIV drug resistant mutations due to delayed detection of treatment failure [5–7]. To avoid this, WHO strongly recommends viral load monitoring as the preferred approach to diagnose and confirm treatment failure [8–11].
Nucleic Acid Testing (NAT) based technologies are considered as the gold standards for HIV-1 viral load estimation because of their high specificity, sensitivity and wide linear range of detection. However, these assays require testing to be performed by well-trained technicians and need at least 8-10 h. With an increasing demand to scale up HIV-1 viral load monitoring, a robust, easy to use and sensitive point-of-care technology (POC) [12] is utmost essential. Numbers of POC viral load assays are in the development pipeline [13, 14] and GeneXpert® HIV-1 Quant Assay (Cepheid Innovations Pvt. Ltd., USA) is one of the recently introduced assays. It is a fully automated integrated system that is routinely used for detecting M. tuberculosis DNA and resistance to Rifampicin [15, 16] as well as other pathogens (MRSA, Norovirus, HBV, HPV [17–21]) including HIV. It is a rapid assay that delivers results within 90 mins. It uses reverse transcriptase polymerase chain reaction (RT-PCR) technology to achieve high sensitivity over the range of 40 to 10,000,000 copies/ml (1.6 to 7 log10copies/ml) and can quantitate all HIV-1 Group M, N and O subtypes [22]. Furthermore, a reduced turnaround time leads to prompt HIV diagnosis and clinical management in adult and pediatric samples. Apart from this, the assay is very simple and safe to perform; their integrated reagents and consumables are robust in usage and can be stored in refrigerators. Due to minimal requirement of infrastructure, it is very easy to scale up the activity of viral load testing nationally. Considering all these features, the Gene Xpert HIV-1 Quant assay serves as a valuable point of care viral load assay that could be useful for monitoring antiretroviral therapy as well as integrated HIV-TB management in developing countries like India [23]. Therefore in the present study, we compared performance of GeneXpert® HIV-1 Quant Assay with the gold standard, routinely used Abbott m2000rt RealTime HIV-1 (Abbott Molecular, Germany) in the resource limited Indian settings in view of the scale up of viral load testing [24]. The performance evaluation of this new assay will ensure its decentralized utility in the clinical set up and will assist in management of patient care efficiently.
Methods
Study design
A systematic validation of the GeneXpert® HIV-1 Quant assay (a point-of-care technology) in view of scaling up HIV-1 viral load in India to monitor the success of national ART programme was carried out.
Ethics statement
The study was approved by the 59th National AIDS Research Institute (NARI) Ethics Committee (Approval No: NARI/EC Protocol No. 2015-10). All study participants provided a written informed consent for collecting demographic information, plasma viral load testing and retention of plasma samples.
Study setting and specimen collection
The study was conducted from June to September 2015 in HIV-1 positive adult individuals. 314 HIV-1 seropositive (ART naïve n = 151, On ART n = 129, suspected ART failure n = 34; individuals were screened to obtain varying viral load ranges) and 20 normal healthy HIV seronegative individuals were enrolled at three ART centers located in Pune (ART centres: Model Colony; YCM Hospital; Sassoon General Hospital).The whole blood specimens were collected in 10 ml EDTA vacutainers (Becton Dickinson, USA), transported to NARI, centrifuged at 405 g for 10 mins, plasma was separated within 6 h, aliquoted and stored at −70 °C until tested.
Viral load estimation
Following manufacturers’ instructions, all plasma specimens were tested by an automated Abbott m2000rt RealTime HIV-1 assay (Abbott Molecular Inc., Germany) within 2 days of collection in batches of 24 and results were expressed as copies/ml [25–27].
Of 314, 219 specimens falling in nine viral load ranges (Table 1) were selected and tested by the HIV-1 Quant assay following the manufacturer’s instructions within 3 days after sample collection. Briefly, the assay was carried out by adding 1.2 mL of plasma to the cartridges which contain reagents for RNA extraction, reverse transcription and real-time cDNA quantification based on the conserved region of 3′-LTR of HIV-1.Additionally, following sets 1) spiked copy controls (prepared by spiking the normal human plasma (NHP) with pooled high HIV-1 viral load plasma [n = 25, 20 - 20, 00,000 copies/ml] and HIV-1 viral culture supernatant (n = 25, 40 - 40, 00,000 copies/ml) and 2) samples stored at three time points (1, 2 and 3 months) were used.
Table 1.
