Abstract
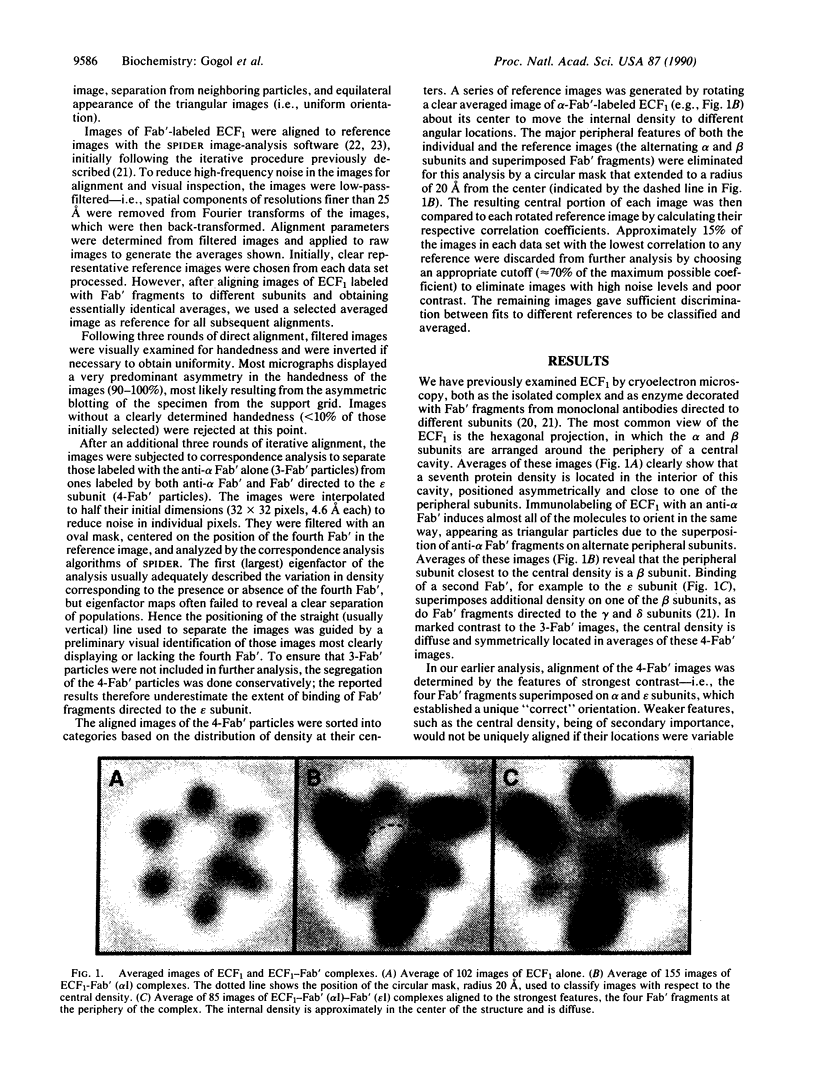
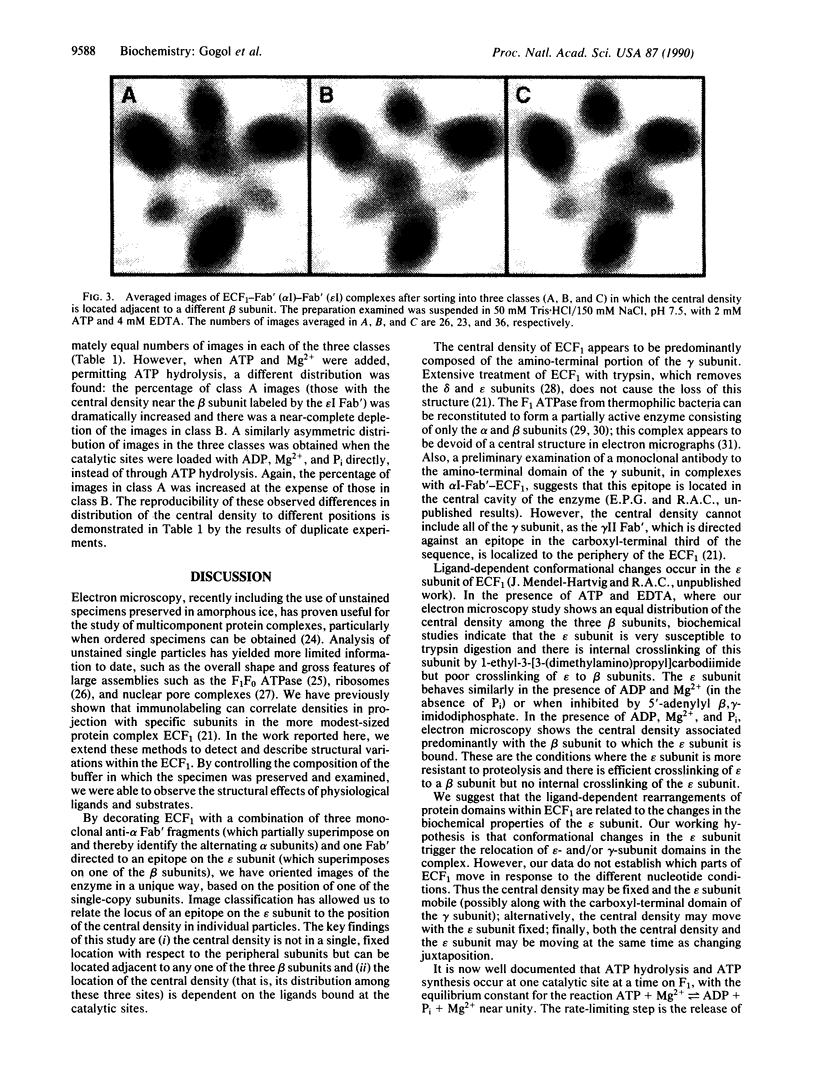
The Escherichia coli F1 ATPase, ECF1, has been examined by cryoelectron microscopy after reaction with Fab' fragments generated from monoclonal antibodies to the alpha and epsilon subunits. The enzyme-antibody complexes appeared triangular due to the superposition of three anti-alpha Fab' fragments on alternating densities of the hexagonally arranged alpha and beta subunits. The Fab' to the epsilon subunit superimposed on a beta subunit. A density was observed near the center of the structure in the internal cavity. The position of this central density with respect to peripheral sites was not fixed. Sorting of images of ECF1 labeled with the combination of three anti-alpha Fab' fragments plus an Fab' directed to the epsilon subunit gave three classes in each of which the central density was closest to a different beta subunit. The distribution of the central density among the three classes was measured for different ligand-binding conditions. When ATP was present in catalytic sites under conditions where there was no enzyme turnover (i.e., without Mg2+ present), there were approximately equal numbers of images in each of three classes. When ATP and Mg2+ were added and ATP hydrolysis was allowed to proceed, almost two-thirds of the images were in the class in which the central density was closest to the beta subunit superimposed by the epsilon subunit. We conclude that domains within the ECF1 structure, either the central mass or a domain including the epsilon subunit, move in the enzyme in response to ligand binding. We suggest that this movement is involved in coupling catalytic sites to the proton channel in the F0 part of the ATP synthase.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akey C. W. Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy. J Cell Biol. 1989 Sep;109(3):955–970. doi: 10.1083/jcb.109.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer P. D. A perspective of the binding change mechanism for ATP synthesis. FASEB J. 1989 Aug;3(10):2164–2178. doi: 10.1096/fasebj.3.10.2526771. [DOI] [PubMed] [Google Scholar]
- Boyer P. D. The unusual enzymology of ATP synthase. Biochemistry. 1987 Dec 29;26(26):8503–8507. doi: 10.1021/bi00400a001. [DOI] [PubMed] [Google Scholar]
- Choate G. L., Hutton R. L., Boyer P. D. Occurrence and significance of oxygen exchange reactions catalyzed by mitochondrial adenosine triphosphatase preparations. J Biol Chem. 1979 Jan 25;254(2):286–290. [PubMed] [Google Scholar]
- Cox G. B., Jans D. A., Fimmel A. L., Gibson F., Hatch L. Hypothesis. The mechanism of ATP synthase. Conformational change by rotation of the beta-subunit. Biochim Biophys Acta. 1984 Dec 17;768(3-4):201–208. doi: 10.1016/0304-4173(84)90016-8. [DOI] [PubMed] [Google Scholar]
