Abstract
Objective
Categorization and risk stratification of endometrial carcinomas is inadequate; histomorphologic assessment shows considerable interobserver variability, and risk of metastases and recurrence can only be derived after surgical staging. We have developed a Proactive Molecular Risk classification tool for Endometrial cancers (ProMisE) that identifies four distinct prognostic subgroups. Our objective was to assess whether molecular classification could be performed on diagnostic endometrial specimens obtained prior to surgical staging and its concordance with molecular classification performed on the subsequent hysterectomy specimen.
Methods
Sequencing of tumors for exonuclease domain mutations (EDMs) in POLE and immunohistochemistry for mismatch repair (MMR) proteins and p53 were applied to both pre- and post-staging archival specimens from 60 individuals to identify four molecular subgroups: MMR-D, POLE EDM, p53 wild type, p53 abn(abnormal). Three gynecologic subspecialty pathologists assigned histotype and grade to a subset of samples. Concordance of molecular and clinicopathologic subgroup assignments were determined, comparing biopsy/curetting to hysterectomy specimens.
Results
Complete molecular and pathologic categorization was achieved in 57 cases. Concordance metrics for pre- vs. post-staging endometrial samples categorized by ProMisE were highly favorable; average per ProMisE class sensitivity(0.9), specificity(0.96), PPV(0.9), NPV(0.96) and kappa statistic 0.86(95%CI, 0.72-0.93), indicating excellent agreement. We observed the highest level of concordance for ‘p53 abn’ tumors, the group associated with the worst prognosis. In contrast, grade and histotype assignment from original pathology reports pre- vs. post-staging showed only moderate levels of agreement (kappa=0.55 and 0.44 respectively); even with subspecialty pathology review only moderate levels of agreement were observed.
Conclusion
Molecular classification can be achieved on diagnostic endometrial samples and accurately predicts the molecular features in the final hysterectomy specimens, demonstrating concordance superior to grade and histotype . This biologically relevant information, available at initial diagnosis, has the potential to inform management (surgery, adjuvant therapy) from the earliest time point in cancer care.
Introduction
Endometrial carcinoma (EC) is the most common gynecologic malignancy worldwide, increasing globally in both incidence and mortality [1-4]. Histotype and grade assignment in EC is unreliable, even among expert pathologists [5-8], leading to inconsistent categorization of tumors within and between cancer centers. Current risk stratification systems used to guide adjuvant therapy are based on these irreproducible histomorphologic features. Additionally, tumor stage can only be assigned after definitive surgery (including hysterectomy and loss of child bearing capacity). For the approximately 14% of women diagnosed with EC under the age of 50 [9], who may be interested in fertility-sparing alternatives, this information comes too late. However all EC patients and not just these younger individuals would benefit from accurate prognostication to determine personalized treatment options (aggressiveness of surgery, chemotherapy, radiation). Our current system is inadequate; patient management, interpretation of clinical trials, and EC research have been hindered by these shortcomings.
There is a need for improved EC subgroup assignment and risk assessment. The Cancer Genome Atlas (TCGA) [10] applied array-based and sequencing methodologies on a large series of endometrioid and serous ECs, and identified four molecular subgroups of EC that were associated with differences in progression free survival. Subsequently, our group and others [11, 12] have demonstrated that pared down pragmatic assays applicable in routine diagnostic practice can be used to identify four molecular subgroups. Although not identical to TCGA categorization, there is significant overlap and these subgroups are also strongly associated with outcomes. In this study we sought to determine whether our new classifier (Proactive Molecular Risk classification tool for Endometrial cancers (ProMisE)) could be applied to endometrial biopsy or curetting specimens containing endometrial cancer that were obtained for diagnostic purposes, and if classification of these samples was concordant with final hysterectomy endometrial samples obtained at definitive surgical staging.
Methods
Cohort selection
To determine an appropriate cohort selection, an a priori power calculation was performed using the distribution of molecular subgroups in the TCGA (∼7% POLE (ultramutated), 28% MSI-high, 39% CNlow and 26% CN-high), to reveal that a sample of size n=47 would be sufficiently large to detect concordance between pre- and post-staging endometrial samples greater than 0.65 (Power = 0.8, α=0.05). Previous studies [13-16] have demonstrated that it is common for grade assignment to change between diagnostic (pre-) and final (post- surgical staging) endometrial specimens (κ=0.65); therefore, we considered the molecular classification tool (ProMisE) to be clinically useful if it improved upon this figure. In order to account for a potential loss of cases due to molecular test failure, we selected 60 women with EC where both diagnostic (pre-) and hysterectomy (post-staging) endometrial specimens were available. With Institutional Review Board approval, we identified 40 cases from our previously described EC hysterectomy cohort [11] that had undergone molecular classification with the ProMisE tool, based on the hysterectomy specimen, for whom there were available pre-surgical staging samples (endometrial biopsies or curettage specimens) that had not undergone molecular classification. These initial 40 cases were selected to ensure representation from all four molecular subgroups. We additionally identified 20 recent cases of EC where both diagnostic and final endometrial specimens were available; for these cases there was no prior knowledge of molecular subgroup. Hysterectomies performed after neoadjuvant treatment were excluded from the study to ensure that there was not disagreement between samples secondary to treatment-induced molecular changes.
The ProMisE molecular classification scheme was used to assign EC specimens (both diagnostic and final hysterectomy within the same individual) to one of four molecular subgroups using methodologies previously described [11, 17]. Testing involved sequential assessment of i) IHC for MMR proteins MLH1, MSH2, MSH6 and PMS2 ii) sequencing for polymerase epsilon (POLE) exonuclease domain mutations (EDMs), and iii) p53 IHC (Figure 1). Agreement of the molecular classification (ProMisE) was then compared between pre- and post-surgical staging specimens.
Figure 1.
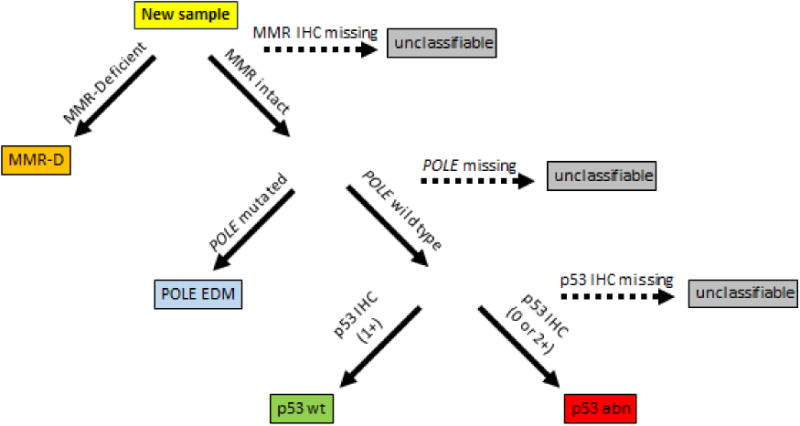
New endometrial cancer samples are tested and categorized according to the above steps; 1st immunohistochemistry (IHC) for the presence of mismatch repair proteins (MLH1, MSH2, MSH6, PMS2) where cases with loss of protein expression classified as MMR deficient (MMR-D). Second, sequencing for the presence of POLE exonuclease domain mutations (POLE EDM). Third, IHC for p53 to distinguish normal expression (IHC score 1) associated with wild type (p53 wt) from null/loss of function mutations (IHC score 0) or missense/gain of function mutations (IHC 2) grouped together as p53 abn.