Viral load ranges selected for validation
| Viral Load ranges | Specimens tested by Abbott Real Time HIV-1 | Specimens tested by GeneXpert HIV-1 Quant |
|---|---|---|
| Not detected | 85 | 27 |
| < 40 | 20 | 24 |
| 40 -200 | 24 | 15 |
| 201 -2500 | 24 | 16 |
| 2501 -5000 | 21 | 16 |
| 5001 -10,000 | 23 | 19 |
| 10,001 -1 L | 59 | 59 |
| 100,001 -5 L | 45 | 26 |
| > 5 L | 13 | 17 |
| Total | 314 | 219 |
Statistical analysis
The viral load values (copies/mL) were transformed to log10 copies/mL and the statistical analysis was performed using Stata/IC 10.1 for Windows (Copyright © 1984-2009). The percent concordance among the nine viral load ranges was determined and reproducibility, inter- and intra-assay variation were studied using coefficient of variation. The Bland-Altman plot was generated to assess the limits of agreement and mean bias (95% CI). The descriptive statistical tests were used to compare the misclassifications rates for different viral load cut-offs as per two internationals viz. [Demographic Health Surveys/AIDS Indicator Surveys (DHSs)/AISs) cut-off 200 copies/ml [28] and WHO cut-off 400 copies/ml] [29] and one national cut-off [NACO cut-off 1000 copies/ml] [30] used for classifying the treatment failures. The coefficient of determination (R2) was used to assess the linear fit for paired readings of log viral load copies by two methods. Additionally, the lower and upper limits of detection (LLD and ULD) were calculated using minimum and maximum values. The uncertainty limits for natural variation in GeneXpert viral load results were also calculated.
Results
The paired plasma specimens collected from 219 study participants (mean age 37.6 years, SD = 9.4) were tested by Abbott m2000 Real Time PCR and GeneXpert HIV-1 Quant assays.
Concordance between Abbott and HIV-1 quant assays
The percentage of concordant samples in both the assays is depicted in Fig. 1 and Table 2. Out of 175 samples detected by the Abbott assay, 8 samples (range: 42-92 copies/mL) were undetectable (false undetected) by the GeneXpert assay. Also, one of the 44 samples undetectable by the Abbott assay, was detectable (42 copies/mL, false detected) in the GeneXpert assay.
Fig. 1.
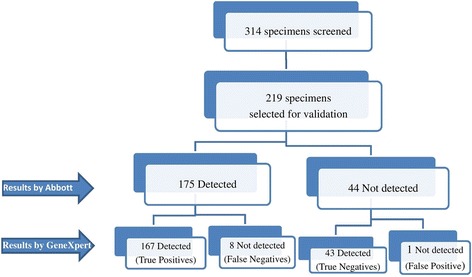
Flow Chart of specimens selected for the validation of GeneXpert HIV-1 Quant
Table 2.
Concordance in different viral load categories of Abbott Real Time HIV-1 and GeneXpert HIV-1 Quant

* Lakhs
The concordance was determined by measuring the percentage of samples containing <40 or ≥40 copies/ml which was 91.32% (200/219 samples). Table 2 shows detailed analysis of the concordance obtained between the two assays. The percent concordance between each of the seven quantifiable categories (40- > 5 L copies/mL) ranged from 82-100%. Out of 167 samples quantifiable by the GeneXpert assay, 86.8% (145/167) of the samples were fitting within the acceptable limit (+/− 0.5 log copies/mL) whereas 13.2% (22/167) were outside this limit. Further analysis of 167 quantifiable samples for studying the correlation between two assays indicated a statistically significant (p < 0.01) Pearson’s correlation coefficient (r = 0.886).
Correlation and bland-Altman analysis
The scatter plot indicated a moderate fit with coefficient of determination: R2 = 0.784 (Fig. 2). Further, the level of agreement studied using the Bland-Altman plots indicated that the differences were within the limits of agreement (−0.909 to 1.15). No systematic trend was observed from lower to higher values. The mean bias was positive showing an overestimation by the HIV-1 Quant assay with an average bias of 0.120 and 95% CI (0.042 to 0.199) (n = 167) (Fig. 3). Overall, results indicated a strong agreement between results of the two assays.
Fig. 2.
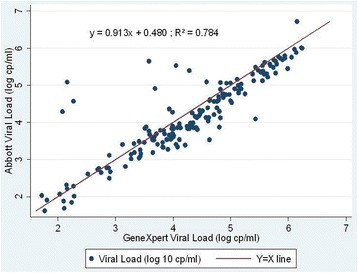
Scatter plot of plasma viral load of Abbott Real Time HIV-1 and GeneXpert HIV-1 Quant
Fig. 3.
