Abstract
The field of big data is generally concerned with the scale of processing at which traditional computational paradigms break down. In medical imaging, traditional large scale processing uses a cluster computer that combines a group of workstation nodes into a functional unit that is controlled by a job scheduler. Typically, a shared-storage network file system (NFS) is used to host imaging data. However, data transfer from storage to processing nodes can saturate network bandwidth when data is frequently uploaded/retrieved from the NFS, e.g., “short” processing times and/or “large” datasets. Recently, an alternative approach using Hadoop and HBase was presented for medical imaging to enable co-location of data storage and computation while minimizing data transfer. The benefits of using such a framework must be formally evaluated against a traditional approach to characterize the point at which simply “large scale” processing transitions into “big data” and necessitates alternative computational frameworks. The proposed Hadoop system was implemented on a production lab-cluster alongside a standard Sun Grid Engine (SGE). Theoretical models for wall-clock time and resource time for both approaches are introduced and validated. To provide real example data, three T1 image archives were retrieved from a university secure, shared web database and used to empirically assess computational performance under three configurations of cluster hardware (using 72, 109, or 209 CPU cores) with differing job lengths. Empirical results match the theoretical models. Based on these data, a comparative analysis is presented for when the Hadoop framework will be relevant and non-relevant for medical imaging.
Keywords: Apache Hadoop, Sun Grid Engine, Verification
1. INTRODUCTION
As imaging datasets and computing grid sizes grow larger, traditional computing's separation of data and computational nodes creates a problem. Moving data from where it is centrally stored to computational nodes can saturate a network with relatively few active processes. Under certain conditions, the bottleneck in the computing architecture becomes the network bandwidth. An inexpensive solution is to locate the data on the computational nodes to avoid the problem of saturating the network by copying data. This is already implemented by some “big data” architectures, e.g., Apache Hadoop [1-5]. Previously, DICOM to NiFTI conversion had been identified as an area where significant gains in scalability could be realized by using big data frameworks [6]. In [6], we use HBase that is built upon Hadoop to logically and physically sort the data by indexed row keys. We propose a novel data allocation policy within HBase to strongly enforce collocation of hierarchically row key for storing slice-wise DICOM data. In this way, a group-wise DICOM retrieval occurs locally without involving the network [7]. However, this creates new questions, e.g.: when does this novel Hadoop/HBase framework perform better than traditional high performance computing clusters like Sun grid engine (SGE) [8]? In this case, there are many parameters of concern, such as the cluster size, machine cores, node memory, distribution of resources, image processing job, etc. [9-12]. This work develops theoretical models to characterize the performance of SGE and Hadoop and verify the models empirically. The theoretical models have two parts. The first is wall-clock time, which represents the total time as experienced by the user. The second part is resource time, which measures elapsed time on each node when a process starts across all nodes. The models are further verified based on a real lab-based cluster environment focusing on custom image processing.
2. METHODS
2.1 Computation modules
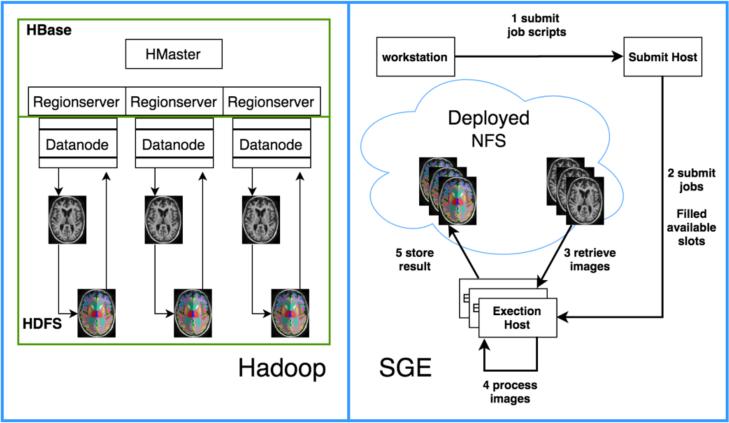
Hadoop and HBase is to enable co-location of data storage and computation while minimizing data transfer, while SGE separates data storage from computation. Figure 1 summarizes both methods’ working flows. It is worth mentioning that there are mainly two kinds of map jobs in Hadoop[13]. A data-local map involves local data within a node. However, it is notable that if no single node contains all requested data for a single job (due to a large request or local storage scarity), the minimal necessary data will be retrieved over the internet. A rack-local or non-local map will retrieve data through other data nodes. Ideally only data-local maps will occur. In reality, around 5% of maps tend to be rack-local as evidenced by previous DICOM to NiFTI conversion experiments, which we had trained on a data / core balanced cluster [6]. The “balanced” means data is distributed equally to every machine, and every machine have the same number of cores. In the rest of the paper, we use “Hadoop” to simplify representing our novel Hadoop/HBase framework.
