Abstract
Protein glycosylation drives many biological processes and serves as markers for disease; therefore the development of tools to study glycosylation is an essential and growing area of research. Mass spectrometry can be used to identify both the glycans of interest and the glycosylation sites to which those glycans are attached, when proteins are proteolytically digested and their glycopeptides are analyzed by a combination of high resolution MS and MS/MS methods. One major challenge in these experiments is collecting the requisite MS/MS data. The digested glycopeptides are often present in complex mixtures and in low abundance, and the most commonly used approach to collect MS/MS data on these species is data dependent acquisition (DDA), where only the most intense precursor ions trigger MS/MS. DDA results in limited glycopeptide coverage. Semi-targeted data acquisition is an alternative experimental approach that can alleviate this difficulty. However, due to the massive heterogeneity of glycopeptides, it is not obvious how to expediently generate inclusion lists for these types of analyses. To solve this problem, we developed the software tool GlycoPep MassList, which can be used to generate inclusion lists for LC-MS/MS experiments. The utility of the software was tested by conducting comparisions between semi-targeted and untargeted data dependent analysis experiments on a variety of proteins, including IgG, a protein whose glycosylation must be characterized during its production as a biotherapeutic. When the GlycoPep MassList software was used to generate inclusion lists for LC-MS/MS experiments, more unique glycopeptides were selected for fragmentation. Generally, ~ 30% more unique glycopeptides can be analyzed per protein, in the simplest cases, with low background. In cases where background ions from proteins or other interferents are high, usage of an inclusion list is even more advantageous. The software is freely publically accessible.
Keywords: N-linked glycosylation, Glycopeptide analysis, Software tool, LC MS/MS, Data dependent acquisition
Introduction
Protein glycosylation is one of the most significant and fundamental post translational modifications (PTMs) in nature [1]. Glycosylation, like other PTMs, leads to the increased diversification of protein structures and functions [2,3]. The most common type of glycosylation, N-linked glycosylation, occurs when the glycan is appended to the side chain of a asparagine following the consensus sequence N-X-T/S, where × is any amino acid except proline [4]. The glycans appended at N-linked glycosylation sites mainly depend on the glycosyltransferase enzyme availability and the microenvironment of the protein [5,6]. The glycans play crucial roles in a variety of biological processes including protein folding [7], protein stabilization [8], immune response [9], cell-environment communication [10], and fertilization [11]. Many times, a particular glycan profile is essential for the glycoprotein to function optimally [9]. Alterations in the glycosylation site, the extent of glycosylation, or the glycosylation profile are associated with a broad spectrum of diseases [12,13], ranging from rheumatoid arthritis [14], Alzheimer’s disease [15] to prostate [16], colorectal [17] and breast cancer [18]. The identification of abnormally glycosylated proteins has biomedical value because these features may serve as disease biomarkers [19–21]. Therefore, in order to gain more understanding in protein structure-function correlations and/or to exploit glycoproteins in biomarker discovery and disease diagnosis, one must be able to characterize the glycosylation on N-linked proteins in an efficient manner.
Analysis of N-linked glycosylation can be done in a variety of ways, but often researchers prefer to obtain the relevant information by analyses of proteolized glycopeptides [22,23]. In these experiments, the glycans remain linked to the site on the protein at which they reside. In this case, the most common workflow involves tryptic digestion of the protein, followed by LC-MS/MS analysis [24]. Past research has demonstrated that in all but the simplest cases, high resolution MS data are not sufficient to accurately identify glycopeptides [25]. MS/MS is necessary to distinguish among various possible glycopeptide assignments for any given MS peak [25,22]. The MS/MS data can be used to determine the glycosylated protein sequence, the glycosylation site, and the glycan composition [26,27].
Collision induced dissociation (CID) is the most commonly used dissociation method for glycopeptide analysis. One of its key advantages is its rapid duty cycle, allowing for data acquisition on multiple co-eluting glycopeptides. However, glycopeptide analysis by CID is still challenging, due to glycopeptides’ intrinsic low abundance. The macroheterogeneity which comes from the difference of glycosylation site occupancy, and the microheterogeneity, which results from the attachment of different glycans to one specific glycosylation site, render each glycoform in a low copy number after proteolysis [28–30]. Consequently, the most commonly used MS/MS data-dependent acquisition (DDA) mode, where the most intense ions in the full MS scan are selected for MS/MS, is not optimal for glycopeptide analyses because the high-abundant precursor ions, which are often non-glycosylated peptides, are redundantly selected for MS/MS, while the relatively low abundant glycopeptide ions may not trigger MS/MS, even if dynamic exclusion is enabled [31]. This experiment results in missed detection of glycopeptides and limited glycopeptide coverage, particularly when the sample has a complex matrix that brings high background corresponding to non-glycosylated peptides.
A significant thrust of research in the area of glycopeptide analysis is, therefore, focusing on the problem of enhancing the number of glycopeptides selected for MS/MS analysis in a given sample. Sample preparation strategies, particularly glycopeptide enrichment, can contribute to this solution by reducing the number of non-glycosylated peptides present in the sample [32,33]. New MS methods are also needed. One such strategy that has not yet been readily adopted, but which theoretically could benefit the field, is a targeted analysis approach, taking advantage of instruments’ ability to selectively conduct MS/MS on ions preloaded onto an inclusion list.
Targeted data acquisition is a well-known strategy that has been used to alleviate the biased ion selection inherent in DDA strategies in other fields, but it has not yet been commonly applied to the field of glycoproteomics. The proteomics field has already demonstrated that targeted data acquisition consumes fewer MS/MS scans on peptides that have high abundance but are not of interest to the investigators [31]. Targeted data acquisition strategies have been successfully employed to study arginine methylation [34], for example. The method, however, does not readily transfer to the field of glycopeptide analysis, primarily because the glycan component on the glycopeptide may be any one of hundreds of different masses. With such a tremendous variety of glycans, it would be a time-consuming process to generate an inclusion glycopeptide mass list every time a new glycoprotein is to be studied.