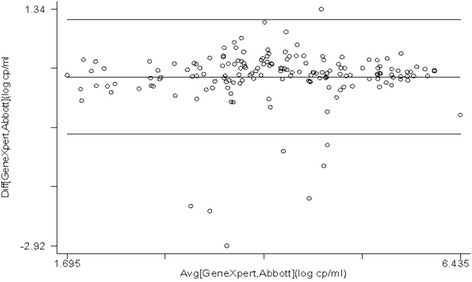
Bland-Altman plot for Abbott Real Time HIV-1 and GeneXpert HIV-1 Quant assay results
Sensitivity, specificity, PPV, NPV and percent misclassification
The misclassification data refers to reporting a “Not detected” viral load result as “detected” and vice versa. Considering the implementation of the validated assay on a larger scale, the data was analyzed in view of three cut-offs, DHSs/AIS; WHO and NACO that consider 200, 400 and 1000 copies/ml respectively, which indicated that the sensitivity remained constant (97%) for the three cutoffs while the specificity differed (100, 97 and 98% respectively). The PPV was 100% for the DHS cut off; 99% for WHO and NACO while the NPV showed an increasing trend (88, 89, 91% respectively) (Table 3). The lowest misclassification rate (2.14%) was observed for the cut-off 200 with 95% CI (0.6%, 5.4%). Overall, the percent misclassification rates for the three cut offs were not statistically different when compared by the proportion test (p = 0.736).
Table 3.
Data on sensitivity, specificity, PPV, NPV & % misclassification as per national and international cut-offs
| Cut-offs classifying treatment failures | sensitivity, specificity, PPV, NPV & % misclassification |
|---|---|
| DHSs and AISs cut-off (>200 copies/ml) |
Sensitivity = 97%, specificity = 100%, PPV = 100%, NPV = 88% % misclassification (95%CI) = 2.14% (0.6%, 5.4%) |
| WHO cut-off (>400 copies/ml) |
Sensitivity = 97%, specificity = 97%, PPV = 99%, NPV = 89% % misclassification (95%CI) = 2.67% (0.9%, 6.1%) |
| NACO cut-off (>1000 copies/ml) |
Sensitivity = 97%, specificity = 98%, PPV = 99%, NPV = 91% % misclassification (95%CI) = 2.67% (0.9%, 6.1%) |
ULD, LLD, reproducibility analysis and uncertainty limits for the natural variation
The viral load results for 10 paired specimens (with 3 repeat tests) were selected from inter-assay and reproducibility analyses was carried out. The Bland-Altman analysis showed mean difference (max among all) of 0.064 log10 copies/ml (95% CI: -0.170, 0.042); which is much lesser than the acceptable limit of 0.5 log10 copies/ml. The LLD and ULD for GeneXpert system were 1.62 log10 copies/ml. (42 copies/ml) and 6.24 log10 copies/ml (1,740,000 copies/ml) respectively.
Precision analysis on spiked copy controls
The inter- and intra-assay variation assessed using copy controls spiked with pooled HIV-1 viral load plasma indicated variation in the GeneXpert assay as 1.42% (range 0.36–2.74%) and 1.24% (range 0.31–3.34%) respectively across the range of viral load copies (20–20 L copies/ml) (Table 4).
Table 4.
Inter and Intra Assay Variation between Abbott Real Time HIV-1 and GeneXpert HIV-1 Quant
| NHP spiked with high VL plasma | NHP spiked with viral stock | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HIV-1 viral load (copies/ml) | Coefficient of variation (% CV) | HIV-1 viral load (copies/ml) | Coefficient of variation (% CV) | ||||||
| Inter-Assay | Intra-Assay | Inter-Assay | Intra-Assay | ||||||
| Abbott | GeneXpert | Abbott | GeneXpert | Abbott | GeneXpert | Abbott | GeneXpert | ||
| 200 | 4.11 | 1.97 | 4.77 | 3.34 | 40 | 1.53 | 6.96 | 4.28 | 2.33 |
| 2000 | 1.08 | 1.45 | 1.42 | 0.31 | 4000 | 1.03 | 3.17 | 0.77 | 10.66 |
| 20,000 | 0.86 | 2.74 | 2.51 | 0.97 | 40,000 | 0.28 | 6.84 | 0.42 | 0.96 |
| 2,00,000 | 0.57 | 0.36 | 1.74 | 1.11 | 4,00,000 | 0.60 | 3.00 | 0.49 | 2.89 |
| 20,00,000 | 0.81 | 0.58 | 0.78 | 0.45 | 40,00,000 | 0.69 | 0.78 | 0.33 | 0.78 |
Additionally, inter- and intra-assay variation assessed using copy controls spiked with HIV-1 viral culture supernatant indicated variation in the GeneXpert assay as 4.15% (range 0.78–6.96%) and 3.52% (range 0.78–10.66%) respectively across the range of viral load copies (40 – 40 L copies/ml) (Table 4).
Carryover analysis
The possibility of carry-over contamination was verified by placing the 20 seronegative sample before or after the seropositive samples. The results indicated that all seronegative samples were negative without any carry over, indicating 100% specificity of the GeneXpert assay. (Data not shown).
Linear regression analysis of viral load on stored plasma samples
The linear regression analysis of the samples stored for 1, 2 and 3 months showed a good fit with no significant difference in the regression coefficient [R2 = 0.920; 0.896 and 0.982 respectively] (Fig. 4).