TMA construction
For all diagnostic endometrial samples (endometrial biopsy, endometrial curettage specimens), a tissue microarray was constructed using 0.6 mm cores in duplicate.
Immunohistochemistry
Methodological details regarding IHC for mismatch repair proteins (MLH1, MSH2, MSH6, PMS2) and for p53 have previously been described [11, 18]. In cases with equivocal or uninterpretable immunohistochemical results based on the TMA slides, immunohistochemistry was repeated on full sections. Scoring was performed by one of three pathologists (CBG, QN, JL). MMR status was interpreted as lost if there was complete absence of staining in the tumor cells with adequate positive staining of internal controls (inflammatory cells or stroma). p53 was interpreted as abnormal if there was complete negative staining (null-pattern) or strong/diffuse staining in >70% of tumor cells (aberrant positive pattern). All other patterns were interpreted as wild-type.
DNA extraction
Methods have previously been described [11, 17]. Briefly, DNA from formalin fixed paraffin embedded (FFPE) tumor blocks were extracted using the Qiagen FFPE tissue kit, and all DNA was quantified using the Qubit fluorometer kit (Life Technologies). To determine somatic status normal DNA was either extracted from available buffy coat or representative normal FFPE blocks.
Sequencing
Targeted primers were designed to cover the POLE EDM exons 9-14. PCR products (150-200bp) were amplified using the Fluidigm 48×48 Access Arrays, as per manufacturers protocol, with input of 100ng FFPE derived DNA, and 50ng high-quality DNA from buffy coat or frozen tumor DNA. DNA barcodes (10bp) with Illumina cluster-generating adapters were added to the libraries, and 96 samples pooled. The library pools were sequenced using the Illumina MiSeq for ultra-deep sequencing. All validated POLE mutations were bi-directionally sequenced twice at minimum using tumor DNA, and once in the normal to validate somatic or germline status using either ultra-deep MiSeq sequencing or Sanger sequencing. Additional details can be found in the previous publications [11, 17].
Histotype and Grade assignment
We had original diagnoses from the host institutions on both diagnostic (biopsy/curettage specimens) and final hysterectomy specimens. In addition, three gynecologic subspecialty pathologists form three independent tertiary care institutions (RS, MK, CHL) reviewed 1-2 representative haematoxylin and eosin stained slides of diagnostic and final hysterectomy specimens with the goal of assigning histotype and grade. For grade, three choices (grade 1, 2, or 3) were considered. For histotype, pathologists were asked to render a diagnosis in one of the following categories: endometrioid, mucinous, serous, clear cell, dedifferentiated, carcinosarcoma, mixed and other. These pathologists were blinded to the original pathology reports and to each other's interpretation.
Statistics
Descriptive statistics were used to characterize the demographic, clinical and pathological data for evaluable cases according to molecular subgroups assigned in both the diagnostic and final hysterectomy specimens. To compare the diagnostic (biopsy/curetting) and final hysterectomy specimens using the original diagnoses assigned at our institution, overall accuracy and Cohen's kappa (κ) statistic were calculated for the ProMisE molecular classifier, grade and histotype. In addition, we computed the average per class sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). To account for the ordinal nature of grade, we additionally computed a weighted kappa with squared weights. Histotype was ultimately grouped in to 4 more encompassing categories: endometrioid, serous, mixed, and other. Interobserver agreement between three subspecialty pathologists was calculated using Fleiss' generalized Kappa coefficient for multiple raters and Krippendorf's alpha for ordinal data for within sample type assessment. Cohen's kappa and a weighted Cohen's kappa was used to compare across sample type and the result was averaged. 95% confidence intervals were computed using a bootstrap approach with 1000 bootstrap samples. There was no comparison of molecular subgroups to clinical outcomes as this has previously been performed [11].
Results
Cohort
Of the 60 cases considered, two were excluded as they were found to have received neoadjuvant therapy before hysterectomy, and one case was excluded secondary to insufficient tumor volume for DNA extraction and POLE sequencing. A total of 57 cases were compared for grade, histotype, and molecular subgroup assignment by ProMisE in both diagnostic (pre-staging) and final hysterectomy (post-staging) specimens. Surgical staging had occurred between 1987 and 2013. Histotype, grade, and pathological details assigned to diagnostic specimens and final hysterectomy specimens were taken from original pathology reports from our center. Details, including patient demographics, tumor grade, and histotype for the diagnostic specimens are shown in Table 1a.Table 1b shows the same parameters in addition to stage, LVSI, myometrial invasion, nodal status, adjuvant therapy, and ESMO risk group as designated on final hysterectomy/post-surgical staging specimens. The distribution of cases shows good representation of all four molecular subgroups. We had enriched this cohort for p53 abn and POLE EDM cases to ensure that these lower frequency subgroups could be adequately evaluated.
Table 1a.
Descriptive statistics of cohort according to ProMisE molecular subgroups as defined by diagnostic (biopsy or curettage) specimen. All percentages given are column percentages.