At least two groups have shown that using inclusions lists for glycopeptide analysis is a promising approach. Yin Wu and colleagues developed a software-based strategy that adds putative glycopeptide ions to an inclusion list for targeted MS/MS experiments [35]. Their strategy relies on using the software GlycoPID to identify glycopeptides in untargeted manner first, then additional ions in the high resolution spectra that may also be glycopeptides are added to an inclusion list on a second, or third, or fourth round of experiments. This iterative fashion of targeted MS/MS experiment clearly achieves enhanced coverage, compared to a single round of untargeted experiments. The disadvantage of this approach is that multiple LC-MS/MS analyses are required for each sample, and the GlycoPID tool must be used for the glycopeptide assignments. GlycoPID is just one of many emerging bioinformatics platforms used for the glycopeptide analysis, and often users may want to use different software (or even manual analysis) to analyze their data. More recently, Froehlich and coworkers developed a mass defect classifier that tentatively identifies potential glycopeptide ions in a first-pass LC-MS analysis, and those ions could then be loaded onto an inclusion list during a re-analysis experiment [36]. This approach may be advantageous to the one built into GlycoPID, because no glycopeptide compositions need to be assigned initially, and users can select any glycopeptide analysis software they choose. This approach still requires an initial data acquisition phase, followed by data analysis, followed by at least one more round of data acquisition.
In the work described herein, we provide a software tool designed to generate inclusion lists for glycopeptides prior to any data acquisition steps. This software allows users to target glycopeptides for MS/MS in a single round of experiments, reducing the sample requirements and analysis time that results from doing multiple LC-MS injections. Any glycopeptide analysis software can be used in conjunction with this tool to interpret the data. The software was designed such that users could input any protein sequence of interest, and the peptides containing N-linked glycosylation sites are coupled to an on-board glycan library, which can be customized by the user, producing appropriate glycopeptide masses that can easily be uploaded into an inclusion list for MS/MS analyses. A preliminary demonstration of the potential application of the software is described, along with some strategies that can potentially maximize the benefit of identifying glycopeptides using this approach.
Materials and Methods
Materials and Reagents
Fetuin from fetal bovine serum, avidin from chicken egg white, and human serum IgG were purchased from Sigma Aldrich (St. Louis, MO). Sequencing grade trypsin was obtained from Promega (Madison, WI). HPLC grade acetonitrile and methanol, ammonium bicarbonate, urea, guanidine hydrochloride (GdnHCl), dithiothreitol (DTT), iodoacetamide (IAM), and formic acid were purchased from Sigma Aldrich (St. Louis, MO). All the reagents were of analytical grade or better and were used without further purification.
Protein Digestion
Between 100 and 400 µg bovine fetuin or chicken avidin was dissolved in 50 mM ammonium bicarbonate buffer (pH 8.0) and denatured by adding urea until the concentration reached 6 M. The disulfides of the denatured proteins were reduced by DTT, which was added to reach a final concentration of 10 mM. The DTT reacted for one hour at room temperature. IAM was added to alkylate the disulfides; its final concentration was 25 mM. It reacted for one hour in the dark at room temperature. The excess IAM was quenched by DTT (final concentration 30 mM) for half an hour at room temperature. Subsequently, the protein solutions were diluted with 50 mM ammonium bicarbonate buffer to reach a urea concentration of 1 M, prior to incubation with trypsin (trypsin/protein, 1/30) at 37°C for 20 hours.
Human IgG, 100 µg, was dissolved in 50 mM ammonium bicarbonate buffer (pH 8.0) and was denatured by the addition of GdnHCl until the solution concentration reached 6 M. The reduction, alkylation and quenching of excess IAM were the same as described above. The resulting protein solution was buffer exchanged with 50 mM ammonium bicarbonate buffer for two times to remove most of the GdnHCl. The buffer exchanged solution was made to a final volume of 100 µL before incubation with trypsin at a trypsin/protein ratio of 1/30 at 37°C for 20 hours. Finally, the digestion was stopped by adding formic acid with a formic acid/digestion solution ratio of 1/100. Each digested protein sample was aliquoted and stored at −20°C until it was analyzed.
LCMS
Glycoprotein samples were separated on a reverse phase C18 capillary column (300 µm i.d. × 5 cm, 100 Å pore size, Micro-Tech, Vista, CA) online using a Waters Acquity high performance liquid chromatography (Milford, MA) prior to mass spectrometric analysis in an LTQ Orbitrap Velos Pro hybrid mass spectrometer (Thermo Scientific, San Jose, CA). About 5 µL of the diluted digestion sample was injected with a mobile phase flow rate of 10 µL/min and gradient elution. Mobile phase A consisted of water with 0.1% formic acid, and mobile phase B consisted of acetonitrile with 0.1% formic acid. For human IgG, the HPLC gradient was as follows: 5% B for 3 min, 5% to 40% B in 37 min, 40% to 90% B in 10 min, 90% B for 10 min, 90% to 5% B in 10 min, and 5% B for 10 min. For the mixture of fetuin and avidin, the same solvents were used, and the HPLC gradient was: 2% B for 5 min, 2% to 45% B in 50 min, 45% to 90% B in 8 min, 90% B for 10 min, 90% to 2% B in 10 min, and 2% B for 10 min. A wash and blank run were applied between each sample to minimize sample carryover.
For mass spectrometric analysis, the positive ion mode was utilized with an ESI source voltage of 3 kV and capillary temperature of 250°C. The MS full scans were at a resolution of 30,000 (for m/z 400). The CID MS/MS scans were collected in a linear ion trap in a data-dependent fashion. The ten most intense precursor ions from inclusion lists (when applicable) or from the MS preview scan were isolated for CID. The parent mass widths for inclusion lists were ±10 ppm. After being selected for MS/MS, with a repeat count of 2 within a repeat duration of 50 s, the precursor ion was dynamically excluded for 180 s. The FTMS had an automatic gain control (AGC) target value of 5e5 with a maximum injection time of 400 ms. For CID MS/MS, the AGC was set with a target value of 104 and a maximum injection time of 50 ms. The isolation mass window for selecting a precursor ion was 2 Da. Normalized collision energy of 30% was applied with an activation time of 10 ms.
The MS and CID data were manually interpreted to obtain the glycopeptide coverage. Every potential glycopeptide ion for each glycoprotein was searched for manually, and its presence was identified when (1) a peak was detected whose m/z was within 10 ppm of the expected m/z; and (2) its retention time matched the retention time of other glycopeptide ions with similar peptide and glycan backbone; and (3) when MS/MS data were available, the product ions were consistent with the glycopeptide.
Software Overview
GlycoPep MassList is a free publicly accessible software tool designed for generation of inclusion lists in targeted analysis of glycopeptides. The theoretical glycopeptide mass computation can be easily performed by specifying the protein sequence, glycan library, charge state, mass range, number of missed cleavages, and the isotope preferred by the users. The software was written in Java (JDK7) and can be run on Windows 7 or newer version of Windows. Java Runtime Environment 7 (JRE 7) is recommended to successfully run the software.