Fig. 4.
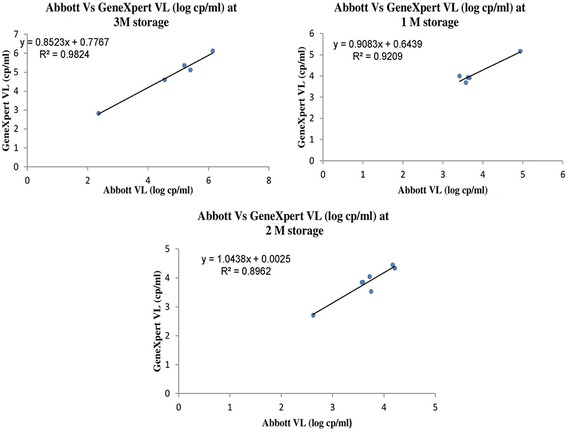
Regression plots for viral load of stored samples for Abbott Real Time HIV-1 and GeneXpert HIV-1 Quant
Discussion
NAT based real-time quantitative PCR’s are the preferred options for high-throughput HIV-1 viral-load testing in developed and developing countries [31]. However, these assays are time consuming and flow separately through three steps - sample preparation, amplification, detection and require specimen transportation, high-end instrumentation, skilled personnel, and dedicated laboratories [22]. Hence as suggested by the recent WHO guidelines [11] and to generate results with “sample-in/answer-out” approach [32], replacing NAT based assays with partially/fully automated POC assays is crucial.
The National ART programme in India was launched in April 2004 in limited number of hospitals. Subsequently, concentrated efforts resulted in expansion of the programme. Currently, the programme has adopted a public health approach for provision of ART & provides comprehensive prevention, care and treatment services, with a standardized and simplified combination of ART regimen free of cost to more than 8.8 lakhs PLHIVs across 528 ART centers and 1080 Link ART centers across the country which is a second highest number in the world. Wider access to ART has led to 29% reduction in estimated annual AIDS related deaths [33].
However, the success of high-quality ART depends on timely monitoring of viral load and confirmation of ART failure. Introduction of a simple and reliable POC viral load assay will deliver ART service more appropriately and significantly in a decentralized manner. This will potentially reduce the cost of transportation of samples and monitoring patients on antiretroviral therapy without compromising the quality of care required for treatment success. The “ASSURED” criteria set by WHO describes the POC assays as affordable, sensitive, specific, user-friendly, robust/rapid, equipment-free, and deliverable techniques. Many such POC technologies [13] are in development and GeneXpert HIV-1 Quant assay is one of them. The system requires single-use disposable GeneXpert cartridge that are self-contained and could be useful for viral load monitoring in resource limited settings. As the patient load in Indian ART centers is low and variable, the viral load testing requirement can be low every day in smaller in district ART centers. Such small number of viral load tests are manageable with POC GeneXpert platform [33] In certain districts because the RNTCP, some GeneXpert POC machines for TB diagnosis have already been placed [34] The load of TB testing along with viral load estimation on the same instrument is possible. Hence, convergence of both the program’s is possible through integrated testing.
Our results indicated that HIV-1 Quant assay compared well with the Abbott plasma viral load assay [27, 32]. The high percentages of concordance (91.32%), correlation (r = 0.886) and minimum differences in the mean viral loads (0.12 log copies/ml, as measured by Bland-Altman analysis) were observed between the two assays. In comparison to the Abbott assay, the results obtained by the GeneXpert assay for low viral load categories showed an agreement of 79% (19/24) for not detected category, 70%(14/20) for <40 copies/ml category and 100% for 40-200 copies/ml considering number of concordant samples within acceptable limit of 0.5 log copies/ml. Further comparison of the GeneXpert assay showed an agreement of (91, 86 and 83%) in the moderate viral load categories (201-2500, 2501-5000 and 5001-10,000 copies/ml). Whereas in the higher viral load categories (10001-1 L, 100,001-5 L and >5 L copies/ml), the agreement was (87, 82 and 90%). In contrast to the previous studies [35, 36], higher agreements in the viral load categories ranging from 40 to >5 L copies/ml were reported in our studies. Furthermore, it is noteworthy to mention that the LLD (42 copies/ml) obtained in our assays coincided well with the LLD of manufacturer (40 copies/ml).
The main strength of this study lies in the analysis of percent misclassifications based on the international and national cut offs used for classifying the treatment failures. The GeneXpert assay was found to be highly sensitive (91 to 95%) and specific (99 to 100%) and the misclassification rates were not statistically different as per three cut-offs (p = 0.736). Also, the study indicated that the kit worked well after 3 months of storage at −70 °C and suggested its robustness in case of the programme implementation while facing hurdles with kit supply, equipment down time, manpower, shortages, etc.