Figure 1.
Hadoop and SGE data retrieval, processing and storage working flow basing on Multi-atlas CRUISE (MaCRUISE) segmentation [14, 15]. The data in an HBase table is approximately balanced to each Regionserver. The Regionserver collocates with a Hadoop Datanode to fully utilize the data collocation and locality[7]. We design our proposed computation models using only the map phase of Hadoop's MapReduce [13]. In this phase, the data is retrieved locally; if the result were moved to reduce phase, more data movement would occur, because the reduce phase does not ensure process local data. Within the map phase, all necessary data is retrieved and saved on a local directory and gets furtherly processed by locally installed binary executables command-line program. After that, the results of processing are uploaded back to HBase. For SGE, the user submits a batch of jobs to a submit host, and this host dispatches the job to execution hosts. Each execution host retrieves the data within a shared NFS and stores the result back to the NFS.
2.2 Theoretical model
We summarize the modeled parameters that affect both wall-clock time and resource time in Table 1.
Table 1.
Theoretical model parameter definition
| Definition | Description |
|---|---|
| I/O speed (VsourceR, VsourceW, VhostR, VhostW) | I/O speed is the data read/write rate on source where data stores and retrieves, and on host where data get processed. Source for SGE is the Network storage accessed by NFS. Source for Hadoop is the hard disk allocated for HBase. |
| Bandwidth (B, Breal): | B is the bandwidth of cluster. In our case, it is based on a gigabit network. Breal is the real bandwidth that is shared by one job. Once there is a network congestion, the value of Breal is actually smaller than or equal to the cluster's given fixed bandwidth B. |
| Disk speed (VdiskR, VdiskW) | Data read/ write speed of local hard drive. |
| Input/output dataset (datain / dataout) | The total data size for one job that is downloaded/uploaded while processing. |
| Core (#core, #allowed_core, corei) | #core is the number of processor cores in the cluster. #allowed_core is the maximum number of allowed concurrent cores can be used without causing network congestion. We use corei to represent the number of individual cores on each machine. |
| Job (#job, jobi) | #job is the total number of jobs. jobi is the number of jobs will be dispatched to each machine. |
| Job time per dataset (Tj) | Processing time per dataset. |
| Round (#Round) | The total number of round for parallel jobs. |
| Region (#region, regioni) | Each part of a Hbase table is split into several regions (blocks of data).The total number of regions # region per Regionserver is approximately balanced[7]. Regioni denotes the number of region on specified machine. |
| Machine ( #machine) | The number of machines (workstations) of the cluster. |
| File ( #file) | All files that are involved in the whole process. |
| Negligible overhead (η) | An insignificant overhead that could be eliminated and not counted. |
| α | α is a binary parameter. It helps the equations explain when do they only valid for Hadoop scenario (α = 1) rather than SGE scenario (α = 0). |
| β | β is a binary parameter. It helps the equations explain when do they only valid for SGE scenario (β = 1) rather than Hadoop scenario (β = 0). |
| γ | γ is an experimental empirical parameter to represent the ratio of rack-local map task for Hadoop scenario, namely the data is loaded/stored via network. Similarly, the value of γ is always 1 for SGE scenario since all data is transferred through network. |
(1). Wall-Clock Time
Wall-clock time is what the user sees and experiences. Equation (1) is an overview wall-clock time model basing on Figure 1. It contains three types of I/O: data retrieval , processing , storage . For SGE, data is loaded from and stored to network storage, so VsourceR and VsourceW is related with average bandwidth. All data I/O in execution host occur in the pre-allocated memory for the job, so VhostR and VhostW can be ignored compared with bandwidth. For Hadoop, the worst case of input data retrieval from HBase and output data storage to HBase directly involves local hard disk [7]. When the input data is processed on a host, it is temporally saved on a place on hard disk, and the output also generated on a temp place of local drive. Thus, VsourceR, VsourceW, VhostR and VhostW are all related with the local disk reading/writing speed (VdiskR/VdiskW).
