Abstract
Macroautophagy (autophagy hereafter) is a process that collects cytoplasmic components, particularly mitochondria, and degrades them in lysosomes. In mammalian systems, basal autophagy levels are normally low but are profoundly stimulated by starvation and essential for survival. Cancer cells up-regulate autophagy and can be more autophagy-dependent than most normal tissues. Genetic deficiency in essential autophagy genes in tumors in many autochthonous mouse models for cancer reduces tumor growth. In K-rasG12D-driven non–small cell lung cancer (NSCLC) and other models, autophagy sustains metabolism and survival. The mechanism by which autophagy promotes tumorigenesis varies in different contexts, but evidence points to a critical role for autophagy in sustaining metabolism, thereby preventing p53 activation, energy crisis, growth arrest, apoptosis, senescence, and activation of the immune response. Autophagy in NSCLC preserves mitochondrial quality and regulates their abundance. By degrading macromolecules in lysosomes, autophagy provides mitochondria with substrates to prevent energy crisis and fatal nucleotide pool depletion in starvation. We review here how autophagy supports mammalian survival and how cancer cells usurp this survival mechanism to maintain mitochondrial metabolism for their own benefit. Insights from these studies provide the rationale and approach to target the autophagy survival pathway for cancer therapy.
Autophagy captures and degrades intracellular proteins and organelles in lysosomes (Mizushima and Komatsu 2011). The autophagy pathway was first identified by yeast genetic approaches to confer survival to nitrogen starvation. Later it became apparent that newborn mice also require autophagy to survive starvation (Kuma et al. 2004; Komatsu et al. 2005). Thus, autophagy is an intracellular nutrient scavenging pathway whereby cells eat parts of themselves and presumably recycle the breakdown products, to sustain survival in starvation.
Basal levels of autophagy are normally low but are profoundly induced by stress and starvation, indicating that rapid autophagy mobilization is part of the integrated stress response (Mizushima et al. 2004). Genetic ablation of autophagy in various settings causes the accumulation of autophagy substrates, particularly damaged proteins and organelles such as abnormal mitochondria (Mizushima and Komatsu 2011). Thus, autophagy is needed for metabolic homeostasis and for protein and organelle quality and numerical control. Without autophagy, survival to stress is compromised, and cells and tissues are subjected to the toxic effects of vast accumulation of autophagy substrates. Because the levels of half the cellular proteins are affected by the functional status of autophagy, this pathway represents a major mechanism for protein turnover and homeostasis (Mathewet al. 2009, 2014).
Unlike normal cells, tumor cells can have high levels of basal autophagy and can be constitutively dependent on autophagy for survival (Degenhardt et al. 2002; Guo et al. 2011; Yang et al. 2011). Autophagy is also induced in hypoxic tumor regions and confers a survival advantage (Degenhardt et al. 2002). These findings suggest that autophagy inhibition may compromise tumor survival and may be a new approach ripe to target for cancer therapy (Amaravadi et al. 2011, 2016). Indeed, genetically engineered mouse models (GEMMs) for cancer where essential autophagy genes are ablated show the functional importance of autophagy for various aspects of tumorigenesis (Amaravadi et al. 2016). Autophagy in these GEMMs for cancer promotes the growth, survival, and malignancy of a broad spectrum of cancers that was reviewed in detail recently (Amaravadi et al. 2011). Herein we provide an overview and discuss the settings and underlying mechanisms by which autophagy promotes tumorigenesis.
AUTOPHAGY IS REQUIRED FOR MAMMALIAN SURVIVAL
First insight into the role of autophagy in sustaining mammalian survival came from studies where an essential autophagy gene was deleted in mice. These mice are born but fail to survive the neonatal starvation period (Kuma et al. 2004; Komatsu et al. 2005). Feeding autophagy-deficient neonatal mice extends survival only briefly indicating that autophagy is essential for mammalian survival to neonatal starvation but also generally. In contrast, adult mice subjected to conditional whole-body deletion of an essential autophagy gene live for 2–3 months, indicating that adult mice are less autophagy-dependent than neonates (Karsli-Uzunbas et al. 2014). Adult mice are physiologically and metabolically different from neonates and possess dedicated nutrient stores that may reduce autophagy dependence. Fasting these adult mice without autophagy, however, is lethal (Karsli-Uzunbas et al. 2014). Thus, the main function of autophagy conserved from yeast to mammals is to confer survival to starvation.