| Total | MMR-D | POLE EDM | p53 wt | p53 abn | |
|---|---|---|---|---|---|
| Age at Surgery | |||||
| mean (SD) | 63.9 (± 13.8) | 61.0 (± 15.6) | 62.1 (± 10.1) | 56.7 (± 15.0) | 74.0 (± 5.8) |
| median | 68.8 | 63.0 | 65.3 | 51.0 | 71.8 |
| missing | 16 | 3 | 3 | 9 | 1 |
| BMI | |||||
| mean (SD) | 30.8 (± 8.2) | 30.2 (± 8.4) | 29.1 (± 7.9) | 38.2 (± 9.6) | 27.7 (± 4.4) |
| median | 28.9 | 29.2 | 29.4 | 39.2 | 27.7 |
| missing | 20 | 4 | 4 | 10 | 2 |
| Grade | |||||
| Grade 1 | 23 (40.4%) | 6 (33.3%) | 4 (40.0%) | 11 (64.7%) | 2 (16.7%) |
| Grade 2 | 11 (19.3%) | 6 (33.3%) | 0 (0.0%) | 3 (17.6%) | 2 (16.7%) |
| Grade 3 | 23 (40.4%) | 6 (33.3%) | 6 (60.0%) | 3 (17.6%) | 8 (66.7%) |
| Histological Subtype | |||||
| Endometrioid | 40 (70.2%) | 13 (72.2%) | 7 (70.0%) | 16 (94.1%) | 4 (33.3%) |
| Serous | 4 (7.0%) | 1 (5.6%) | 0 (0.0%) | 0 (0.0%) | 3 (25.0%) |
| Mixed* | 5 (8.8%) | 2 (11.1%) | 2 (20.0%) | 0 (0.0%) | 1 (8.3%) |
| Other** | 8 (14.0%) | 2 (11.1%) | 1 (10.0%) | 1 (5.9%) | 4 (33.3%) |
|
| |||||
| Total | 57 (100%) | 18 (31.6%) | 10 (17.5%) | 17 (29.8%) | 12 (21.1%) |
Mixed* includes mixed endometrioid and clear cell (1), mixed endometrioid and clear cell and mucinous (1) and mixed endometrioid and serous (3).
Other** includes high grade NOS (4), dedifferentiated (1), undifferentiated (1), carcinosarcoma (1), and large cell neuroendocrine (1).
Table 1b.
Descriptive statistics of cohort according to ProMisE molecular subgroups as defined by post-staging hysterectomy specimen. All percentages given are column percentages.
| Total | MMR-D | POLE EDM | p53 wt | p53 abn | |
|---|---|---|---|---|---|
| Age at Surgery | |||||
| mean (SD) | 63.9 (± 13.8) | 63.5 (± 15.0) | 62.4 (± 9.6) | 54.2 (± 15.5) | 74.3 (± 6.0) |
| median | 68.8 | 71.5 | 65.3 | 49.7 | 73.8 |
| missing | 16 | 3 | 2 | 10 | 1 |
| BMI | |||||
| mean (SD) | 30.8 (± 8.2) | 30.6 (± 7.7) | 28.4 (± 6.8) | 37.1 (± 11.0) | 27.8 (± 4.) |
| median | 28.9 | 30.0 | 27.0 | 40.0 | 27.9 |
| missing | 20 | 4 | 3 | 11 | 2 |
| Grade | |||||
| Grade 1 | 16 (28.1%) | 4 (25.0%) | 3 (27.3%) | 9 (47.4%) | 0 (0.0%) |
| Grade 2 | 17 (29.8%) | 6 (37.5%) | 1 (9.1%) | 8 (42.1%) | 2 (18.2%) |
| Grade 3 | 24 (42.1%) | 6 (37.5%) | 7 (63.6%) | 2 (10.5%) | 9 (81.8%) |
| Histological Subtype | |||||
| Endometrioid | 44 (77.2%) | 14 (87.5%) | 9 (81.8%) | 18 (94.7%) | 3 (27.3%) |
| Serous | 8 (14.0%) | 1 (6.2%) | 1 (9.1%) | 0 (0.0%) | 6 (54.5%) |
| Mixed* | 3 | 1 (6.2%) | 1 (9.1%) | 0 (0.0%) | 1 (9.1%) |
| Other** | 1 | 0 (0.0%) | 0 (0.0%) | 1 (5.3%) | 0 (0.0%) |
| Stage | |||||
| IA | 21 (55.3%) | 6 (50.0%) | 2 (22.2%) | 6 (75.0%) | 7 (77.8%) |
| IB | 10 (26.3%) | 3 (25.0%) | 6 (66.7%) | 1 (12.5%) | 0 (0.0%) |
| III | 6 (15.8%) | 2 (16.7%) | 1 (11.1%) | 1 (12.5%) | 2 (22.2%) |
| IV | 1 (2.6%) | 1 (8.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| missing | 19 | 4 | 2 | 11 | 2 |
| LVSI | |||||
| No | 21 (55.3%) | 5 (41.7%) | 5 (55.6%) | 6 (75.0%) | 5 (55.6%) |
| Yes | 17 (44.7%) | 7 (58.3%) | 4 (44.4%) | 2 (25.0%) | 4 (44.4%) |
| missing | 19 | 4 | 2 | 11 | 2 |
| Myometrial Invasion | |||||
| None | 8 (21.1%) | 1 (8.3%) | 2 (22.2%) | 3 (37.5%) | 2 (22.2%) |
| <50% | 14 (36.8%) | 6 (50.0%) | 0 (0.0%) | 3 (37.5%) | 5 (55.6%) |
| >50% | 16 (42.1%) | 5 (41.7%) | 7 (77.8%) | 2 (25.0%) | 2 (22.2%) |
| missing | 19 | 4 | 2 | 11 | 2 |
| Nodal Status | |||||
| Not Tested | 2 (5.3%) | 0 (0.0%) | 2 (22.2%) | 0 (0.0%) | 0 (0.0%) |
| Tested Negative | 33 (86.8%) | 11 (91.7%) | 7 (77.8%) | 8 (100.0%) | 7 (77.8%) |
| Tested Positive | 3 (7.9%) | 1 (8.3%) | 0 (0.0%) | 0 (0.0%) | 2 (22.2%) |
| missing | 19 | 4 | 2 | 11 | 2 |
|
| |||||
| Total | 57 (100%) | 16 (28.1%) | 11 (19.3%) | 19 (33.3%) | 11 (19.3%) |
Mixed* includes mixed endometrioid with undifferentiated (1), and mixed serous with high grade (1) and low grade(1) endometrioid
Other** includes carcinosarcoma (1)
Concordance of molecular classifier
Table 2 shows the overall concordance (Table 2a) and the concordance metrics (Table 2b) including average per molecular subgroup sensitivity, specificity, PPV, NPV and kappa statistic for the ProMisE molecular classifier comparing diagnostic and final surgical samples, with the latter held to be the “gold standard”. Kappa statistic of 0.86 (95%CI) was consistent with a “near perfect” level of agreement [19], fulfilling our goal of improvement over previously published data showing poor concordance between pre- and post-operative samples, when assessed for the conventional histopathological parameters of grade and histotype [20-22]. Also shown are the concordance metrics within each molecular subgroup (Table 2c.). Sensitivity, specificity, positive predictive values and negative predictive values are all highly favorable across subgroups (≥.9 except sensitivity for p53 wt (0.84) and POLE (0.82) subgroup, and positive predictive value for MMR-D (0.83)) with perfect or near perfect metrics within the p53 abn subgroup.
Table 2a.