| (1) |
Wall-clock time summary
Equation (2) is a summarized model for wall-clock time. Twc_network is the time for jobs to load data from the network for both SGE and Hadoop scenarios. Twc_local is for jobs that load data locally, and it only serves for the Hadoop scenario.
| (2) |
Wall-clock time for jobs only loading data locally Twc_local
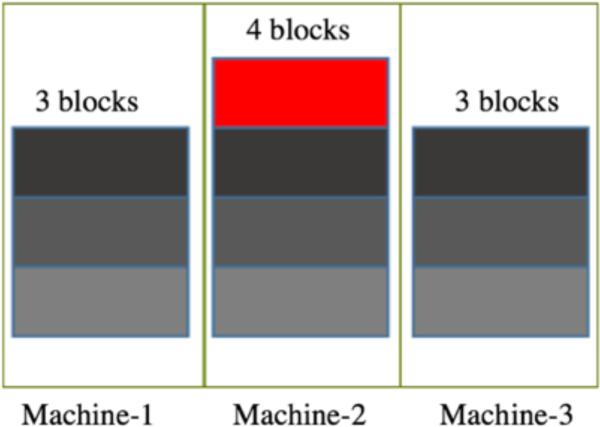
Firstly, we make an assumption for the model Twc_local that if the Hadoop map task is a data-local map task, all data retrieval/storing are happened locally, and the minimal necessary data movement from other nodes over network due to large request are ignored in this model. Therefore, #Round is determined by data and core allocation of the cluster - i.e., if the cluster is a core-balanced, the wall-clock time of all tasks is defined by the machine with the most data as Figure 2 illustrates. If the cluster is core-unbalanced, we need to take into consideration the ratio between number of jobs will be dispatched for each machine and the cores on that machine as in equation (3) presented. For the number of jobs per machine, according to the input dataset's row key in HBase, we can know the dataset belongs to which region of HBase table. The place of region stands for the node that the job will be run [7]. Then for each machine, once we find the maximum ratio of , the “short plate” machine will decide the value of #Round.
| (3) |
If we need to process all HBase table's data, we can easily use the distribution of table's regions to find the number of local jobs for each machine as equation (4) demonstrated.
| (4) |
Figure 2.
Assume there are 10 jobs are ready to process. Each block represents the input dataset for each jobs. Three machines are all have same number of cores. If there is no data transfer, all data processing time is defined by the medium's processors, if all datasets have the same processing speed.