Glycan Library of the Software
To rapidly and effectively generate potential glycopeptide masses, a default glycan library, consisting of 340 glycans, was integrated into the software. These glycans include high mannose, hybrid, and complex glycans that are found in nature. The glycans were categorized into five groups: 1. high mannose and pauci-mannose glycans; 2. complex or hybrid glycans without sialic acids; 3. glycans with NeuAc; 4. glycans with NeuGc 5. glycans with [PO3] or [SO3]. For the sialylated glycans, those containing NeuAc were separated from the ones containing NeuGc because NeuGc cannot be biosynthesized by humans [37]. Thus, when analyzing human derived glycoproteins, NeuGc-containg glycans are not present and do not need to be considered.
Although the native glycan library is extensive and contains biologically relevant glycans from a wide range of sources, it does not cover all the N glycans that are present in nature. Therefore, the software was designed so that users can upload their own glycan libraries. The users can also use one or multiple groups of glycans from the software together with their own glycan libraries.
Implementation of the Software for the Experiments Described Here
The glycopeptide m/z’s that were selected for inclusion list experiments in the examples shown herein were generated using these procedures: (1) No missed cleavages were calculated. (2) It was assumed that the glycopeptide did not have any post translational modification other than glycosylation. (3) The glycopeptide m/z’s from charge state +2 to +8 were considered. (4) The glycopeptide m/z’s in the range of m/z 800 to 2000 were selected. Procedures (1) and (2) were used to reduce the number of entries on inclusion list, yet keep the valuable entries. This increased the specificity of ion selection and decreased the probability of random matches between the masses on the list and the masses detected from the sample background that did not correspond to glycopeptide ions. While we have included the option to calculate glycopeptides with miscleavages, initial testing has shown that this option does not increase the number of unique glycopeptides subjected to MS/MS experiments, particularly when the protein is optimally digested initially. Procedure (3) was applied so that the glycopeptides with a variety of sizes could be identified using one search. The mass range (m/z 800 to 2000) was selected based on our experience on MS-based glycopeptide analysis: Almost all glycopeptides are observed in at least one charge state above m/z 800. While these procedures may not be universal best practices, depending on the experiment to be performed, they demonstrated themselves to be advantageous in numerous test cases we ran. In addition to these procedures, we observed that it is worthwhile to select the precursor ion type carefully. The higher abundance 13C mass of the glycopeptide was used instead of the lower abundance monoisotopic mass. In the experiments using human IgG, the first 13C masses were used. In the experiments using a fetuin and avidin mixture, the second 13C masses were used. These isotopes were selected based on the size of the peptide. The larger the peptide, the more likely that a higher isotope will be the most abundant peak in the isotopic cluster.
Results and Discussion
Software Development and Implementation
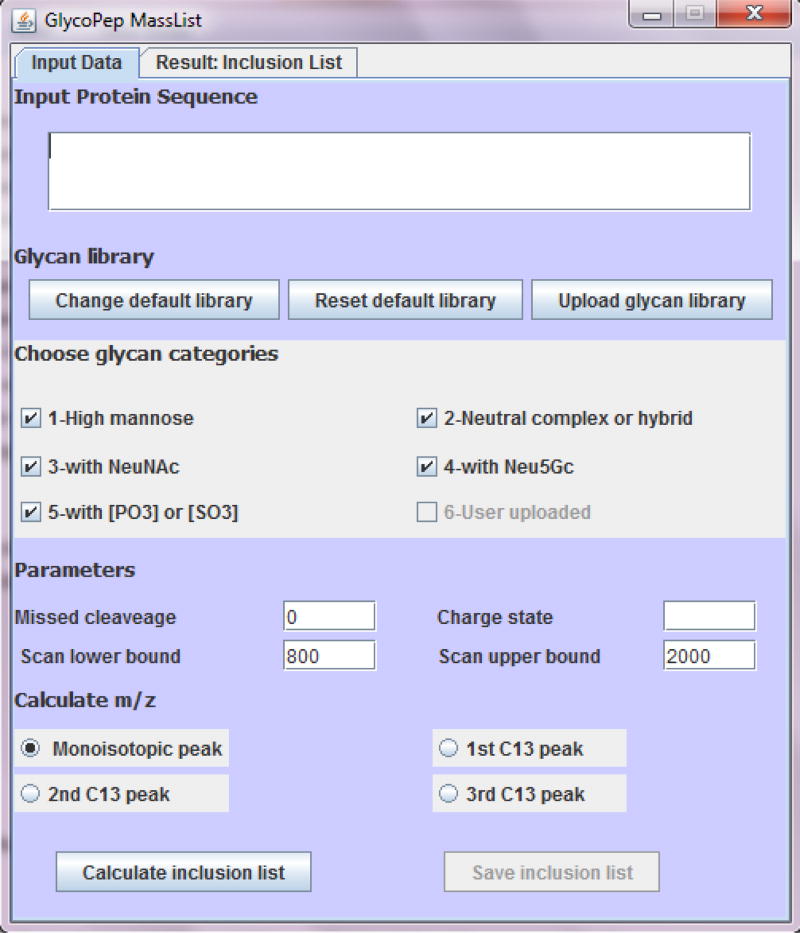
The software described herein is a free tool to calculate theoretical glycopeptide masses and generate inclusion lists for targeted data acquisition on glycopeptides. A screenshot of the user interface is shown in Fig. 1. In order to calculate the glycopeptide mass, users input the required information such as protein sequence, glycan library, number of missed cleavages (up to 3), charge state (from 1 to 8) and mass range (400 to 2000). The users can also choose to calculate monoisotopic mass or the mass of one of the higher isotopes (up to the third 13C mass), depending on the glycopeptide of interest.
Fig. 1.