Assay precision and reproducibility were investigated by spiking the seronegative plasma with high viral load plasma/culture supernatant. Our data indicated that the GeneXpert assay showed excellent intra-assay and inter-assay reproducibility at high viral loads (2000-20,000,00 copies/ml), with %CV ranging between 0.31-1.11, 0.36-1.45 except for inter-assay variation for 20,000 copies/ml (%CV 2.74). However, higher variations were observed at low viral loads, ie; at 200 copies/ml (%CV 3.34, 1.97) respectively which was found to be less than the Abbott assay. Thus, variation observed at low viral load results agrees with previously reported study by Swenson et al., in 2014 where comparisons were based on 4221 paired samples [37], These variations could be attributed to the discrepancies in different testing platforms in quantifying subtype C samples as reported earlier [38]. Apart from variations observed while using such platforms, the parameters like type of sample and its integrity were assessed in one of the study wherein good precision as well as accuracy was reported for another POC (Liat HIV Quant (IQuum) platform) in comparison with other platforms (Roche CAP/CTMv2 and Abbott RealTime HIV-1) [39]. In our study, sero-negative samples were included to assess false positives and the carry over effect, which was found to be 100% specific in classifying negative samples as negative with no carryover effect, even if a negative sample preceded a positive specimen.
The main advantages of the GeneXpert HIV-1 viral load assay lies in its simplicity (ease of performance due to its integrated cartridge based closed system), rapid turnaround time (90 min as compared to other viral load platform that require 8-12 h), compactness (equipment is smaller than other platforms), cost effectiveness (17 USD which is cheaper as compared to other assays) and the large portfolio of different assays to manage patients with different variants of HIV-1, TB, and HIV-TB co-infection in the longer run [13, 15]. As nearly 1.1 lakh PLHIVs are reported to have HIV-TB co-infection. India shares one fourth of the global TB burden and one person dies of TB, every 2 min. Keeping in mind, the morbidity due to HIV-TB co-infections, the Indian government has launched the Revised National TB Control Program (RNTCP) in collaboration with NACO (National AIDS Control Organization) and CTD (Central TB Division) that uses WHO recommended Directly Observed Treatment Short Course (DOTS) strategy that is operational in 632 districts/reporting units. It provides daily anti-TB treatment (ATT) for PLHIV at all ART centers, as a single window approach [23, 33, 34]. Prioritization of rapid molecular Cartridge Based GeneXpert assay for all PLHIV with presumptive TB to ensure early diagnosis of TB will help on convergence of load per day for TB where less number of machines is available at district level. Furthermore, both the activities can be best utilized if two programs converge. However, the decision to use one assay or the other could depend on assay-independent parameters such as laboratory space, number of samples and workflow. As per the study design, the predefined viral load ranges quantified using the gold standard Abbott assay obviate the risk of any potential bias for sample selection as it covers varied viral load ranges from low to high viral copies. Furthermore, the inherent capacity of the system for processing 4 samples at a time could serve as a limitation to provide viral load testing for patient care at ART centers that have higher number of patient visits.
Our results suggest that the GeneXpert cartridge based test as an alternative method for viral load monitoring in resource-limited settings, specifically from the rural and remote parts of the country. Our study ascertains the validity and performance of the GeneXpert HIV-1 Quant assay as a point of care technology which could be helpful as reference to other scientists for validating similar POC instruments. Thus, our study results can be an important tool for clinicians, patients, policy makers etc. to scale up the ambitious UNAIDS 90-90-90 treatment target with timely diagnosis of ART failures as well as for management of HIV-TB patients [40]. Further studies of operational issues to apply these findings within a clinical setting on a large scale will be useful in translating into policy decisions that will benefit India and the global community.
Conclusion
The viral load results of GeneXpert HIV-1 Quant and Abbott HIV-1 m2000 Real Time PCR were comparable. The differences between the HIV-1 viral load estimates for both the assays were within the limits of agreement. Thus, the GeneXpert HIV-1 Quant can be used as a Point of care assay for viral load estimation in resource limited settings. The outcomes from this study appraise the national and international programs for adoption and placement of such integrated platforms for PLHIV and HIV-TB patient care.
Acknowledgements
We thank the World Health Organization Country office, India and Cepheid India Pvt. Ltd. for their technical support. We would also like to thank National AIDS Control Organization (NACO) for facilitating study enrollments. We are grateful to other staff of NARI clinics, ART centers (NARI, YCM and BJMC) and Virology Department, NARI for their help while carrying out this study.
Funding
The study was supported by Cepheid India Pvt. Ltd. in consultation with WHO Country office for India, New Delhi, India.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available as in India, raw data can be only shared by institutes under Government of India (GoI) if prior regulatory [Health Ministry’s Screening Committee (HMSC)] approvals are obtained. Therefore we are unable to share the data for this purpose. However, the dataset can be available from the corresponding author on reasonable request.