Wall-clock time for job containing data network transfer Twc_network
As equation (5) presented, all data in SGE is transferred through the network, and jobs are equally distributed to the free cores of machines, namely jobs do not care where data is. Hence the value of #Round for SGE is simply decided by . Ideally, there is no data transfer in the Hadoop MapReduce approach. However, as mentioned in section 2.1, there are about 5% rack-local maps in our trained experiment so that it also involves network transfer. And for Hadoop, disk reading / writing speed should be considered in the time model due to the data retrieval, processing and storage working flow.
| (5) |
Network saturation release point
Under a fixed bandwidth, data traveling through a network can affect the number of running cores. We made an assumption that the value of allowed concurrent running cores #allowed_core without arising network congestion is showed in equation (6). If the total number of running cores #core that a cluster can provide is more than #allowed_core, heavier data loading may cause network saturation and make real bandwidth Breal for each job smaller than or equal to the given fixed cluster bandwidth B. Under network congestion circumstances, job completion time gets delayed because job has to spend more time waiting for data movement by network I/O. The relationship of average Breal of all jobs and cluster's B within one round parallel job processing cycle is introduced in equation (7). Moreover, we define the network saturation release point (NSRP) is at the point when #allowed_core equals to #core. Equation (8) summarizes the Breal before and after a NSRP.
| (6) |
| (7) |
| (8) |
(2). Resource Time
Resource time is time elapsed time on each node when a process starts across all nodes as displayed in equation (9). For SGE, the resource time is decided by the sum of all job's processing time and data transfer through network. Breal may affect the resource time of SGE since when network saturation occurs, the core has to wait for data being loaded to the node. Ideally, there is few data movements for Hadoop/HBase, except the small proportion for rack-local maps. The resource time usage for Hadoop is determined by total job time, data retrieval via the network in rack-local map and local disk reading/writing.
| (9) |
2.3 Experiment Design
Three parallel experiment environments are setup for both Hadoop and SGE. The experiment design does not aim to share the benefit of processing group hierarchical related imaging data similar in [6]. However, our goal is to verify the behavior of our Hadoop scenario that maximize the localization of data retrieval/processing/storage for a job. We make the experiment simpler that compressing 3,310 T1 images to the .gz format. Each job compresses only one NiFTI image with 2GB memory available and generate one compressed images. All T1 images for Hadoop scenario are saved into a newly created HBase table. We estimate achievable empirical average bandwidth as 70 Mb/second; disk read speed as 100 Mb/second with write speed as 65 Mb/second. The total input size of the images is 70.7 GB and the processing generates 21.5 GB of compressed files as output. To explore the impacts of processing time, we manually increase the processing time by adding a sleep function without any data retrieval to make the job length of the experiment take an additional 15 – 105 seconds on a fixed dataset to mimic different job processing speed. Also, we vary cluster size to assess the scalability of SGE and Hadoop cluster from 6 – 21 machines.
Table 2 presents the detail of the experiment setup. Each machine was used as a Hadoop Datanode and HBase RegionServer for data locality [7]. All machines were also configured using SGE. There is an additional Machine for both methods serving as cluster master.
Table 2.
Hadoop v.s. SGE experiment cluster setup with same memory allocation and fixed datasets.
| Core allocation | Hadoop data allocation | Estimated NSRP | Experiment type |
|---|---|---|---|
| 209 cores ( 3 machines with 32 cores, 10 machines with 3 cores, 7 machines with 12 cores, 1 machine with 11 cores). | 43 regions (20 machines with 2 regions, 1 machine with 32 cores has 3 regions). | Until Tj reaches 85 s. | T1 NiFTI image compression (5s), and manually increase the processing time by adding a sleep function (10s, 25 s, 40 s, 55 s, 70 s, 85 s, 100 s, 115 s respectively) |
| 132 cores balanced on 11 machines (12 cores/machine). | 34 regions (10 machines with 3 regions, 1 machine with 4 regions ). | Until Tj reaches 54 s | |
| 72 cores balanced on 6 machines (12 cores/machines). | 11 regions (5 machines with 2 regions, 1 machines with 1 regions ). | Until Tj reaches 29 s |
Our goals are to empirically verify 1) if each of the scenarios can match the wall-clock / resource time theoretical models basing on estimated network saturation release point NSRP; 2) test if the cluster can present a scalable performance, i.e., when SGE has more cores, the saturation length will be longer than seen with less cores with the increasing of data processing time per job (Tj), 3) how balanced/unbalanced data and core allocation can affect both computing architectures.