- Cross R. L., Grubmeyer C., Penefsky H. S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate enhancements resulting from cooperative interactions between multiple catalytic sites. J Biol Chem. 1982 Oct 25;257(20):12101–12105. [PubMed] [Google Scholar]
- Cunningham D., Cross R. L. Catalytic site occupancy during ATP hydrolysis by MF1-ATPase. Evidence for alternating high affinity sites during steady-state turnover. J Biol Chem. 1988 Dec 15;263(35):18850–18856. [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Energy-transducing H+-ATPase of Escherichia coli. Purification, reconstitution, and subunit composition. J Biol Chem. 1979 Sep 10;254(17):8230–8236. [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):2009–2015. [PubMed] [Google Scholar]
- Frank J., Verschoor A., Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981 Dec 18;214(4527):1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Friedl P., Hoppe J., Gunsalus R. P., Michelsen O., von Meyenburg K., Schairer H. U. Membrane integration and function of the three F0 subunits of the ATP synthase of Escherichia coli K12. EMBO J. 1983;2(1):99–103. doi: 10.1002/j.1460-2075.1983.tb01388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavilanes-Ruiz M., Tommasino M., Capaldi R. A. Structure-function relationships of the Escherichia coli ATP synthase probed by trypsin digestion. Biochemistry. 1988 Jan 26;27(2):603–609. doi: 10.1021/bi00402a016. [DOI] [PubMed] [Google Scholar]
- Gogol E. P., Aggeler R., Sagermann M., Capaldi R. A. Cryoelectron microscopy of Escherichia coli F1 adenosinetriphosphatase decorated with monoclonal antibodies to individual subunits of the complex. Biochemistry. 1989 May 30;28(11):4717–4724. doi: 10.1021/bi00437a031. [DOI] [PubMed] [Google Scholar]
- Gogol E. P., Lücken U., Bork T., Capaldi R. A. Molecular architecture of Escherichia coli F1 adenosinetriphosphatase. Biochemistry. 1989 May 30;28(11):4709–4716. doi: 10.1021/bi00437a030. [DOI] [PubMed] [Google Scholar]
- Gresser M., Cardon J., Rosen G., Boyer P. D. Demonstration and quantitation of catalytic and noncatalytic bound ATP in submitochondrial particles during oxidative phosphorylation. J Biol Chem. 1979 Nov 10;254(21):10649–10653. [PubMed] [Google Scholar]
- Grubmeyer C., Penefsky H. S. The presence of two hydrolytic sites on beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1981 Apr 25;256(8):3718–3727. [PubMed] [Google Scholar]
- Kagawa Y., Ohta S., Otawara-Hamamoto Y. Alpha 3 beta 3 complex of thermophilic ATP synthase. Catalysis without the gamma-subunit. FEBS Lett. 1989 May 22;249(1):67–69. doi: 10.1016/0014-5793(89)80017-1. [DOI] [PubMed] [Google Scholar]