Abbreviations
- AIDS
Acquired immunodeficiency syndrome
- AISs
AIDS indicator surveys
- ART
Antiretroviral therapy
- ATT
Antitubercular treatment
- CAP-CTM
COBAS® AmpliPrep/COBASO TaqMan®
- CI
Confidence interval
- CTD
Central TB division
- CV
Coefficient of variation
- DHSs
Demographic and health surveys
- EC
Ethics committee
- HBV
Hepatitis B virus
- HIV
Human immunodeficiency virus
- HPV
Human papilloma virus
- LLD
Lower limit of detection
- MRSA
Methicillin-resistant Staphylococcus aureus
- NACO
National AIDS Control Organization
- NAT
Nucleic acid amplification technique
- NPV
Negative predictive value
- PCR
Polymerase chain reaction
- PLHIV
People living with HIV/AIDS
- POC
Point of care
- PPV
Positive predictive value
- RNTCP
Revised national TB control program
- SD
Standard deviation
- TB
Tuberculosis
- ULD
Upper limit of detection
- UNAIDS
United Nations programme on HIV/AIDS
- WHO
World Health Organization
Authors’ contributions
SK proposed the idea; SK and RG together designed the study. SJ, VD and VC performed viral load assays. RG, SS, SJ, PK and RL analyzed and interpreted the results. All authors contributed in data collection, analysis and manuscript preparation. SJ and PK drafted the manuscript. RG, SK, DK, NG and BR strengthened the manuscript with inputs. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The study was reviewed and approved by the Institutional Review Board (IRB) of National AIDS Research Institute (NARI). The study was approved by the 59th National AIDS Research Institute (NARI) Ethics Committee (Approval No: NARI/EC Protocol No. 2015-10). Written informed consent was obtained from each participant for plasma viral load testing and collecting demographic information. None of the enrolled participants were below 18 years of age. Individual records were coded and accessed only by research staff. All participants were provided HIV-1 viral load reports tested by Abbott Real Time PCR assay.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Smita Kulkarni, Email: skulkarni@nariindia.org, Email: skulkarni@icmr.org.in.
Sushama Jadhav, Email: sushamajadhav9@gmail.com.
Priyanka Khopkar, Email: priyankakhopkar@gmail.com.
Suvarna Sane, Email: suvarnasane@gmail.com.
Rajkumar Londhe, Email: rajkumarlondhe@gmail.com.
Vaishali Chimanpure, Email: vaishali.chimanpure@gmail.com.
Veronica Dhilpe, Email: verong2014@gmail.com.
Manisha Ghate, Email: mghate@nariindia.org.
Rajendra Yelagate, Email: rajendrayelagate@gmail.com.
Narayan Panchal, Email: narayanpanchal@gmail.com.
Girish Rahane, Email: girish.rahane@gmail.com.
Dilip Kadam, Email: deelipkadam@gmail.com.
Nitin Gaikwad, Email: nitin_thorax@yahoo.co.in.
Bharat Rewari, Email: rewarib@who.int.
Raman Gangakhedkar, Email: rgangakhedkar@nariindia.org.
References
- 1.Treatment 2015 initiative. A document published by UNAIDS. Available at: http://www.unaids.org/sites/default/files/sub_landing/files/JC2484_treatment-2015_en.pdf. Accessed 8 May 2017.
- 2.Global AIDS update, 2016. A document published by UNAIDS. Available at: http://www.unaids.org/sites/default/files/media_asset/global-AIDS-update-2016_en.pdf. Accessed 8 May 2017.
- 3.National AIDS Control Programme Phase IV (2012-2017) Strategy. A document published by Department of AIDS Control. Available at: naco.gov.in/sites/default/files/NACP-IVStrategyDocument.pdf. Accessed on 8 May 2017.
- 4.Disease specific XII plan HIV/AIDS Document published by Indian Council of Medical Research (ICMR). Available at: http://icmr.nic.in/Publications/hpc/PDF/Annexure%2019.pdf. Accessed 8 May 2017.
- 5.Ferradini L, Jeannin A, Pinoges L, et al. Scaling up of highly active antiretroviral therapy in a rural district of Malawi: an effectiveness assessment. Lancet. 2006;367(9519):1335–1342. doi: 10.1016/S0140-6736(06)68580-2. [DOI] [PubMed] [Google Scholar]
- 6.Chaiwarith Rm, Charoenyos N, Sirisanthana T, et al. Discontinuation of secondary prophylaxis against Penicilliosis marneffei in AIDS patients after HAART. AIDS. 2007;21:365–367. doi: 10.1097/01.aids.0000253374.19966.f9. [DOI] [PubMed] [Google Scholar]
- 7.Rutherford ME, Mulholland K, Hill PC. How access to health care relates to under five mortality in sub–Saharan Africa: systematic review. Tropical Med Int Health. 2010;15(5):508–519. doi: 10.1111/j.1365-3156.2010.02497.x. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. Consolidated guidelines on the use of Anti-retroviral drugs for treatment and preventing HIV infection. Available at: http://apps.who.int/iris/bitstream/10665/85321/1/9789241505727_eng.pdf. Accessed 8 May 2017.