Comparison of ProMisE molecular classification of diagnostic samples (rows) and post-staging hysterectomy samples (columns).
| Post-staging Samples | ||||||
|---|---|---|---|---|---|---|
| MMR-D | POLE EDM | p53 wt | p53 abn | Total | ||
| Diagnostic Samples | MMR-D | 11 | 1 | 2 | 0 | 14 |
| POLE EDM | 0 | 10 | 1 | 0 | 11 | |
| p53 wt | 1 | 0 | 19 | 0 | 20 | |
| p53 abn | 0 | 1 | 0 | 11 | 12 | |
|
| ||||||
| Total | 12 | 12 | 22 | 11 | 57 | |
Table 2b.
Comparison of overall concordance statistics (with 95% confidence intervals) based on ProMisE molecular classification of diagnostic samples and post-staging samples.
| Overall Concordance Statistics | |
|---|---|
| Overall Accuracy | 0.89 (0.78-0.96) |
| Cohen's kappa | 0.86 (0.72-0.93) |
| No Information Rate (NIR) | 0.39 |
| P-Value (Accuracy> NIR) | 0 |
Table 2c.
Comparison of concordance statistics (with 95% confidence intervals) for each ProMisE molecular subgroups.
| Average | MMR-D | POLE EDM | p53 wt | p53 abn | |
|---|---|---|---|---|---|
| Sensitivity | 0.9 | 0.94 (0.72-1) | 0.82 (0.52-0.95) | 0.84 (0.62-0.94) | 1 (0.74-1) |
| Specificity | 0.96 | 0.93 (0.81-0.97) | 0.98 (0.89-1) | 0.97 (0.87-1) | 0.98 (0.89-1) |
| Pos Pred Value | 0.9 | 0.83 (0.61-0.94) | 0.9 (0.6-0.99) | 0.94 (0.73-1) | 0.92 (0.65-1) |
| Neg Pred Value | 0.96 | 0.97 (0.87-1) | 0.96 (0.86-0.99) | 0.92 (0.8-0.97) | 1 (0.92-1) |
| Prevalence | 0.28 (0.18-0.41) | 0.19 (0.11-0.31) | 0.33 (0.22-0.46) | 0.19 (0.11-0.31) | |
| Detection Rate | 0.26 (0.17-0.39) | 0.16 (0.09-0.27) | 0.28 (0.18-0.41) | 0.19 (0.11-0.31) | |
| Detection Prev | 0.32 (0.21-0.44) | 0.18 (0.1-0.29) | 0.3 (0.2-0.43) | 0.21 (0.12-0.33) | |
| Accuracy | 0.95 | 0.93 (0.83-0.97) | 0.95 (0.86-0.98) | 0.93 (0.83-0.97) | 0.98 (0.91-1) |
| Balanced Acc | 0.93 | 0.93 | 0.9 | 0.91 | 0.99 |
Interrogation of cases discordant on molecular classification
In total there were 6 of 57 cases with discordant results between diagnostic and final surgical sample as assessed using ProMisE, only one of which was within the p53 abn subgroup. Table 3 summarizes the six discordant cases, which are presented in more detail, case by case, below.
Table 3. Discordant cases according to ProMisE subclassfication.
| Case | Specimen | Pathology | MMR IHC | POLE | p53 | Initial Assignment | Reassessment | Final ProMisE | Retest |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Diagnostic | CAH + gr 1 EM | intact | no mut | abn | p53 abn | Retest: No POLE mut found | p53 abn | Discordant |
| Hysterectomy | gr 1 EM, stage IB | intact | P286S* | abn | POLE EDM | Retest validates | POLE EDM | ||
| 2 | Diagnostic | gr 2 EM | MSH6 loss | no mut | wt | MMR-D | Retest: Δ to intact | p53 wt | Concordant |
| Hysterectomy | gr 2 EM, stage IB | intact | no mut | wt | p53 wt | Retest: confirms intact | p53 wt | ||
| 3 | Diagnostic | gr 1 EM | MSH2/MSH6 loss | no mut | wt | MMR-D | Retest: changed to intact | p53 wt | Concordant |
| Hysterectomy | gr 2 EM, stage IA | intact | no mut | wt | p53 wt | Retest confirms intact | p53 wt | ||
| 4 | Diagnostic | gr 3 undiff | MSH2/MSH6 loss | P441L | abn | MMR-D | Retest confirms MSH2/MSH6 loss | MMR-D | Concordant |
| Hysterectomy | gr 3 SC, stage IB | intact | P441L | abn | POLE EDM | Retest:Δ to MSH2/MSH6 loss | MMR-D | ||
| 5 | Diagnostic | gr 1 EM | intact | no mut | wt | p53 wt | Retest confirms intact | p53 wt | Discordant |
| Hysterectomy | gr 3 EM, stage IB | MLH1/PMS2 loss | no mut | wt | MMR-D | Retest confirms loss of MLH1 | MMR-D | ||
| 6 | Diagnostic | gr 1 EM | intact | P286R | wt | POLE EDM | Retest validates | POLE EDM | Discordant |
| Hysterectomy | gr 1 EM, stage IA | intact | no mut | wt | p53 wt | Retest: No POLE mut found | p53 wt |
In case 1 a POLE EDM was detected at low allelic frequency (8% then 1% on retest) in the final hysterectomy sample (grade 1 endometrioid, early stage), but could not be detected in the diagnostic endometrial biopsy (grade 1 endometrioid). This patient had a very favorable outcome, and was a long term survivor. Sequencing results from our Fluidigm panel (3 genes in addition to POLE) revealed mutations in TP53: G360R,and PPP2R1A:R183W, and we suspect her TP53 mutation is secondary; consistent with a POLE EDM/ultramutated tumor, and this EC is appropriately categorized as POLE EDM although without whole genome or even exome sequencing ‘ultramutator’ phenotype cannot be determined accurately.
Cases 2 and 3 are similar, both demonstrating loss of MMR proteins on endometrial biopsy specimen leading to classification as MMR-D, however the final hysterectomy shows all proteins intact. We re-tested the diagnostic samples, using full sections rather than tissue microarrays, and were able to demonstrate presence of all four MMR proteins thus changing their classification to p53 wt and concordant with final hysterectomy. For Case 4, MSH2/MSH6 loss was noted on endometrial biopsy specimen and confirmed on re-testing. MMR IHC had been interpreted as intact originally on hysterectomy however on re-testing with whole sections revealed MSH2/MSH6 loss thus ProMisE classification is also concordant in this case upon review of whole sections. These misclassifications are therefore attributable to the small samples present on tissue microarrays, with 0.6 mm cores.