User interface for GlycoPep MassList. Users input the protein sequence, glycan library, charge state, mass range, number of missed cleavages, and select the mass (monoisotopic mass or 13C mass) to be calculated. In silico tryptic digestion is then performed on the protein and all the potential glycosylated peptides are reported by the software. The output is displayed under “Result: Inclusion List”. One or multiple groups of glycans can be selected, or custom glycan libraries can be uploaded, or a combination of manually input libraries and those in the database can be used
After the appropriate data are input, the software executes an in silico tryptic digestion on the protein sequence, and all the peptides that have a potential N-linked glycosylation site (NXS/T, X≠P) are extracted. In the current form of the software, cysteines are, by default, considered to be alkylated by iodoacetamide (IAM), a widely used alkylation reagent in protein digestion. Next, theoretical glycopeptide m/z’s, based on all the combinations of the glycosylated peptides and the user-specified glycans, are computed. The results are displayed under “Result: Inclusion List”. Each theoretical glycopeptide appears on one line, with the glycopeptide composition shown on the left and m/z on the right.
After the software was developed and carefully tested, two sets of CID experiments were conducted, in order to compare the performance of a semi-targeted data acquisition strategy (using the inclusion list) and the conventionally used data dependent acquisition strategy (without inclusion lists), where the top 10 most intense precursor ions are selected for MS/MS.. The two experiments are henceforth referred to as the “Inclusion” experiment, or the “Top 10” experiment, respectively.
Analysis of Human IgG
Plasma-derived human IgG is an important class of antibody [38–40]. The glycosylation site located at the Fc region has an effect on the interaction of IgG with Fc gamma receptor (FcγR) [41]. Aberrant glycosylation on this site is related to diseases [42]. For example, a lower degree of galactosylation was observed in patients with rheumatoid arthritis [8]. When comparing the IgG glycosylation profiles of diseased and healthy states, one must identify all glycoforms present, otherwise the glycosylation differences, due to the samples themselves, will not be known. IgG is also a very important recombinantly expressed biotherapeutic [43]. The efficacy and side effect of IgG therapeutics are closely related to their glycosylation [43,44]. For instance, antibodies with high mannose glycans are cleared faster in human serum [45]. Glycans containing NeuGc can cause an unwanted immune response [44]. Thus, it is important to ensure the glycan profile fidelity across different expression vehicles, conditions, and batches.
Hence, human IgG was used to compare the efficiency of the new “Inclusion List” software and strategy, to the traditional, untargeted, Top 10 approach. Utilizing the software for the Inclusion experiment requires three steps: Choosing an appropriate glycan library, building the inclusion list, and then conducting the LC-MS experiment. Each step is elaborated upon, briefly, here. When the software is used for other applications, a similar workflow should be followed.
An appropriate glycan list must be chosen. The N linked glycopeptides from IgG Fc region mainly contain complex-type biantennary glycans. These glycans are mostly core-fucosylated; they have up to two galactose residues; they may be sialylated and bisected by an N-acetylglucosamine (GlcNAc) residue [46]. We compiled a glycan library ideal for IgG analyses by researching the literature associated with human IgG glycosylation [45,38–40,47]. The glycan list for these experiments contains 52 glycans, and it can be found in Supporting Information Table 1. All these glycans are either reported previously for IgG or are reasonable additional glycans that may be present, based on the rules of glycotransferase processing.
After building the glycan library, which can now be used for any IgG experiment, GlycoPep MassList was used to generate the inclusion masses for the targeted data acquisition. The IgG protein sequence and the compiled glycan library for human IgG were uploaded to obtain the theoretical glycopeptide masses. Since EEQYNSTYR and EEQFNSTFR glycopeptides are commonly observed in tryptic digests of IgG, they were both included. To compare the Inclusion experiment with the commonly used Top 10 experiment, LC-MS runs with an inclusion list and without the inclusion list were performed back-to-back on the same day using the same digested protein sample. In the Inclusion experiment, the inclusion list, populated with the first 13C masses of the theoretical glycopeptides, was imported into the instrument software, while in the Top 10 experiment, no inclusion list was applied.
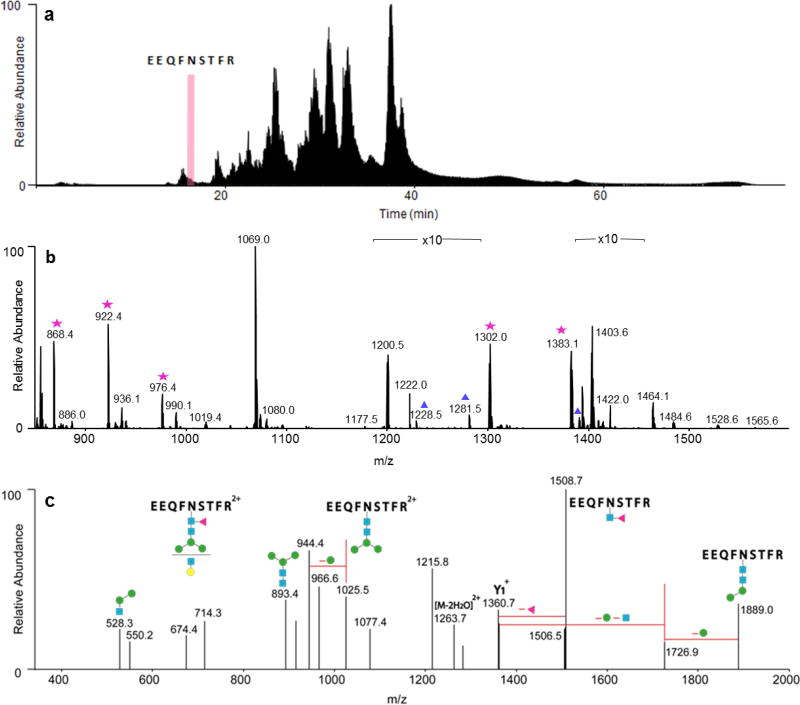
The resulting MS data are depicted in Fig. 2. The tryptic glycopeptides EEQYNSTYR and EEQFNSTFR eluted at about 6.5 min and 16.5 min, respectively. The elution of EEQFNSTFR is indicated by a pink bar on the total ion chromatogram (TIC) in Fig. 2a. The MS data for this glycopeptide rich region of the chromatogram, containing multiple glycopeptides from the EEQFNSTFR site is shown in Fig. 2b. The high abundance glycopeptides in this spectrum, labeled with stars, were selected for CID in both the Inclusion and Top 10 experiments, while the low abundance ones, labeled with triangles, were only selected for CID in the Inclusion experiment.
Fig. 2.