- Lightowlers R. N., Howitt S. M., Hatch L., Gibson F., Cox G. B. The proton pore in the Escherichia coli F0F1-ATPase: a requirement for arginine at position 210 of the a-subunit. Biochim Biophys Acta. 1987 Dec 17;894(3):399–406. doi: 10.1016/0005-2728(87)90118-6. [DOI] [PubMed] [Google Scholar]
- Lücken U., Gogol E. P., Capaldi R. A. Structure of the ATP synthase complex (ECF1F0) of Escherichia coli from cryoelectron microscopy. Biochemistry. 1990 Jun 5;29(22):5339–5343. doi: 10.1021/bi00474a019. [DOI] [PubMed] [Google Scholar]
- Matsuno-Yagi A., Hatefi Y. Studies on the mechanism of oxidative phosphorylation. Positive cooperativity in ATP synthesis. J Biol Chem. 1990 Jan 5;265(1):82–88. [PubMed] [Google Scholar]
- Miwa K., Yoshida M. The alpha 3 beta 3 complex, the catalytic core of F1-ATPase. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6484–6487. doi: 10.1073/pnas.86.17.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Altendorf K. Bacterial adenosine 5'-triphosphate synthase (F1F0): purification and reconstitution of F0 complexes and biochemical and functional characterization of their subunits. Microbiol Rev. 1987 Dec;51(4):477–497. doi: 10.1128/mr.51.4.477-497.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E. ATP synthesis by oxidative phosphorylation. Physiol Rev. 1988 Jan;68(1):177–231. doi: 10.1152/physrev.1988.68.1.177. [DOI] [PubMed] [Google Scholar]
- Stewart M., Vigers G. Electron microscopy of frozen-hydrated biological material. Nature. 1986 Feb 20;319(6055):631–636. doi: 10.1038/319631a0. [DOI] [PubMed] [Google Scholar]
- Vik S. B., Cain B. D., Chun K. T., Simoni R. D. Mutagenesis of the alpha subunit of the F1Fo-ATPase from Escherichia coli. Mutations at Glu-196, Pro-190, and Ser-199. J Biol Chem. 1988 May 15;263(14):6599–6605. [PubMed] [Google Scholar]
- Wagenknecht T., Grassucci R., Frank J. Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J Mol Biol. 1988 Jan 5;199(1):137–147. doi: 10.1016/0022-2836(88)90384-1. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- Yoshimura H., Endo S., Matsumoto M., Nagayama K., Kagawa Y. Hexagonal structure of two-dimensional crystals of the alpha 3 beta 3 complex thermophilic ATP synthase. J Biochem. 1989 Dec;106(6):958–960. doi: 10.1093/oxfordjournals.jbchem.a122981. [DOI] [PubMed] [Google Scholar]
- al-Shawi M. K., Parsonage D., Senior A. E. Thermodynamic analyses of the catalytic pathway of F1-ATPase from Escherichia coli. Implications regarding the nature of energy coupling by F1-ATPases. J Biol Chem. 1990 Mar 15;265(8):4402–4410. [PubMed] [Google Scholar]
- von Meyenburg K., Jørgensen B. B., Nielsen J., Hansen F. G., Michelsen O. The membrane bound ATP synthase of Escherichia coli: a review of structural and functional analyses of the atp operon. Tokai J Exp Clin Med. 1982;7 (Suppl):23–31. [PubMed] [Google Scholar]