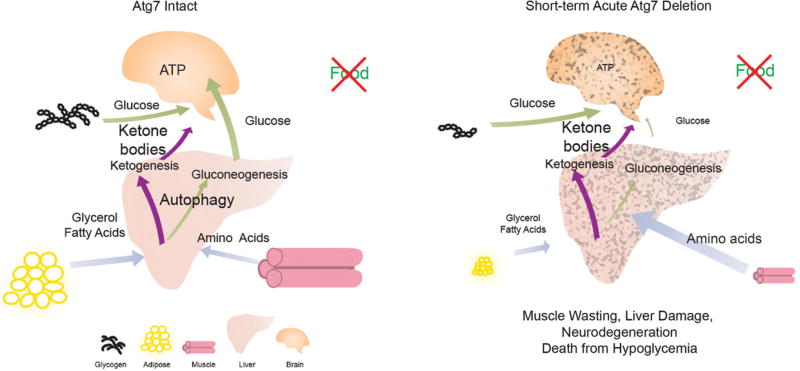
How does autophagy promote survival to starvation at the cellular and organismal level? During fasting, normal adult mice activate autophagy at the cellular level to sustain cell survival and they mobilize nutrient stores at the organismal level to sustain circulating nutrients essential for survival (Karsli-Uzunbas et al. 2014). For example, glycogen is degraded to generate circulating glucose (Fig. 1). Lipid in white adipose tissue (WAT) is also degraded to produce glycerol and fatty acids that are used as substrates for ketogenesis and gluconeogenesis in the liver (Fig. 1). With prolonged starvation, muscle protein is degraded to produce amino acids for use by the liver to synthesize ketone bodies and glucose. This prioritized mobilization of circulating nutrients is aimed at maintaining circulating substrates for brain metabolism, the absence of which is fatal (Fig. 1).
Figure 1.
Mechanism by which autophagy promotes the survival of adult mice during fasting. During fasting, healthy adult mice activate autophagy and degrade glycogen to maintain circulating glucose and mobilize lipid stores to produce glycerol and fatty acids for ketogenesis and gluconeogenesis. As a last resort, muscle degradation by atrophy-associated E3 ligases may supply amino acids for ketogenesis and gluconeogenesis (Karsli-Uzunbas et al. 2014). As a result, glucose homeostasis is maintained by autophagy in mice to surviving fasting. However, fasting adult mice with acute autophagy ablation causes excessive depletion of nutrient stores including lipid and glycogen, accelerated muscle catabolism, and liver damage, which causes severe systemic hypoglycemia, brain damage, and mouse death. (Modified, with permission, from Karsli-Uzunbas et al. 2014, © AACR.)
In contrast, adult mice without autophagy owing to conditional deletion of the essential autophagy gene Atg7 have a systemic metabolic defect that is fatal during fasting (Karsli-Uzunbas et al. 2014). The inability to activate autophagy at the cellular level causes rapid depletion of glycogen and WAT lipid stores, and extensive muscle catabolism (cachexia) (Fig. 1). The accelerated depletion of nutrient stores causes autophagy-deficient mice to fail to maintain circulating glucose levels, resulting in death within 16 h from hypoglycemia (Karsli-Uzunbas et al. 2014). Autophagy in the liver appears necessary to sustain circulating glucose levels and brain function (Ezaki et al. 2011; Karsli-Uzunbas et al. 2014). Thus, autophagy in mammals is essential to supply circulating nutrients for systemic metabolism during fasting. The inability of cells to use autophagy may create a systemic metabolic deficit, accelerating catabolism of nutrient stores (Karsli-Uzunbas et al. 2014). Alternatively, autophagy may be directly involved in the secretion of essential metabolic substrates into the circulation, the absence of which increases dependency on nutrient stores. These findings also raise the interesting possibility that suppression of autophagy may promote cachexia, a significant medical problem for advanced cancer patients (Karsli-Uzunbas et al. 2014).