Example data from Inclusion experiment. a Ion chromatogram of the IgG glycoprotein digest, indicating where one of the peptides, EEQFNSTFR, elutes. b MS data for the highlighted region in a. Stars: Glycopeptide selected for MS/MS by Inclusion and Top 10. Triangles: Glycopeptides selected for MS/MS only by Inclusion but not by Top 10. c The CID spectrum of a glycopeptide that was only selected by Inclusion. The ion, m/z 1281.52, corresponds to EEQFNSTFR+[Hex]4[HexNAc]3Fuc]1. Its composition can be confirmed by the product ions in c. Blue squares: N-acetylhexosamines. Red triangle: fucose. Green and yellow circles: hexoses
One glycopeptide identified only by Inclusion but not Top 10 is shown in Fig. 2c. This low abundance glycopeptide was only observed in the +2 charge state in the high resolution MS data. Because of its relatively low abundance, this precursor ion was not selected for CID in the Top 10 experiment. Nevertheless, in the Inclusion experiment, this ion was selected for CID. As clearly shown in Fig. 2c, the CID data still produced sufficient data to confirm the glycopeptide assignment.
All the detected glycopeptides, selected by Inclusion and/or Top 10, were identified. The data for each experiment are fully reported in Supplemental Information Table 2. Most of the detected glycopeptides from each site were core fucosylated. A minor portion of them were sialylated. The sialylated glycopeptides eluted about one minute later than the non-sialylated ones, which agreed with previous reports [39].
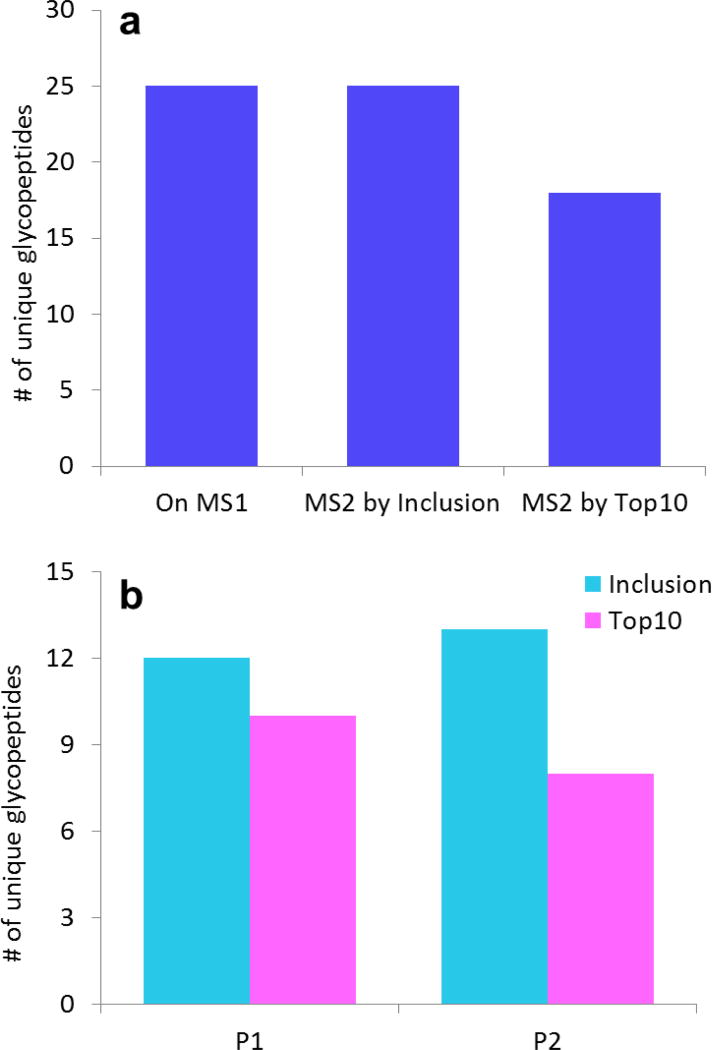
The glycopeptide coverage summary for the Inclusion and Top 10 experiments is shown in Fig. 3. It is worth noting from Fig. 3a that all the unique glycopeptides observed in the high resolution spectrum with ion intensities more than 200 counts (the signal threshold for CID) were selected for CID by the Inclusion experiment. Additionally, Inclusion outperformed Top 10 by selecting seven more unique glycopeptides for CID, which contributed to a notable 39% higher glycopeptide coverage.
Fig. 3.
IgG glycopeptides selected for CID by Inclusion or Top 10 experiments. a Comparison of the total number of unique glycopeptides observed in the high resolution MS data vs. those that were selected for CID during Inclusion or Top 10. Top 10 got 100% coverage and selected seven more glycopeptides (39% more) than Top 10. b The number of unique glycopeptides from each site selected for MS/MS by Inclusion and Top 10. P1=EEQYNSTYR. P2=EEQFNSTFR. For P2, a greater advantage was observed because the chromatogram had more co-eluting interferents in this region that were selected for MS/MS by the Top 10 experiment
The number of unique glycopeptides from each site selected for CID by the Inclusion and Top 10 experiments are shown in Fig. 3b. Inclusion showed higher efficiency than Top 10 on both glycosylated peptides analyzed. The Inclusion approach was especially advantageous on the site, EEQFNSTFR. In this case, Top 10 selected only 8 unique glycopeptides for MS/MS, while Inclusion selected 13, showing a prominent advantage of 62% higher coverage. Although the advantage was narrower on site, EEQYNSTYR, where 12 unique glycopeptides were triggered CID by Inclusion, and 10 by Top 10, Inclusion still showed 20% higher coverage.
The reason the Inclusion approach was more advantageous on one of these two sites is likely related to what else was co-eluting when the glycopeptides were being selected for CID. The glycopeptide, EEQYNSTYR, eluted at about 6.5 min, while there was a limited number of non-glycosylated peptides co-eluting. The limited background from the non-glycosylated peptides allowed the traditional Top 10 approach to pick up almost all of the glycopeptides for CID. The other glycopeptide, EEQFNSTFR, eluted at about 16.5 min, when the non-glycosylated peptides also started eluting. As the non-glycosylated peptides both outnumbered the glycopeptides, and were present in higher abundance, the efficiency of the traditional Top 10 experiment was substantially reduced: More duty cycle was wasted on the background peaks instead of glycopeptide peaks. On the contrary, the Inclusion experiment was able to prioritize the duty cycle on the highly possible glycopeptide peaks pre-assigned on the inclusion list. This resulted in the Inclusion experiment working especially well for the EEQFNSTFR peptide, compared to EEQYNSTYR.