Case 5 remained discordant after re-review and repeat whole section MMR testing, and the discordant results were due to tissue sampling. In the endometrial sampling, there was only a low-grade endometrioid adenocarcinoma which had retention of MLH1 and PMS2. The hysterectomy however, had a low-grade endometrioid adenocarcinoma as well as dedifferentiated carcinoma, and this latter component, which was not sampled in the endometrial biopsy, showed loss of MLH1 and PMS2, as is commonly seen in dedifferentiated carcinoma of the endometrium (Figure 2).
Figure 2.
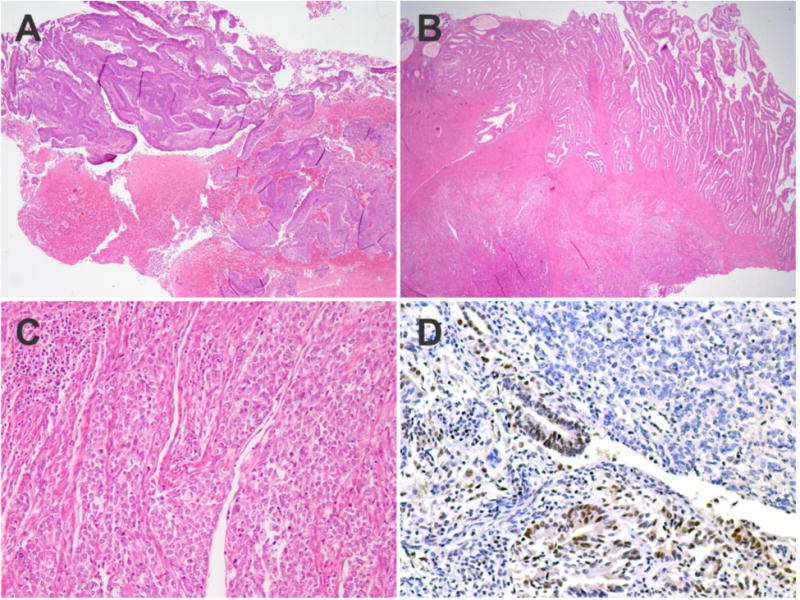
Histopathologic features of endometrial carcinoma with discrepant mismatch repair protein results on endometrial sampling and hysterectomy. The endometrial sampling consists of only low-grade endometrioid adenocarcinoma (A). On the hysterectomy, the superficial portion of the tumor contains the low-grade endometrioid adenocarcinoma while the deeper portion is higher grade with solid architecture (B). In the solid areas, the nuclei are enlarged, irregular and the cells are mildly discohesive; peritumoral lymphocytes are also present at the leading edge of the tumor (C). Immunohistochemical staining for MLH1 shows retained staining in the low-grade glandular component and loss of staining in the high-grade solid component (D).
Case 6 shows discordance in POLE EDM results, with mutations found in the diagnostic biopsy sample at 18% frequency (23% on retesting) but no POLE EDMs found in the final hysterectomy sample. Both diagnostic and hysterectomy samples were grade 1 endometrioid tumors, with minimal myometrial invasion in the hysterectomy specimen.
In summary, 3 of the 6 discrepancies between diagnostic sample and hysterectomy are attributable to the use of TMAs with the small sample size and were easily resolved with the more use of whole section immunostaining, as would be done in clinical practice. In 2 cases there was failure to detect POLE EDM (in the diagnostic sample in one case and the hysterectomy specimen in the other) because of low tumor cellularity or frequency of the mutant allele, and again these discordant results do not reflect inferiority of the biopsy/curetting specimen compared to the hysterectomy specimen for molecular classification. Instead, they reflect as yet unsolved issues around detection and interpretation of low frequency POLE EDM. There was thus a single tumor where the diagnostic specimen had failed to sample a high-grade (dedifferentiated) component of the tumor. This case (case 5) was a true sampling error as the biopsy was not reflective of the final tumor with respect to grade, histotype or molecular classification.
Concordance of grade and histotype in diagnostic endometrial vs. final hysterectomy samples
The overall concordance and concordance metrics for grade and histotype, based on the original pathology reports, comparing diagnostic pre-surgical staging samples to final hysterectomy samples, are shown in Table 4. Kappa statistics for simplified (4-category) histotype was 0.44(0.23-0.65) (Table 4a.) and for grade a weighted kappa of 0.7 (0.5-0.83) (Table 4b.) were comparable to what has been reported previously, and was worse than the high level of reproducibility seen with molecular subclassification.
Table 4a.
Comparison of histology assessment (using simplified categories) of diagnostic samples (rows) and post-staging hysterectomy samples (columns) from original pathology reports.
| Post-staging Samples | ||||||
|---|---|---|---|---|---|---|
| Endometrioid | Serous | Mixed | Other | Total | ||
| Diagnostic Samples | Endometrioid | 37 | 0 | 2 | 1 | 40 |
| Serous | 0 | 4 | 0 | 0 | 4 | |
| Mixed | 3 | 1 | 1 | 0 | 5 | |
| Other | 4 | 3 | 0 | 1 | 8 | |
|
| ||||||
| Total | 44 | 8 | 3 | 2 | 57 | |
Table 4b.
Comparison of overall concordance statistics (with 95% confidence intervals) based on histotype assessment (using simplified categories) of diagnostic samples and post-staging samples from original pathology reports.
| Overall Concordance Statistics | |
|---|---|
| Overall Accuracy | 0.75 (0.62-0.86) |
| Cohen's kappa† | 0.44 (0.23-0.65) |
| No Information Rate (NIR) | 0.77 |
| P-Value (Accuracy> NIR) | 0.69 |
Please note kappa must be interpreted with caution due to symmetrical imbalance of row and column marginals in table 4a.
Concordance of grade and histotype between gynecologic subspecialty pathologists
Table 5a shows the average concordance metrics for grade and histotype between the three gynecologic pathologists as evaluated within the 48 diagnostic (pre-surgical staging) and final hysterectomy (post-surgical staging) samples available for review. Concordance remains low; kappa for grouped grade (grade1/2 vs. grade 3) (0.74) and simplified histotype (0.51),even when assigned by experts. Finally comparing each subspecialty pathologists diagnoses for grade and histotype in diagnostic vs. final hysterectomy specimens i.e., WITHIN an individual patient there was on average kappa of 0.56 and 0.57 respectively (Table 5b).
Table 5a.
Interobserver agreement between three subspecialty pathologists, computed using Fleiss's kappa.
| Diagnostic Samples | Post-Staging Samples | |
|---|---|---|
| Simplified Histology | 0.51 (n=48) | 0.59 (n=48) |
| Grouped Grade | 0.74 (n=47) | 0.73 (n=46) |
Table 5b.