As autophagy is essential for glucose homeostasis in fasted adult mice, is there a role also in the fed state? A small fraction of the adult mice conditionally deleted for Atg7 die soon after deletion from streptococcus infection (Karsli-Uzunbas et al. 2014). In in vitro experiments autophagy plays a role in host defense by eliminating intracellular bacteria including streptococcus, and this finding indicates that this may also be relevant in vivo. Most adult mice conditionally deleted for Atg7 die 2–3 months postdeletion (Karsli-Uzunbas et al. 2014). This contrasts neonatal mice that fail to tolerate Atg7 deletion (Kuma et al. 2004; Komatsu et al. 2005). Neurodegeneration is the main cause of death of Atg7 conditionally ablated adult mice (Karsli-Uzunbas et al. 2014), which is not surprising given that autophagy deficiency targeted to the brain also causes neurodegeneration (Hara et al. 2006; Komatsu et al. 2006). What is surprising is that neurodegeneration upon conditional deletion of Atg7 in the whole body is greatly accelerated in comparison to tissue-specific deletion (Karsli-Uzunbas et al. 2014). This suggests that autophagy deficiency in non-neuronal tissues also contributes to neurodegeneration, perhaps through defective systemic metabolism. These Atg7-ablated adult mice have a deficit in circulating arginine levels, and it will be of great interest to explore if this contributes to the neurodegeneration phenotype (Karsli-Uzunbas et al. 2014). Another important unanswered question is why tissues like the brain are more autophagy-dependent than other tissues, such as lung. In summary, switching off autophagy in adult mice is tolerated for weeks in the fed state.
AUTOPHAGY PROMOTES TUMORIGENESIS IN GEMMs FOR CANCER
The first clue that autophagy is important for the survival of tumor cells came from the observation that, in stark contrast to normal cells, some cancer cells have high levels of basal autophagy (Guo et al. 2011; Yang et al. 2011). Up-regulation of autophagy in cancer cells is caused by direct activation of the transcription factors of the microphthalmia-associated transcription factor (MiTF)/TFE family that control autophagy and lysosomal biogenesis or by removal of a repressive phosphorylation on the autophagy initiation machinery (Perera et al. 2015; Wong et al. 2015). Moreover, deletion of an essential autophagy gene compromises the survival of those tumor cells in vitro and in tumors in vivo (Guo et al. 2011; Yang et al. 2011). This prompted many laboratories to examine the role of autophagy in GEMMs for cancer where essential autophagy genes are deleted in tumor cells arising spontaneously in the context of a functional immune system. In most of these models, loss of autophagy reduces tumor growth, survival, and proliferation (Amaravadi et al. 2016). In models of K-ras-driven lung and pancreatic cancer, loss of autophagy limits tumor progression to benign disease, which is attributed in part to amplified activation of p53. The requirement for autophagy for tumor growth and survival is also apparent with p53 deletion. In lung cancer GEMMs, deletion of Atg7 dramatically alters tumor pathology from carcinomas to that of benign oncocytomas, which are human tumors that massively accumulate defective mitochondria (Guo et al. 2013; Strohecker et al. 2013). As defective mitochondria are a major autophagy substrate, this indicates that benign human tumors manifest a phenotype of defective autophagy, perhaps explaining their benign status. Indeed, genomic and biological characterization of human renal oncocytomas revealed a metabolic defect stemming from pathogenic mitochondrial genome mutations and defective intracellular trafficking, lysosome, and autophagy function (Joshi et al. 2015). These benign tumors represent a natural setting where tumor progression is halted due to deficient metabolism, which can be caused by loss of mitochondrial energy homeostasis and quality control.
TUMORS ARE MORE AUTOPHAGYDEPENDENT THAN MOST NORMAL TISSUES
These collective findings suggest that autophagy inhibition may be a therapeutic approach for some cancers (Amaravadi et al. 2016). As autophagy is important for some normal tissues, this raised the question as to whether the selective dependency of tumors on autophagy is sufficient to produce a therapeutic window. To address this, the essential autophagy gene Atg7 was deleted conditionally in the whole body of mice with established K-ras-driven lung cancer. Conditional, systemic autophagy loss has remarkable antitumor activity, more so than tumor-specific loss (Karsli-Uzunbas et al. 2014). This suggests that host autophagy in addition to tumor cell autonomous autophagy contributes to tumor growth. Indeed, stromal cell autophagy in models of pancreatic cancer leads to secretion of alanine that is used by tumor cells as metabolic fuel (Sousa et al. 2016). Blockade of this tumor metabolism-promoting metabolic cross talk between stroma and tumor is an additional anticancer benefit of autophagy inhibition (Amaravadi et al. 2016). Whether systemic autophagy beyond stroma also contributes to tumor growth remains to be explored.
AUTOPHAGY RECYCLES MACROMOLECULES
Although the experiments above begin to address the role of autophagy in sustaining whole-body mammalian metabolism, what happens at the cellular level is still an open question. How does autophagy promote survival of cells in starvation? Is this through recycling of cellular components? Do cancer cells do this differently from normal cells and tissues? To begin to address these points we developed an assay to directly measure autophagy-mediated recycling of macromolecules into cancer cell metabolism to find out whether recycling occurs, and if so, what it is used for.