2.4. Datasets
The experiment uses 3,310 T1 images retrieved from a secure, shared web database application for MRI data that was gathered from healthy subjects/volunteers and subjects/volunteers with ADHD; the Tennessee Twin Study based on psychopathology risk and its subjects constitute a portion of the neuroimaging sub-study of the Baltimore Longitudinal Study on Aging.
3. RESULTS
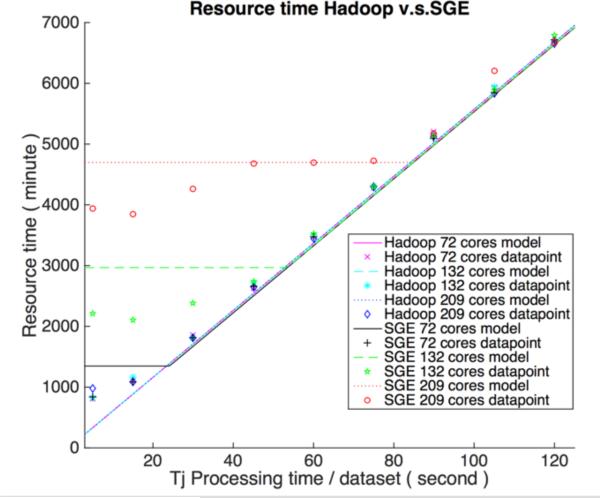
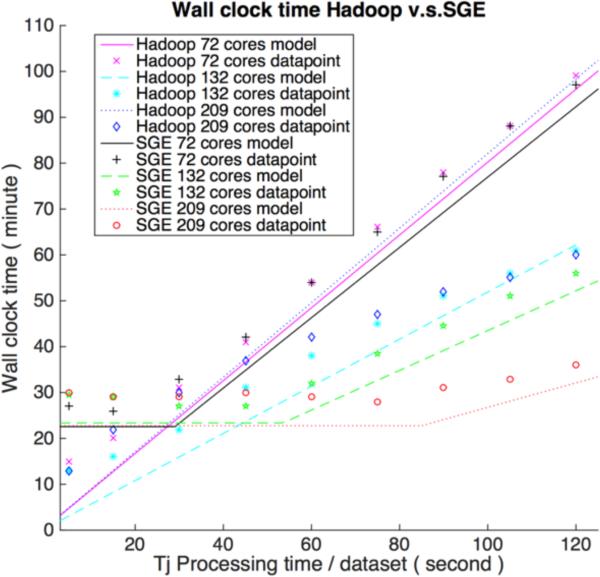
Figure 3 presents the verification result for Hadoop and SGE on wall clock time. The wall-clock time usage for SGE is a bit over what would be expected by theoretical model and could be explained by non-modeled overhead in SGE, such as job dispatching. When cores increase under the fixed number of jobs and size of datasets, the network saturation persists longer for SGE, and the wall clock time is limited because of data transfer. The turning points (NSRP) for SGE match the theoretical point when allowed maximum running cores without causing network saturation is larger than the cluster's cores, which can also be verified by result (when dataset processing time is 30 s for SGE 72 cores scenario, 60 s for SGE 132 cores scenario and 90 s for SGE 209 cores). When the cluster has 132 cores, Hadoop's time becomes gradually longer than SGE; the reason is that data in HBase is not perfectly distributed. According to the experiment setup in Table 2, the total processing time is decided by the node which has one more region. Our model can also predict Hadoop 72 cores scenario since the data / core allocation is approximately balanced. Hadoop 209 cores result is more complex and does not perfectly match theoretical model. Neither core nor data allocation for this scenario are balanced. Through the result, we can see that when most jobs are running on the limited machines, Hadoop randomly dispatches some jobs to idle cores, thus the final wall-clock time is smaller than theoretical models suggest. The final number of rack-local maps for Hadoop 209 cores is almost 30%, which is much larger than our previously observed model parameter (5%) based on a balanced cluster. On the other hand, SGE balanced the job and data distribution; namely, the average number of running cores is greater than seen with Hadoop.