Concordance between diagnostic samples and post-staging hysterectomy specimen pathologic assignment computed using Cohen's kappa. Level of agreement for each pathologist is shown as well as averages.
| Pathologist 1 | Pathologist 2 | Pathologist 3 | Average | |
|---|---|---|---|---|
| Simplified Histology | 0.49 | 0.56 | 0.62 | 0.56 |
| Grouped Grade | 0.56 | 0.56 | 0.57 | 0.57 |
Discussion
Inadequacies in our current system of endometrial cancer classification and risk stratification have prompted a call to action [23-26], to identify new biologically informative tools. This study confirms the previously reported lack of reproducibility of conventional histopathological assessment, both between observers, and in comparing biopsy/curetting to hysterectomy specimens. Attempts at comprehensive treatment guidelines [24, 27], interpretation of past and future clinical trials, and EC research are severely limited by our inability to consistently classify this disease. The tremendous advances made in research and treatment of other cancers have not been realized in EC.
Although TCGA represented a positive step towards informative classification, the methods used were impractical. Research teams from Leiden and our own center have subsequently developed lower cost methods applicable to formalin-fixed paraffin-embedded (FFPE) specimens to identify four prognostically distinct molecular subgroups of EC [11, 12]. For ProMisE, the molecular classification tool we have developed, we are following the Institute of Medicine guidelines [28] for the development of ‘omics based testing, and have completed the ‘discovery’ and ‘confirmation’ stages with anticipated completion of the final ‘validation’ stage in late 2016. We can then embark on clinical trials to determine how this tool can change clinical care and the costs and benefits to patients (outcomes, health economic) associated.
Reproducibility of any classifier is of critical importance, and one aspect of reproducibility is the potential to give a definitive classification based on diagnostic specimens e.g. biopsy or curetting's. Almost all women ultimately diagnosed with endometrial carcinoma have some sort of assessment of their endometrium-either by office biopsy or dilatation and curettage, which both identifies an EC and informs proceeding to the next step of surgical staging. This specimen is usually abundant/high volume and is immediately fixed often resulting in superior quality immunohistochemistry compared to final hysterectomy specimens, which may sit at room temperature for a variable period of time before processing. MMR and p53 proteins have relatively short half-lives and their detection is therefore dependent on prompt fixation, as is also true for detection of estrogen receptor and HER2 in breast cancer. There have been few studies looking at traditional pathological parameters and molecular features in endometrial biopsies [20-22, 29], but to our knowledge we are the first group to explore molecular classification in these specimens. Our results provide indirect evidence that addresses one of the questions raised by the aforementioned studies demonstrating only moderate agreement of histotype and grade assignment in diagnostic versus hysterectomy specimens i.e. is the lack of reproducibility primarily due to inadequate tumor sampling or to the inter- and intraobserver variability of grade and histotype assignment. We had only one case where there was clearly a sampling issue, with a high-grade dedifferentiated component not present in the biopsy specimen. This suggests that the observed problems with imperfect concordance between diagnostic and hysterectomy specimens reflects the inherent lack of reproducibility of grade and histotype rather than true sampling differences.
The obvious advantage to successful molecular classification with ProMisE in diagnostic specimens is earlier availability of prognostic information. Knowledge about a woman's risk of having metastatic disease, recurrence, and/or death may impact the urgency and comprehensiveness of surgical staging, and/or adjuvant therapy. Globally, there is a wide range of surgical practice, ranging from delay of hysterectomy (progesterone treatment) to preserve fertility, maintaining ovaries to preserve endogenous hormonal production and avoid associated comorbidities [30-33], pelvic +/- para-aortic lymph node assessment (complete, sampling, sentinel, or none), washings, omental/upper abdominal assessment (complete, biopsy, none). Each component of these surgical procedures has a cost: to the patient (fertility, cardiovascular disease, perioperative risk of injury, lymphedema) or the health care system (pathology processing and interpretation, operating room time). As we move towards personalized medicine, determining the ‘best’ surgical procedure would be a tremendous start. In our current system risk stratification is achievable only AFTER surgical staging (myometrial invasion and stage are major components). Although adjuvant therapy can be guided from the post-staging information and risk group assignment, these again are limited by the irremediable irreproducibility of histotype and grade [5-8]. This could change with use of the ProMisE molecular classifier. Next steps include testing the application of ProMisE in the context of a clinical trial examining outcomes (survival parameters, patient reported outcomes/quality of life) and health economic implications as compared to current standard of care.
In summary, use of this molecular classifier is a pragmatic option for classifying all endometrial cancers at the time of initial diagnosis. The techniques described herein are practical and achievable at any cancer center. Endometrial biopsy or curettage specimens are routinely obtained during the work up and evaluation of endometrial cancers and if/when cancer is diagnosed the ProMisE molecular tool can be applied. Processing of the sample is done as it is currently, requiring no special handling, as steps can be performed on FFPE material. Our experience with more than 3000 clinical cases supports the use of endometrial biopsies or curetting specimens for MMR assessment as they are promptly fixed, with better antigen preservation than the corresponding hysterectomy specimen. We continue to look for surrogates for sequencing of POLE but given the advances in technologies and rapidly decreasing costs of sequencing we do not see this as a tremendous obstacle, particularly as we focus on a single gene, with targeted primers providing restricted coverage e.g. only POLE exonuclease domain exons 9-14.
Discordant cases will be encountered, as they were in this series. Although far more consistent than grade or histotype assignment, there remains the possibility that the molecular classifier tool could assign a woman with EC to the inappropriate subgroup, based on analysis of the biopsy. Importantly, incorrect assignment appears to be very unlikely in the subgroup with the worst outcomes (p53 abn), suggesting we are unlikely to miss someone who may need more comprehensive surgery and additional chemotherapy and/or radiotherapy. Three of six cases with discordant results in this study can be explained based on the use of a tissue microarray as a research tool; in these three cases this resulted in an error in MMR assessment in either the diagnostic or hysterectomy specimen and these errors were easily resolved through use of whole sections for immunostaining, as would be done in routine practice. In two cases there were discrepancies attributable to low frequency POLE EDM mutations or possibly low tumor cellularity, resulting in the diagnostic specimen in one case and the hysterectomy specimen in the other being considered to have intact POLE. It is not clear at the present time what the allelic mutation frequency should serve as a threshold for diagnosis of POLE EDM and further work is required to address this important issue, but it does not reflect issues related to use of diagnostic compared to hysterectomy specimens. In a single case there was failure to sample a dedifferentiated component of a tumor in the biopsy, a rare EC variant associated with mutations in chromatin remodelling genes and frequent MMR protein [34, 35]. In theory, mixed carcinomas may also pose a challenge to classify using ProMiSE, however, truly biologically mixed tumors are exceptionally rare [36] and most cases diagnosed as mixed carcinoma are actually due to morphologic ambiguity rather than admixtures of molecularly distinct clones [37].