- 9.Hosseinipour MC, Kumwenda JJ, Weigel R, Brown LB, Mzinganjira D, Mhango B, Eron JJ, Phiri S, van Oosterhout JJ. Second-line treatment in the Malawi antiretroviral programme: high early mortality, but good outcomes in survivors, despite extensive drug resistance at baseline. HIV Med. 2010;11(8):510–518. doi: 10.1111/j.1468-1293.2010.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomohiro K, Siti Q, Adiana MW, et al. HIV-1 transmitted drug resistance mutations among antiretroviral therapy- Naïve individuals in Surbaya, Indonesia. AIDS Res Ther. 2015;12:5. doi: 10.1186/s12981-015-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach - Second edition. 2016. Available at: http://www.who.int/hiv/pub/arv/arv-2016/en/. Accessed 8 May 2017. [PubMed]
- 12.Holland CA, Kiechle FL. Point-of-care molecular diagnostic systems — past, present and future. Curr Opin Microbiol. 2005;8:504–509. doi: 10.1016/j.mib.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Mohan Kumar Haleyur Giri Setty and Indira K. Hewlett, “Point of Care Technologies for HIV,” AIDS Res Treat. 2014;2014:Article ID 497046,20. doi:10.1155/2014/497046. [DOI] [PMC free article] [PubMed]
- 14.Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29(5):240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang AM, Li F, Liu XH, Xia L, Lu SH. Application of gene Xpert mycobacterium tuberculosis DNA and resistance to rifampicin assay in the rapid detection of tuberculosis in children. Zhonghua Er Ke Za Zhi. 2016;54(5):370–374. doi: 10.3760/cma.j.issn.0578-1310.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Do CG, Tran ND, Dang TMH, et al. Prospective evaluation of GeneXpert for the diagnosis of HIV-negative pediatric TB cases. BMC Infect Dis. 2015;15:70. doi: 10.1186/s12879-015-0814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurice B, Hedi M, Emilie P, et al. Value of Xpert MRSA/SA blood culture assay on the gene Xpert® Dx system for rapid detection of Staphylococcus aureus and coagulase-negative staphylococci in patients with staphylococcal bacteremia. Diagn Microbiol Infect Dis. 2013;75(Issue 2):139–143. doi: 10.1016/j.diagmicrobio.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Halligan E, Edgeworth J, Bisnauthsing K, Bible J, Cliff P, Aarons E, Klein J, Patel A, Goldenberg S. Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: a comparative diagnostic and clinical utility study. Clin Microbiol Infect. 2014;20(8):O460–O467. doi: 10.1111/1469-0691.12476. [DOI] [PubMed] [Google Scholar]
- 19.Gaby Vercauteren. Evaluation of Quality performance and operation of diagnostics, World Health Organization (WHO), UN Meeting with manufacturers, 2013. Available at: https://extranet.who.int/prequal/sites/default/files/documents/19_Vercauteren.pdf. Accessed 8 May 2017.
- 20.Jack C, Cuschieri K, Denton K, et al. Performance of the Xpert HPV assay in women attending for cervical screening. Papillomavirus Res. 2015;1:32–37. doi: 10.1016/j.pvr.2015.05.002. [DOI] [Google Scholar]
- 21.Einstein MH, Smith KM, Davis TE, et al. Clinical evaluation of the cartridge-based GeneXpert human Papillomavirus assay in women referred for Colposcopy. J Clin Microbiol. 2014;52(6):2089–2095. doi: 10.1128/JCM.00176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gueudin M, Baron A, Alessandri-Gradt E, Lemée V, Mourez T, Etienne M, Plantier JC. Performance evaluation of the new HIV-1 quantification assay, Xpert HIV-1 viral load, on a wide panel of HIV-1 variants. J Acquir Immune Defic Syndr. 2016;72(5):521–526. doi: 10.1097/QAI.0000000000001003. [DOI] [PubMed] [Google Scholar]
- 23.Guidelines on prevention and management of TB in PLHIV at ART center. Available at: http://tbcindia.nic.in/showfile.php?lid=3253. Accessed 8 May 2017.