Targeted Data Analysis of Fetuin/Avidin Mixture
To further test the utility of the use of targeted inclusion lists for glycopeptide, we used a more complex sample involving two proteins with multiple glycosylation sites. More specifically, a mixture that had 15 pmol fetuin and 1.7 pmol avidin was used. This glycoprotein mixture was chosen for two reasons. First, these well studied glycoproteins have very different glycosylation profiles. Fetuin has complex type glycans, while avidin has mainly high mannose and hybrid type glycans [48]. Hence, the glycan library for this experiment incorporated all the three types of glycans. Secondly, the avidin protein was purposely added in a low abundance, since identifying low-abundant glycopeptides is a particularly large challenge in the field of glycoproteomics. We were interested in determining how the Inclusion and Top 10 strategies compared when the glycoprotein of interest was a small component of the overall sample.
In this experiment, the glycan library was designed differently from the first set of experiments as well. Unlike the case of human IgG, where most glycans of the glycan library are highly possible oligosaccharides for human IgG, most glycans in this library did not correspond to the ones in the glycoprotein sample used. Consequently, most m/z’s on the inclusion list did not correspond to the glycopeptide m/z’s in the sample. The goal of this set of experiments was to begin to understand the impact of using large inclusion lists; could these lists still select more glycopeptide ions for MS/MS than the commonly used Top 10 approach?
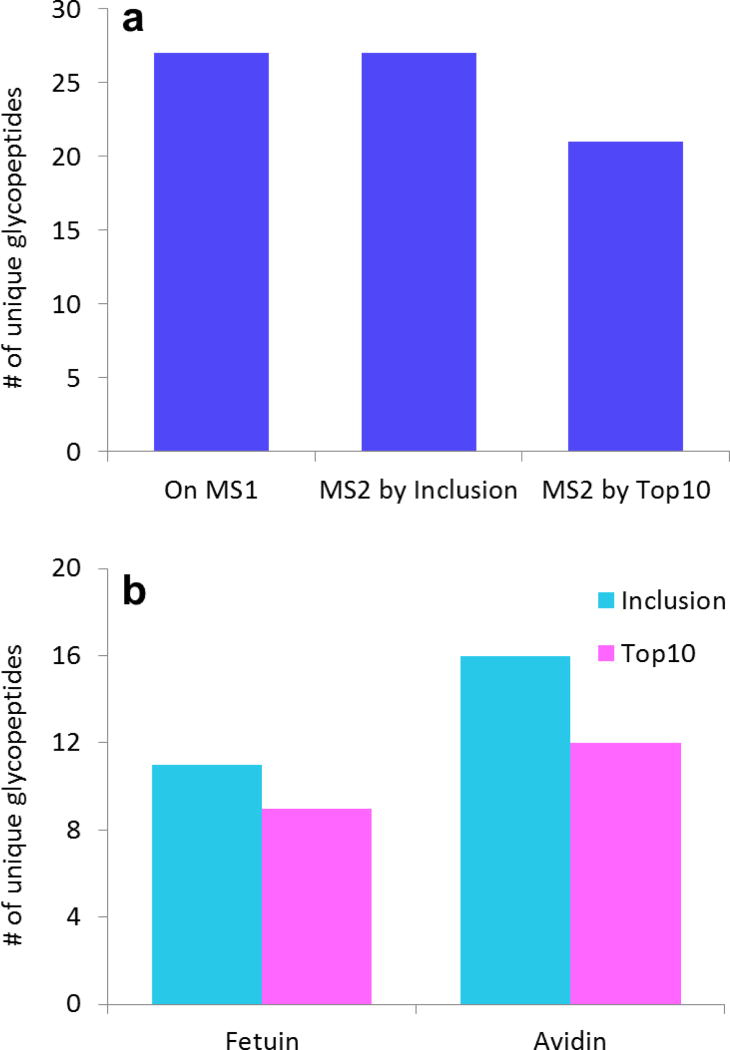
The glycopeptide coverage summary for the Inclusion and Top 10 experiments is shown in Fig. 4. Fig. 4a summarizes the complete glycopeptide coverage for all the glycoforms at all the glycosylation sites, and, as expected, the Inclusion approach was superior to the Top 10 experiment in selecting more glycopeptide ions for CID. Similar to the results of human IgG, all the 27 unique glycopeptides observed in the high resolution mass data that had ion intensities above the signal threshold for CID were selected by Inclusion for MS/MS experiments, while only 21 unique peptides were selected by Top 10. This difference corresponds to a 29% increase in coverage.
Fig. 4.
Result summary from the fetuin/avidin mixture. a All the unique glycopeptides observed in the high resolution spectrum were selected for MS/MS by Inclusion. Inclusion had six more unique glycopeptides (29% more) selected for MS/MS than Top 10. b The number of unique glycopeptides from each protein selected for MS/MS by Inclusion and Top 10. The Inclusion approach was most efficacious for avidin because the Top 10 experiments were less effective when many glycopeptides were present in a high background
To understand the circumstances that benefit the Inclusion approach, we also compared the results for each protein separately. These are displayed in Fig. 4b. In this figure, one can clearly see that the Inclusion approach was most beneficial for the more complex, lower-abundant protein, avidin. For avidin, 16 unique glycopeptides were selected for MS/MS in Inclusion, compared to 12 unique ones for Top 10, representing a 33% higher coverage. For fetuin, Inclusion was still superior, but the gain was smaller. Here, Inclusion vs. Top 10 resulted in 11 unique glycopeptides vs. 9 unique glycopeptides, a 22% higher coverage.
The results for the fetuin/avidin experiments are consistent with those from IgG. When glycopeptides are present in lower abundance, as is the case in the avidin experiment, or when other non-glycosylated peptides are co-eluting, as is the case in the IgG experiment, the Inclusion approach shows its greatest benefit. These findings are not terribly surprising; in fact, we fully expected this to be the case because Top 10 experiments are well-known to leave out MS/MS data on low-abundant species. What is somewhat surprising is that the Inclusion approach worked much better, even on relatively simple samples. These findings suggest that Inclusion approach will show an even greater benefit when challenged with more complex samples, such as low-abundant glycopeptides present in high background of interfering ions.