With regards to the order of testing, we ultimately decided that MMR testing first made sense as we believed this information would prompt early referral to the hereditary cancer program for testing for Lynch syndrome and information from that test may impact on the patient's decision regarding management e.g. foregoing a trial of medical therapy with high dose progesterone. An outstanding question is how to handle cases that are positive for more than one of the classifiers. This is particularly important for tumors with POLE EDM or MMR-D and p53 abn. Such ‘double feature’ cases only account for 3-4% of EC, and based on available information the POLE EDM or MMR-D appears to be more important than p53 abn when both are present [11, 38-40], more cases need to be evaluated to confirm this.
Weaknesses in this series included using tissue microarrays rather than whole sections and possible compromise of quality of DNA extracted from archival cases (two cases from over 15 years ago, majority within last 10 years). As for any biomarker done on small samples, care must be taken to ensure that there is sufficient tumor-derived DNA before proceeding to testing. In cases where there are any concerns about DNA sample quality based on the biopsy, testing should be repeated on the definitive surgical specimen. Both of these problems would be expected to decrease reproducibility of the classifier, if they had any effect. We did not have a large number of non-serous/non-endometrioid cases; such tumors are rare and an example is the dedifferentiated carcinoma (case 5 in Table 3). It remains to be seen whether ProMisE can be applied to these tumors, or whether they should be recognized as distinct diseases. Further interrogation of uncommon EC histotypes is needed.
We look forward to an era of consistent molecular subclassification of EC, stratifying future clinical trials by molecular subgroups to provide earlier and more reliable prognostic information to patients and their physicians. We will need to determine how best to incorporate molecular tools with current practice, focusing on information we have from the time of diagnosis (e.g. grade but not stage, and patient phenotype; age, BMI). There may be additional clinical or molecular parameters that enable us to further discern differences in outcomes within these molecular subgroups, and we anticipate more and more personalized approaches to EC research and management. We are emboldened by the demonstration of this tool's utility on diagnostic endometrial samples obtained prior to surgical staging, expanding the potential clinical impact of this tool.
Table 4c.
Comparison of grade assessment of diagnostic samples (rows) and post-staging samples (columns) from original pathology reports.
| Post-staging Samples | |||||
| Grade 1 | Grade 2 | Grade 3 | Total | ||
| Diagnostic Samples | Grade 1 | 12 | 9 | 2 | 23 |
| Grade 2 | 3 | 7 | 1 | 11 | |
| Grade 3 | 1 | 1 | 21 | 23 | |
|
| |||||
| Total | 16 | 17 | 24 | 57 | |
Table 4d.
Comparison of overall concordance statistics (with 95% confidence intervals) based on assessment of grade of diagnostic samples and post-staging samples from original pathology reports.
| Overall Concordance Statistics | |
|---|---|
| Overall Accuracy | 0.7 (0.57-0.82) |
| Weighted kappa | 0.7 (0.5-0.83) |
| No Information Rate (NIR) | 0.42 |
| P-Value (Accuracy> NIR) | 0 |
Highlights.
Molecular classification can be performed on diagnostic endometrial specimens and is highly concordant with hysterectomy.
Biologically relevant information from time of diagnosis can inform treatment decisions, stratify trials, and direct women to hereditary counselling.
Molecular classification of endometrial cancers offers reproducible categorization and has been demonstrated to identify distinct prognostic subgroups
Acknowledgments
Dr.McAlpine has received funding from the Canadian Institute of Health Research (PPP-144251). We are extremely grateful for generous donations of the family and friends of Sarabjit Gill who established a research fund at the British Columbia Cancer Foundation. We thank Dr. Judith Pike for her assistance in the collection of clinical data.
Footnotes
COI statement: US Provisional Patent Application No. 62192230 has been filed for a molecular based classifier for endometrial cancer; licensee ‘BC Cancer Agency’.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arnold M, Pandeya N, Byrnes G, Renehan AG, Stevens GA, Ezzati M, et al. Global burden of cancer attributable to high body-mass index in 2012: a population-based study. The Lancet Oncology. 2015;16:36–46. doi: 10.1016/S1470-2045(14)71123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Jung KW, Won YJ, Kong HJ, Oh CM, Lee DH, Lee JS. Prediction of cancer incidence and mortality in Korea, 2014. Cancer research and treatment: official journal of Korean Cancer Association. 2014;46:124–30. doi: 10.4143/crt.2014.46.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang LN, McConechy MK, Kobel M, Han G, Rouzbahman M, Davidson B, et al. Histotype-genotype correlation in 36 high-grade endometrial carcinomas. The American journal of surgical pathology. 2013;37:1421–32. doi: 10.1097/PAS.0b013e31828c63ed. [DOI] [PubMed] [Google Scholar]
- 6.Gilks CB, Oliva E, Soslow RA. Poor interobserver reproducibility in the diagnosis of high-grade endometrial carcinoma. The American journal of surgical pathology. 2013;37:874–81. doi: 10.1097/PAS.0b013e31827f576a. [DOI] [PubMed] [Google Scholar]
- 7.Han G, Sidhu D, Duggan MA, Arseneau J, Cesari M, Clement PB, et al. Reproducibility of histological cell type in high-grade endometrial carcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:1594–604. doi: 10.1038/modpathol.2013.102. [DOI] [PubMed] [Google Scholar]
- 8.Guan H, Semaan A, Bandyopadhyay S, Arabi H, Feng J, Fathallah L, et al. Prognosis and reproducibility of new and existing binary grading systems for endometrial carcinoma compared to FIGO grading in hysterectomy specimens. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2011;21:654–60. doi: 10.1097/IGC.0b013e31821454f1. [DOI] [PubMed] [Google Scholar]
- 9.British Columbia Cancer Registry CSaO. Population Oncology. Endometrial cancer rates < 50 yo in British Columbia. :2013–2014. [Google Scholar]
- 10.Cancer Genome Atlas Research N. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talhouk A, McConechy MK, Leung S, Li-Chang HH, Kwon JS, Melnyk N, et al. A clinically applicable molecular-based classification for endometrial cancers. British journal of cancer. 2015;113:299–310. doi: 10.1038/bjc.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelloo E, Bosse T, Nout RA, MacKay HJ, Church DN, Nijman HW, et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2015 doi: 10.1038/modpathol.2015.43. [DOI] [PubMed] [Google Scholar]