- 24.Arnaw K, Sumana MN. Assasying the need of commercial plasma viral load testing in resource limited settings. J Evol Med Dent Sci. 2015;4(Issue 75):13122–13134. [Google Scholar]
- 25.Khopkar P, Mallav V, Chidrawar S, Kulkarni S. Comparative evaluation of the Abbott HIV-1 RealTime™ assay with the standard Roche COBAS® Amplicor™ HIV-1 monitor® test, v1.5 for determining HIV-1 RNA levels in plasma specimens from Pune, India. J Virol Methods. 2013;191(1):82–87. doi: 10.1016/j.jviromet.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 26.Mor O, Gozlan Y, Wax M, Mileguir F, Rakovsky A, Noy B, Mendelson E, Levy I. Evaluation of the RealTime HIV-1, Xpert HIV-1, and Aptima HIV-1 quant Dx assays in comparison to the NucliSens EasyQ HIV-1 v2.0 assay for quantification of HIV-1 viral load. J Clin Microbiol. 2015;53(11):3458–3465. doi: 10.1128/JCM.01806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceffa S, Luhanga R, Andreotti M, et al. Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 real time molecular assays for monitoring HIV-1 viral load and detecting HIV-1 infection. J Virol Methods. 2016;229:35–39. doi: 10.1016/j.jviromet.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 28.AIS guidelines on survey. Available at: http://dhsprogram.com/What-We-Do/Survey-Types/AIS.cfm. Accessed 8 May 2017.
- 29.WHO guidelines on HIV testing. Available at: http://www.who.int/hiv/pub/guidelines/india_art.pdf. Accessed 8 May 2017.
- 30.National guidelines for HIV testing published by NACO. Available at: http://www.naco.gov.in/sites/default/files/National_Guidelines_for_HIV_Testing_21Apr2016.pdf. Accessed 8 May 2017.
- 31.François J. Picard and Michel G. Bergeron. Rapid molecular theranostics in infectious diseases. Ther Focus DDT. 2002;7,No. 21. Available at: http://www.academia.edu/3577512/Rapid_Molecular_Theranostic_in_Infectious_Diseases. Accessed 8 May 2017. [DOI] [PubMed]
- 32.Rouet F, Rouzioux C. The measurement of HIV-1 viral load in resource-limited settings: how and where? Clin Lab. 2007;53(3-4):135–148. [PubMed] [Google Scholar]
- 33.NACO- Assessment of ART Center in India, National Report – September 2015 A document published by NACO. Available at: http://naco.gov.in/sites/default/files/ART%20Assessment%20National%20Report-Final%2028092015.pdf.
- 34.Index-TB Guidelines on extra-pulmonary tuberculosis for India. A document published by ICMR, DOTS, WHO. Available at: http://www.icmr.nic.in/guidelines/TB/Index-TB%20Guidelines%20-%20green%20colour%202594164.pdf.
- 35.Yan CS, Hanafi I, Kelleher AD, et al. Lack of correlation between three commercial platforms for the evaluation of human immunodeficiency virus type 1(HIV-1) viral load at the clinically critical lower limit of quantification. J Clin Virol. 2010;49:249–253. doi: 10.1016/j.jcv.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Sollis KA, Smit PW, Fiscus S, Ford N, Vitoria M, Essajee S, et al. Systematic review of the performance of HIV viral load technologies on plasma samples. PLoS One. 2014;9(2):e85869. doi: 10.1371/journal.pone.0085869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swenson LC, Cobb B, Geretti AM, Harrigan PR, Poljak M, Seguin-Devaux C, Verhofstede C, Wirden M, Amendola A, Boni J, Bourlet T, Huder JB, Karasi JC, ZidovecLepej S, Lunar MM, Mukabayire O, Schuurman R, Tomazic J, Van Laethem K, Vandekerckhove L, Wensing AM. Comparative performances of HIV-1 RNA load assays at lowviral load levels: results of an international collaboration. J Clin Microbiol. 2014;52:517–523. doi: 10.1128/JCM.02461-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muenchhoff M, Madurai S, Hempenstall AJ, Adland E, Carlqvist A, Moonsamy A, Jaggernath M, Mlotshwa B, Siboto E, Ndung’u T, Goulder PJ. Evaluation of the NucliSens EasyQ v2.0 assay in comparison with the Roche Amplicor v1.5 and the Roche CAP/CTM HIV-1 test v2.0in quantification of C-clade HIV-1 in plasma. PLoS One. 2014;9:e103983. doi: 10.1371/journal.pone.0103983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott L, Gous N, Carmona S, Stevens W. Laboratory evaluation of the Liat HIV quant (IQuum) whole-blood and plasma HIV-1 viral load assays for point-of-care testing in South Africa. J Clin Microbiol. 2015;53:1616–1621. doi: 10.1128/JCM.03325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.UNAIDS 90-90-90 Initiative, A document published by UNAIDS. Available at: www.unaids.org/sites/default/files/media_asset/90-90-90_en_0.pdf. Accessed 8 May 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available as in India, raw data can be only shared by institutes under Government of India (GoI) if prior regulatory [Health Ministry’s Screening Committee (HMSC)] approvals are obtained. Therefore we are unable to share the data for this purpose. However, the dataset can be available from the corresponding author on reasonable request.