For Figure 3 (B), because only little data movement was allowed, Hadoop with 72 and 132 cores falls on the theoretical ideal resource usage line much faster than SGE with the same number of cores. Even on an unbalanced cluster, Hadoop with 209 cores can also match the theoretical trend. Variation in available network bandwidth is a potential explanation for the 132 core SGE not matching the theory-derived expected result for the 5-45 seconds/dataset range. The result reveals the real average bandwidth Breal is faster than assumed on 70 Mb/s in section 2.3. That the SGE cluster with more cores was saturated longer also matches theoretical models, and once the dataset processing time is greater than NSRP, all three SGE scenarios fall on the Hadoop model.
Figure 3 (B).
Resource time performance for Hadoop and SGE with different cores
4. CONCLUSION
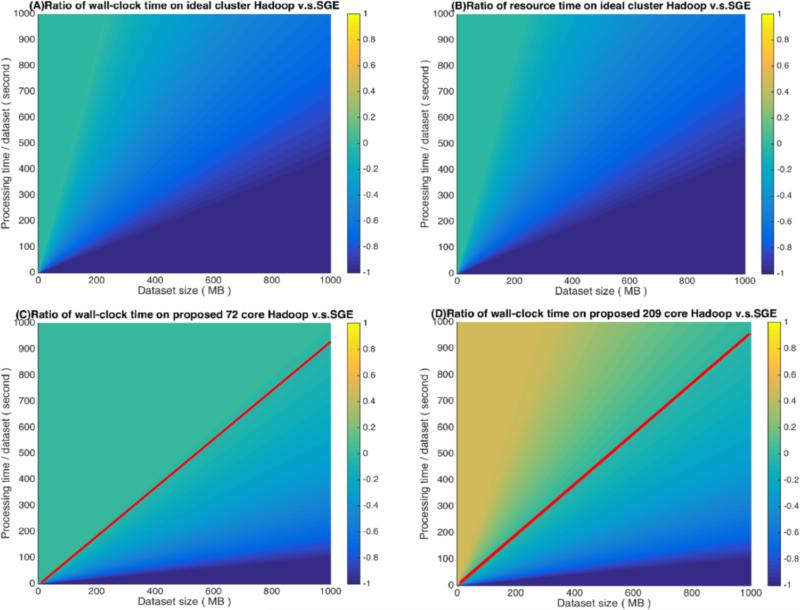
The theoretical model indicates a trend of wall-clock time spent for a particular image processing job. For instance, converting a T1 NiFTI image to MNI using Aladin registration takes at least 2 minutes, so about 400 jobs can run concurrently before network saturation occurs. Thus, SGE cannot saturate the network under the experiments introduced in section 2. The theoretical model also conveys multiple relationships among job numbers, network environment and cluster setup. In Figure 4, the common logarithm ratio (log10) ratio for wall-clock and resource time performance transition of Hadoop's divide SGE's, and the log10 ratio is get scales in range from [−1,1].
Figure 4.
Time ratio of Hadoop v.s. SGE based on core / data balanced and unbalanced cluster. The balancing can only affect the total job running time. (A) Ratio of total running time on a data / core balanced cluster. (B) Ratio of resource time either on a core / data balanced cluster. (C) Ratio of total running time on a proposed 72 cores grid setup (approximate balanced cluster). (D) Ratio of total running time on a proposed 209 cores grid setup (core / data unbalanced cluster). The red lines in (C/D) indicate the parameters for which Hadoop and SGE result in equivalent performance for the specified setup.