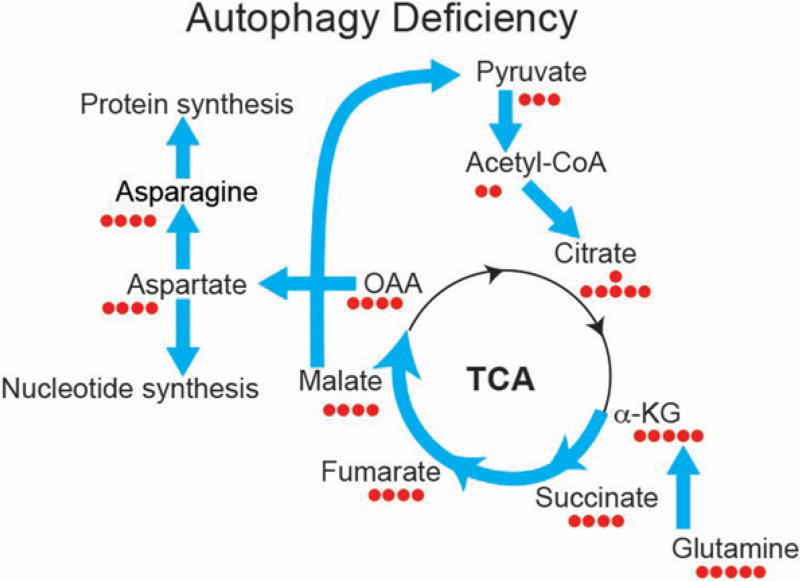
Cancer cells derived directly from the tumors arising in the GEMMs for lung cancer are autophagy dependent as isogenic Atg7-deficient tumor cells are more sensitive to starvation than Atg7 wild-type tumor cells (Guo et al. 2013; Strohecker et al. 2013). To address why autophagy deficiency causes sensitivity to starvation, individual nutrients were added back to determine which were essential for survival. We found that glutamine supplementation in starvation conditions is most effective at rescuing the survival of autophagy-deficient tumor cells (Guo et al. 2013; Strohecker et al. 2013). Because glutamine is a major anapleurotic substrate for the tricarboxylic acid (TCA) cycle (Zong et al. 2016), this suggests that autophagy may recycle intracellular macromolecules to provide amino acid substrates such as glutamine to maintain mitochondrial function (Fig. 2). To test this hypothesis, macromolecules of Atg7 wild-type and deficient tumor cells were stably labeled with heavy isotopes of carbon and nitrogen. The label was then briefly chased out of metabolic pathways with cold media, and then the cells were challenged with starvation to force autophagy-mediated recycling specifically in the Atg7 competent cells and not in the Atg7-deficient cells. Reappearance of heavy isotope labeled metabolites into central carbon metabolism was then assessed by liquid chromatography and mass spectrometry (Guo et al. 2016).
Figure 2.
Defective autophagy in tumor cells alters metabolic flux from glutamine to aspartate and to de novo nucleotide synthesis. K-ras-driven tumor cells depend on autophagy to supply the substrate glutamine and its derivatives to sustain mitochondria metabolism for starvation survival. To compensate, Atg7-deficient tumor cells significantly increase glutamine flux to the tricarboxylic acid (TCA) cycle, de novo nucleotide synthesis, and nicotinamide adenine dinucleotide phosphate (NADPH) generation by up-regulating flux from malate, pyruvate, acetyl-CoA, and citrate for antioxidant defense (Guo et al. 2016). OAA, oxaloacetate; α-KG, α-ketogluturate.
Upon challenge with starvation, labeled metabolites were enriched in the Atg7 competent compared with deficient tumor cells, particularly in TCA cycle intermediates and amino acids synthesized in mitochondria (Guo et al. 2016). What surprised us was that labeling of metabolism was also apparent in the Atg7-deficient tumor cells, although it was significantly less than Atg7 wild-type cells. This indicates that autophagy does recycle macromolecules in starvation, that this is specific to mitochondrial metabolism, but that there also exists autophagy-independent mechanisms for recycling (Guo et al. 2016). What those autophagy-independent mechanisms of recycling are is not known but proteasome-mediated or other forms of lysosome-mediated degradation may be responsible.