For the experiments described here, the number of ions added to the inclusion list ranged between 233 ions for the IgG experiments to 1507 ions for the experiments where both fetuin and avidin were simultaneously examined. Clearly, the large number of ions on the inclusion list, compared to other more targeted approaches, did not limit the instrument’s ability to pick all the relevant glycopeptides for MS/MS analysis. One key reason for the success of these experiments is the fact that we used a relatively small mass width (10 ppm) for the ions on the inclusion list. The mass width (in Da) multiplied by the number of ions on the inclusion list roughly determines the amount of spectral space that is being queried in a given experiment. For example, with 1500 ions on the inclusion list, and a selection criterion of +/− 10 ppm, the spectral space being queried is approximately 0.03 Da multiplied by 1500 ions, or 45 Da, in the range of m/z 800–2000. In other words, about four percent of the available spectral points between m/z 800 and m/z 2000 are being queried under these circumstances. Either increasing the mass tolerance for the ions on the inclusion list (to larger than 10 ppm) or increasing the number of ions queried, increases the percentage of the spectral space being queried; therefore, these changes can decrease the value of using an inclusion list. The instrument control software on the mass spectrometer used for these studies limits the inclusion list to 2000 ions, so there is also a fixed limit to the number of ions that can be queried.
Conclusion
MS/MS data is necessary to accurately identify glycopeptides. Nonetheless, due to the relatively low signals of glycopeptides in MS scans, many glycopeptide ions are not selected for MS/MS during a single DDA experiment. This problem prompted us to develop the software, GlycoPep MassList, to facilitate Inclusion experiments for glycopeptide analysis. This software tool can rapidly generate inclusion lists for glycoproteins, so that targeted glycopeptide analyses can be performed.
To test the application of this software and the Inclusion strategy, two experiments were conducted. In both experiments, the Inclusion strategy outperformed the traditional Top 10 experiment by substantial margins. Furthermore, the experiments herein demonstrated that the Inclusion approach is particularly advantageous when the glycoprotein of interest is present in low abundance, and/or when it co-elutes with many non-glycosylated peptides, and/or when a variety of glycoforms are appended at the same glycosylation site. While the software was tested in several experiments where CID data were collected, the tool is agnostic towards the type of dissociation method used, and it could readily be applied to trigger ETD or HCD experiments as well.
The GlycoPep MassList software is freely available to any interested researchers.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01GM103547 to Heather Desaire.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Dwek RA. Glycobiology: Toward understanding the function of sugars. Chem Rev. 1996;96(2):683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 2.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17(6):666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 3.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat Methods. 2005;2(11):817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 4.Harvey DJ. Proteomic analysis of glycosylation: structural determination of N- and O-linked glycans by mass spectrometry. Expert Rev Proteomics. 2005;2(1):87–101. doi: 10.1586/14789450.2.1.87. [DOI] [PubMed] [Google Scholar]
- 5.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291(5512):2351–2356. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 6.Go EP, Rebecchi KR, Dalpathado DS, Bandu ML, Zhang Y, Desaire H. GlycoPep DB: a tool for glycopeptide analysis using a “smart search”. Anal Chem. 2007;79(4):1708–1713. doi: 10.1021/ac061548c. [DOI] [PubMed] [Google Scholar]
- 7.Wormald MR, Dwek RA. Glycoproteins: glycan presentation and protein-fold stability. Structure. 1999;7(7):R155–R160. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 8.Nagae M, Yamaguchi Y. Function and 3D structure of the N-glycans on glycoproteins. Int J Mol Sci. 2012;13(7):8398–8429. doi: 10.3390/ijms13078398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolarich D, Lepenies B, Seeberger PH. Glycomics, glycoproteomics and the immune system. Curr Opin Chem Biol. 2012;16(1–2):214–220. doi: 10.1016/j.cbpa.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Leymarie N, Griffin PJ, Jonscher K, Kolarich D, Orlando R, McComb M, Zaia J, Aguilan J, Alley WR, Altmann F, Ball LE, Basumallick L, Bazemore-Walker CR, Behnken H, Blank MA, Brown KJ, Bunz S-C, Cairo CW, Cipollo JF, Daneshfar R, Desaire H, Drake RR, Go EP, Goldman R, Gruber C, Halim A, Hathout Y, Hensbergen PJ, Horn DM, Hurum D, Jabs W, Larson G, Ly M, Mann BF, Marx K, Mechref Y, Meyer B, Moeginger U, Neusuess C, Nilsson J, Novotny MV, Nyalwidhe JO, Packer NH, Pompach P, Reiz B, Resemann A, Rohrer JS, Ruthenbeck A, Sanda M, Schulz JM, Schweiger-Hufnagel U, Sihlbom C, Song E, Staples GO, Suckau D, Tang H, Thaysen-Andersen M, Viner RI, An Y, Valmuv L, Wada Y, Watson M, Windwarder M, Whittal R, Wuhrer M, Zhu Y, Zou C. Interlaboratory study on differential analysis of protein glycosylation by mass spectrometry: the ABRF glycoprotein research multi-institutional study 2012. Mol Cell Proteomics. 2013;12(10):2935–2951. doi: 10.1074/mcp.M113.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grass J, Pabst M, Chang M, Wozny M, Altmann F. Analysis of recombinant human follicle-stimulating hormone (FSH) by mass spectrometric approaches. Anal Bioanal Chem. 2011;400(8):2427–2438. doi: 10.1007/s00216-011-4923-5. [DOI] [PubMed] [Google Scholar]
- 12.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5(7):526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 13.Gornik O, Lauc G. Glycosylation of serum proteins in inflammatory diseases. Dis Markers. 2008;25(4–5):267–278. doi: 10.1155/2008/493289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saroha A, Biswas S, Chatterjee BP, Das HR. Altered glycosylation and expression of plasma alpha-1-acid glycoprotein and haptoglobin in rheumatoid arthritis. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879(20):1839–1843. doi: 10.1016/j.jchromb.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Perdivara I, Deterding LJ, Cozma C, Tomer KB, Przybylski M. Glycosylation profiles of epitope-specific anti-beta-amyloid antibodies revealed by liquid chromatography-mass spectrometry. Glycobiology. 2009;19(9):958–970. doi: 10.1093/glycob/cwp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimura T, Shinohara Y, Tissot B, Pang PC, Kurogochi M, Saito S, Arai Y, Sadilek M, Murayama K, Dell A, Nishimura S, Hakomori SI. Glycosylation status of haptoglobin in sera of patients with prostate cancer vs. benign prostate disease or normal subjects. Int J Cancer. 2008;122(1):39–49. doi: 10.1002/ijc.22958. [DOI] [PubMed] [Google Scholar]