- 13.Karateke A, Tug N, Cam C, Selcuk S, Asoglu MR, Cakir S. Discrepancy of pre- and postoperative grades of patients with endometrial carcinoma. European journal of gynaecological oncology. 2011;32:283–5. [PubMed] [Google Scholar]
- 14.Sany O, Singh K, Jha S. Correlation between preoperative endometrial sampling and final endometrial cancer histology. European journal of gynaecological oncology. 2012;33:142–4. [PubMed] [Google Scholar]
- 15.Wang XY, Pan ZM, Chen XD, Lu WG, Xie X. Accuracy of tumor grade by preoperative curettage and associated clinicopathologic factors in clinical stage I endometriod adenocarcinoma. Chinese medical journal. 2009;122:1843–6. [PubMed] [Google Scholar]
- 16.Ben-Shachar I, Pavelka J, Cohn DE, Copeland LJ, Ramirez N, Manolitsas T, et al. Surgical staging for patients presenting with grade 1 endometrial carcinoma. Obstetrics and gynecology. 2005;105:487–93. doi: 10.1097/01.AOG.0000149151.74863.c4. [DOI] [PubMed] [Google Scholar]
- 17.McConechy MK, Talhouk A, Leung S, Chiu DS, Yang W, Senz J, et al. Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016 doi: 10.1158/1078-0432.CCR-15-2233. [DOI] [PubMed] [Google Scholar]
- 18.McConechy MK, Talhouk A, Li-Chang HH, Leung S, Huntsman DG, Gilks CB, et al. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecologic oncology. 2015 doi: 10.1016/j.ygyno.2015.01.541. [DOI] [PubMed] [Google Scholar]
- 19.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- 20.Batista TP, Cavalcanti CL, Tejo AA, Bezerra AL. Accuracy of preoperative endometrial sampling diagnosis for predicting the final pathology grading in uterine endometrioid carcinoma. European journal of surgical oncology: the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2016 doi: 10.1016/j.ejso.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Mitchard J, Hirschowitz L. Concordance of FIGO grade of endometrial adenocarcinomas in biopsy and hysterectomy specimens. Histopathology. 2003;42:372–8. doi: 10.1046/j.1365-2559.2003.01603.x. [DOI] [PubMed] [Google Scholar]
- 22.Helpman L, Kupets R, Covens A, Saad RS, Khalifa MA, Ismiil N, et al. Assessment of endometrial sampling as a predictor of final surgical pathology in endometrial cancer. British journal of cancer. 2014;110:609–15. doi: 10.1038/bjc.2013.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. The Lancet Oncology. 2014;15:e268–78. doi: 10.1016/S1470-2045(13)70591-6. [DOI] [PubMed] [Google Scholar]
- 24.Bendifallah S, Canlorbe G, Collinet P, Arsene E, Huguet F, Coutant C, et al. Just how accurate are the major risk stratification systems for early-stage endometrial cancer? British journal of cancer. 2015 doi: 10.1038/bjc.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendifallah S, Darai E, Ballester M. Predictive Modeling: A New Paradigm for Managing Endometrial Cancer. Annals of surgical oncology. 2016;23:975–88. doi: 10.1245/s10434-015-4924-2. [DOI] [PubMed] [Google Scholar]
- 26.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094–108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 27.Colombo N, Creutzberg C, Amant F, Bosse T, Gonzalez-Martin A, Ledermann J, et al. ESMO-ESGOESTRO Consensus Conference on Endometrial Cancer: Diagnosis, Treatment and Follow-up. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2016;26:2–30. doi: 10.1097/IGC.0000000000000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omenn GS. On best practices: the Institute of Medicine scheme for developing, validating, and demonstrating clinical utility of omics-based diagnostic and predictive tests. Proteomics Clinical applications. 2013;7:748–55. doi: 10.1002/prca.201300041. [DOI] [PubMed] [Google Scholar]
- 29.Stelloo E, Nout RA, Naves LC, Ter Haar NT, Creutzberg CL, Smit VT, et al. High concordance of molecular tumor alterations between pre-operative curettage and hysterectomy specimens in patients with endometrial carcinoma. Gynecologic oncology. 2014;133:197–204. doi: 10.1016/j.ygyno.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 30.Parker WH, Feskanich D, Broder MS, Chang E, Shoupe D, Farquhar CM, et al. Long-term mortality associated with oophorectomy compared with ovarian conservation in the nurses' health study. Obstetrics and gynecology. 2013;121:709–16. doi: 10.1097/AOG.0b013e3182864350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker WH, Broder MS, Chang E, Feskanich D, Farquhar C, Liu Z, et al. Ovarian conservation at the time of hysterectomy and long-term health outcomes in the nurses' health study. Obstetrics and gynecology. 2009;113:1027–37. doi: 10.1097/AOG.0b013e3181a11c64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocca WA, Grossardt BR, Shuster LT. Oophorectomy, estrogen, and dementia: a 2014 update. Molecular and cellular endocrinology. 2014;389:7–12. doi: 10.1016/j.mce.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuster LT, Rhodes DJ, Gostout BS, Grossardt BR, Rocca WA. Premature menopause or early menopause: long-term health consequences. Maturitas. 2010;65:161–6. doi: 10.1016/j.maturitas.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karnezis AN, Hoang LN, Coatham M, Ravn S, Almadani N, Tessier-Cloutier B, et al. Loss of switch/sucrose non-fermenting complex protein expression is associated with dedifferentiation in endometrial carcinomas. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2016;29:302–14. doi: 10.1038/modpathol.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoang LN, Lee YS, Karnezis AN, Tessier-Cloutier B, Almandani N, Coatham M, et al. Immunophenotypic features of dedifferentiated endometrial carcinoma Insights from BRG1/INI1-deficient tumors. Histopathology. 2016 doi: 10.1111/his.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobel M, Meng B, Hoang LN, Almadani N, Li X, Soslow RA, et al. Molecular Analysis of Mixed Endometrial Carcinomas Shows Clonality in Most Cases. The American journal of surgical pathology. 2016;40:166–80. doi: 10.1097/PAS.0000000000000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soslow RA. Endometrial carcinomas with ambiguous features. Seminars in diagnostic pathology. 2010;27:261–73. doi: 10.1053/j.semdp.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Hussein YR, Weigelt B, Levine DA, Schoolmeester JK, Dao LN, Balzer BL, et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2015;28:505–14. doi: 10.1038/modpathol.2014.143. [DOI] [PubMed] [Google Scholar]
- 39.Stelloo E, Nout RA, Osse EM, Jurgenliemk-Schulz IJ, Jobsen JJ, Lutgens LC, et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer - combined analysis of PORTEC cohorts. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016 doi: 10.1158/1078-0432.CCR-15-2878. [DOI] [PubMed] [Google Scholar]
- 40.Talhouk A, M MK, L S, Y W, L A, S J, et al. A clinically applicable molecular-based classification system for endometrial cancers. Journal of Clinical Oncology. 2016 ASCO poster presention: Abstract No: 5518. [Google Scholar]