AUTOPHAGY-MEDIATED RECYCLING PROMOTES CANCER METABOLISM
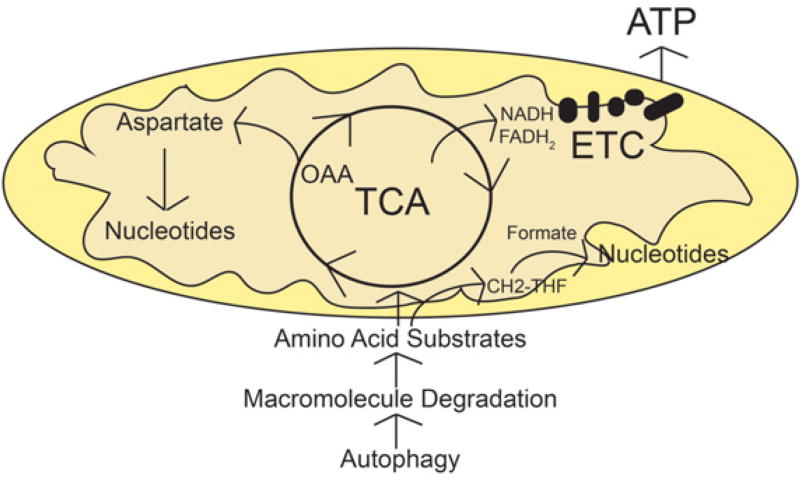
If autophagy is degrading macromolecules in starvation to supply substrates for mitochondrial metabolism, what are the substrates used for? Isotope tracing helped answer this question. Carbon flux from glutamine is accelerated in Atg7-deficient tumor cells in starvation where it flows from the TCA cycle to pyruvate and to aspartate (Fig. 3). Using glutamine carbon to synthesize pyruvate can produce nicotinamide adenine dinucleotide phosphate (NADPH) via malic enzyme for use in generating reduced glutathione for antioxidant defense (Guo et al. 2016). Aspartate is a major biosynthetic product of oxaloacetate (OAA) from the TCA cycle and is a precursor for de novo nucleotide biosynthesis (Guo et al. 2016). Finally, the general supply of substrates to mitochondria through autophagy can drive energy production (Guo et al. 2016). These findings pointed to a role for autophagy-supplied mitochondrial substrates in redox homeostasis, nucleotide biosynthesis, and energy homeostasis (Fig. 4).
Figure 3.
Autophagy recycles macromolecules to provide mitochondrial substrates for nucleotide synthesis and energy homeostasis. Tumor cells rely on mitochondria metabolism for their proliferation and tumorigenesis. In starvation, autophagy is essential to mediate substrate recycling to supply tricarboxylic acid (TCA)-associated amino acids, which are further metabolized through the TCA cycle for energy production and de novo nucleotide biosynthesis. OAA, oxaloacetate; ETC, electron transport chain.
Figure 4.
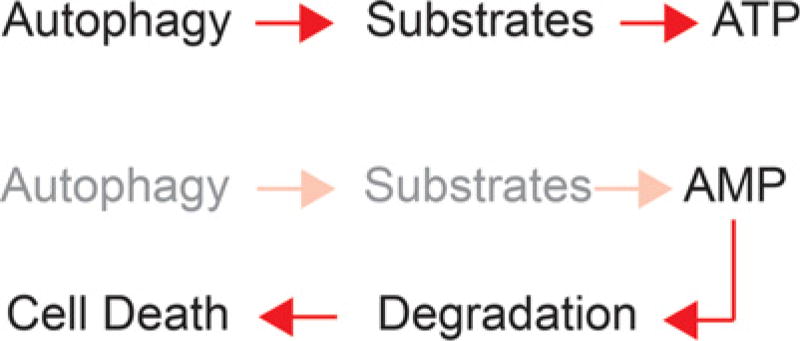
Autophagy-supplied substrates prevent energy crisis and fatal nucleotide depletion. Substrates derived from autophagy are required to generate ATP via tricarboxylic acid (TCA) cycle–driven oxidative phosphorylation in starvation. In the absence of autophagy, defective de novo nucleotide synthesis and increased nucleotide degradation cause nucleotide pool depletion and cell death.
Atg7 deficiency increases reactive oxygen species (ROS), which is extinguished by glutamine supplementation in starvation (Guo et al. 2016). Scavenging ROS, however, fails to rescue survival of starved Atg7-deficient tumor cells. Thus, autophagy-supplied substrates function in maintaining redox homeostasis, although restoring redox balance alone is insufficient to rescue survival in starvation.