We assume there are 5000 jobs as input. As Figure 4 (A) and (B) represents the ratio when core/data allocation are balanced (the ideal scenario for Hadoop), which are based on an assumed lab-based cluster (20 machines, 300 cores, gigabit bandwidth (70 Mb/s), hard drive (Read 100 Mb/s, Write 65 Mb/s), 2GB memory for each jobs). And the ratio reveals SGE can perform similarly with Hadoop when the dataset size is relative small (200 MB) but running time is long, at around 1000 s. However, Hadoop performs much better, at least two-fold, when dataset size reaches over 500 MB and the processing time is around 1000 s either wall-clock time or resource time. We can also appreciate that when job processing time is very long (i.e., over 100 minutes), the resource time of both approaches are close, but Hadoop can still win on the time data transfer takes.
On a data / core unbalanced cluster, the wall-clock time for Hadoop is affected. Figure 4(C) and (D) presume the same data allocation proportion as the Hadoop with 72 cores and 209 cores scenario in Table-2. The ratio value smaller than ‘0’ can be treated as a break down time for SGE as the red line indicated, and the user should try to move from their ‘traditional’ grid framework to a Hadoop ‘big data’ framework. The smaller cluster is more balanced so the performance is better than in the Hadoop 209 scenario. This is because for bigger clusters, data is approximately balanced, but here there is one machine that contains 3 cores, which is much smaller than the average. So here the wall-clock time section distribution is different when compared with Figure 4(A). However, based on our experiment, Hadoop can randomly send some waiting jobs to other free cores (Figrue 3(A) Hadoop 209 cores scenario), so the performance result should be considered a worst case. Additional investigation into factors with imbalanced clusters is warranted.
Figure 3 (A).
Wall-clock time performance for Hadoop and SGE with different cores
Acknowledgement
This work was funded in part by NSF CAREER IIS 1452485. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of NSF. This study was in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. This project was supported in part by ViSE/VICTR VR3029 and the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06.
References
- 1.Apache Hadoop Project Team Apache Hadoop. 2016 http://hadoop.apache.org/
- 2.Borthakur D. The hadoop distributed file system: Architecture and design. Hadoop Project Website. 2007;11:21. 2007. [Google Scholar]
- 3.Yang C-T, Chen L-T, Chou W-L, et al. Implementation of a medical image file accessing system on cloud computing. :321–326. [Google Scholar]
- 4.Jai-Andaloussi S, Elabdouli A, Chaffai A, et al. Medical content based image retrieval by using the Hadoop framework. :1–5. [Google Scholar]
- 5.Taylor RC. An overview of the Hadoop/MapReduce/HBase framework and its current applications in bioinformatics. BMC bioinformatics. 2010;11(Suppl 12):S1. doi: 10.1186/1471-2105-11-S12-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao S, Plassard A, Landman B, et al. Cloud Engineering Principles and Technology Enablers for Medical Image Processing-as-a-Service. Vancouver, Canada: Apr 4-7, 2017. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apache HBase Team Apache hbase reference guide. 2016 http://hbase.apache.org/book.html.
- 8.Gentzsch W. Sun grid engine: Towards creating a compute power grid. :35–36. [Google Scholar]
- 9.Appuswamy R, Gkantsidis C, Narayanan D, et al. Scale-up vs Scale-out for Hadoop: Time to rethink? :20. [Google Scholar]
- 10.Medernach E. Workload analysis of a cluster in a grid environment. :36–61. [Google Scholar]
- 11.Sadashiv N, Kumar SD. Cluster, grid and cloud computing: A detailed comparison. :477–482. [Google Scholar]
- 12.Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. Journal of digital imaging. 2004;17(3):205–216. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. Communications of the ACM. 2008;51(1):107–113. [Google Scholar]
- 14.Huo Y, Plassard AJ, Carass A, et al. Consistent cortical reconstruction and multi-atlas brain segmentation. NeuroImage. 2016 doi: 10.1016/j.neuroimage.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huo Y, Carass A, Resnick SM, et al. Combining multi-atlas segmentation with brain surface estimation. :97840E–97840E-8. doi: 10.1117/12.2216604. [DOI] [PMC free article] [PubMed] [Google Scholar]