- 17.Qiu Y, Patwa TH, Xu L, Shedden K, Misek DE, Tuck M, Jin G, Ruffin MT, Turgeon DK, Synal S, Bresalier R, Marcon N, Brenner DE, Lubman DM. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J Proteome Res. 2008;7(4):1693–1703. doi: 10.1021/pr700706s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake PM, Schilling B, Niles RK, Prakobphol A, Li B, Jung K, Cho W, Braten M, Inerowicz HD, Williams K, Albertolle M, Held JM, Iacovides D, Sorensen DJ, Griffith OL, Johansen E, Zawadzka AM, Cusack MP, Allen S, Gormley M, Hall SC, Witkowska HE, Gray JW, Regnier F, Gibson BW, Fisher SJ. Lectin chromatography/mass spectrometry discovery workflow identifies putative biomarkers of aggressive breast cancers. J Proteome Res. 2012;11(4):2508–2520. doi: 10.1021/pr201206w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10:473–510. doi: 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Dube DH, Bertozzi CR. Glycans in cancer and inflammation-potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg D, Bern M, Parry S, Sutton-Smith M, Panico M, Morris HR, Dell A. Automated N-glycopeptide identification using a combination of single- and tandem-MS. J Proteome Res. 2007;6(10):3995–4005. doi: 10.1021/pr070239f. [DOI] [PubMed] [Google Scholar]
- 23.Desaire H. Glycopeptide analysis, recent developments and applications. Mol Cell Proteomics. 2013;12(4):893–901. doi: 10.1074/mcp.R112.026567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalpathado DS, Desaire H. Glycopeptide analysis by mass spectrometry. Analyst. 2008;133(6):731–738. doi: 10.1039/b713816d. [DOI] [PubMed] [Google Scholar]
- 25.Desaire H, Hua D. When can glycopeptides be assigned based solely on high-resolution mass spectrometry data? Int J of Mass Spectrom. 2009;287(1–3):21–26. [Google Scholar]
- 26.Mechref Y. Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr Protoc Protein Sci. 2012;12(11):1–11. doi: 10.1002/0471140864.ps1211s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan L, Liu M. CID, ETD and HCD fragmentation to study protein post-translational modifications. Mod Chem & Appl. 2012;1(1):e102. [Google Scholar]
- 28.Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11(3):1958–1968. doi: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Glinskii OV, Glinsky VV. Glycobioinformatics: current strategies and tools for data mining in MS-based glycoproteomics. Proteomics. 2013;13(2):341–354. doi: 10.1002/pmic.201200149. [DOI] [PubMed] [Google Scholar]
- 30.Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9(8):747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt A, Gehlenborg N, Bodenmiller B, Mueller LN, Campbell D, Mueller M, Aebersold R, Domon B. An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol Cell Proteomics. 2008;7(11):2138–2150. doi: 10.1074/mcp.M700498-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu T, Qian W-J, Gritsenko MA, II DGC, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4(6):2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madera M, Mechref Y, Novotny MV. Combining lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides. Anal Chem. 2005;77(13):4081–4090. doi: 10.1021/ac050222l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hart-Smith G, Low JK, Erce MA, Wilkins MR. Enhanced methylarginine characterization by post-translational modification-specific targeted data acquisition and electron-transfer dissociation mass spectrometry. J Am Soc Mass Spectrom. 2012;23(8):1376–1389. doi: 10.1007/s13361-012-0417-8. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Mechref Y, Klouckova I, Mayampurath A, Novotny MV, Tang H. Mapping site-specific protein N-glycosylations through liquid chromatography/mass spectrometry and targeted tandem mass spectrometry. Rapid Commun Mass Spectrom. 2010;24(7):965–972. doi: 10.1002/rcm.4474. [DOI] [PubMed] [Google Scholar]
- 36.Froehlich JW, Dodds ED, Wilhelm M, Serang O, Steen JA, Lee RS. A classifier based on accurate mass measurements to aid large scale, unbiased glycoproteomics. Mol Cell Proteomics. 2013;12(4):1017–1025. doi: 10.1074/mcp.M112.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varki A. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol. 2001;116(33):54–69. doi: 10.1002/ajpa.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F. Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics. 2008;8(14):2858–2871. doi: 10.1002/pmic.200700968. [DOI] [PubMed] [Google Scholar]
- 39.Selman MHJ, Derks RJE, Bondt A, Palmblad M, Schoenmaker B, Koeleman CAM, van de Geijn FE, Dolhain RJEM, Deelder AM, Wuhrer M. Fc specific IgG glycosylation profiling by robust nano-reverse phase HPLC-MS using a sheath-flow ESI sprayer interface. J Proteomics. 2012;75(4):1318–1329. doi: 10.1016/j.jprot.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Zauner G, Selman MHJ, Bondt A, Rombouts Y, Blank D, Deelder AM, Wuhrer M. Glycoproteomic analysis of antibodies. Mol Cell Proteomics. 2013;12(4):856–865. doi: 10.1074/mcp.R112.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. 2014;5:1–17. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shade K-T, Anthony R. Antibody glycosylation and inflammation. Antibodies. 2013;2(3):392–414. [Google Scholar]
- 43.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8(3):226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 44.Ghaderi D, Taylor RE, Padler-Karavani V, Diaz S, Varki A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat Biotechnol. 2010;28(8):863–867. doi: 10.1038/nbt.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah B, Jiang XG, Chen L, Zhang Z. LC-MS/MS peptide mapping with automated data processing for routine profiling of N-glycans in immunoglobulins. J Am Soc Mass Spectrom. 2014;25(6):999–1011. doi: 10.1007/s13361-014-0858-3. [DOI] [PubMed] [Google Scholar]
- 46.Reusch D, Haberger M, Maier B, Maier M, Kloseck R, Zimmermann B, Hook M, Szabo Z, Tep S, Wegstein J, Alt N, Bulau P, Wuhrer M. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles--part 1: separation-based methods. MAbs. 2015;7(1):167–179. doi: 10.4161/19420862.2014.986000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wuhrer M, Stam JC, van de Geijn FE, Koeleman CA, Verrips CT, Dolhain RJ, Hokke CH, Deelder AM. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics. 2007;7(22):4070–4081. doi: 10.1002/pmic.200700289. [DOI] [PubMed] [Google Scholar]
- 48.Oliver RWA, Greent BN, Harvey DJ. The use of electrospray ionization MS to determine the structure of glycans in intact glycoproteins. Biochem Soc Trans. 1996;24(3):917–927. doi: 10.1042/bst0240917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.