Starving Atg7-deficient tumor cells severely depletes nucleotide pools, particularly adenosine nucleotides, consistent with a requirement for autophagy to supply substrates for aspartate and de novo nucleotide synthesis (Guo et al. 2016). Supplementation of autophagy-deficient tumor cells with nucleosides during starvation rescues survival, indicating that autophagy prevents fatal nucleotide pool depletion. In starvation, autophagy also prevents loss of mitochondrial respiration, ATP loss and AMP accumulation, and AMP kinase activation (Guo et al. 2016). Thus, a major function of autophagy is to supply substrates for mitochondrial metabolism to maintain energy homeostasis.
The depletion of nucleotide pools and energy crisis in starved autophagy-deficient tumor cells may be related. In yeast, autophagy loss also causes AMP accumulation and energy crisis in starvation, triggering AMP degradation to escape energy crisis in the short term (Walther et al. 2010). In tumor cells, as in yeast, the requirement for autophagy to supply substrates for de novo nucleotide synthesis in starvation may also be caused by energy crisis and AMP degradation as a means to escape energy crisis (Fig. 4). Indeed, nucleotide degradation products accumulate to high levels in starved Atg7-deficient but not Atg7 wild-type tumor cells (Guo et al. 2016). Autophagy therefore becomes essential to provide metabolic substrates to prevent energy crisis upon nutrient deprivation and thereby the subsequent nucleotide pool depletion.
CONCLUSION
From the study of genetically engineered mice, it is clear that autophagy is important for systemic adult mammalian metabolism. During fed conditions, autophagy is less critical, conferring loss of WAT, select tissue damage, susceptibility to infection, and neurodegeneration after 2–3 months. Autophagy is, however, critical to prevent fatal hypoglycemia during fasting. We discovered that autophagy is a component of nutrient sharing among tissues, as rapid depletion of WAT and glycogen in mice with conditional loss of Atg7 is also associated with excessive muscle catabolism. The metabolic defect at the cellular level creates a systemic metabolic deficit, accelerating exhaustion of nutrient stores and compensatory destruction of muscle, which is ultimately fatal.
Many tumors up-regulate autophagy transcriptionally and posttranscriptionally and depend on it for macromolecular recycling and survival, a requirement that is particularly acute in stress in the tumor microenvironment and during starvation. Indeed, the antitumor activity of caloric restriction is enhanced by deficiency in autophagy (Lashinger et al. 2016). In K-ras-driven non–small cell lung cancer, macromolecular recycling by autophagy suppresses oxidative stress, but preventing energy crisis and nucleotide pool depletion are essential for survival to stress. As some tumors are more autophagy-dependent than most normal tissues, it will be important to determine the therapeutic utility of autophagy inhibition in cancer therapy (Amaravadi et al. 2016).
Acknowledgments
J.Y.G. is supported by the National Institutes of Health (NIH) award K22 CA190521. E.W. is supported by the NIH under award numbers R01CA163591, R01CA130893, R01CA188096, and R01CA193970, the Robert Wood Johnson Foundation, and P30CA72720 to the Rutgers Cancer Institute of New Jersey.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
E.W. is on the Scientific Advisory Board of Forma Therapeutics.
References
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Chen G, Lindsten T, White E. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cell. 2002;2:193–203. doi: 10.1016/s1535-6108(02)00126-5. [DOI] [PubMed] [Google Scholar]
- Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Teng X, Laddha SV, Ma S, Van Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD, White E. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Joshi S, Tolkunov D, Aviv H, Hakimi AA, Yao M, Hsieh JJ, Ganesan S, Chan CS, White E. The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep. 2015;13:1895–1908. doi: 10.1016/j.celrep.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lashinger LM, O’Flanagan CH, Dunlap SM, Rasmussen AJ, Sweeney S, Guo JY, Lodi A, Tiziani S, White E, Hursting SD. Starving cancer from the outside and inside: Separate and combined effects of calorie restriction and autophagy inhibition on Ras-driven tumors. Cancer Metab. 2016;4:18. doi: 10.1186/s40170-016-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol Cell. 2014;55:916–930. doi: 10.1016/j.molcel.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T, Novo M, Rossger K, Letisse F, Loret MO, Portais JC, Francois JM. Control of ATP homeostasis during the respiro-fermentative transition in yeast. Mol Syst Biol. 2010;6:344. doi: 10.1038/msb.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PM, Feng Y, Wang J, Shi R, Jiang X. Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nat Commun. 2015;6:8048. doi: 10.1038/ncomms9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’Antonio G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell. 2016;61:667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]