Abstract
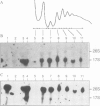
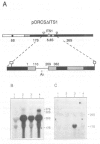
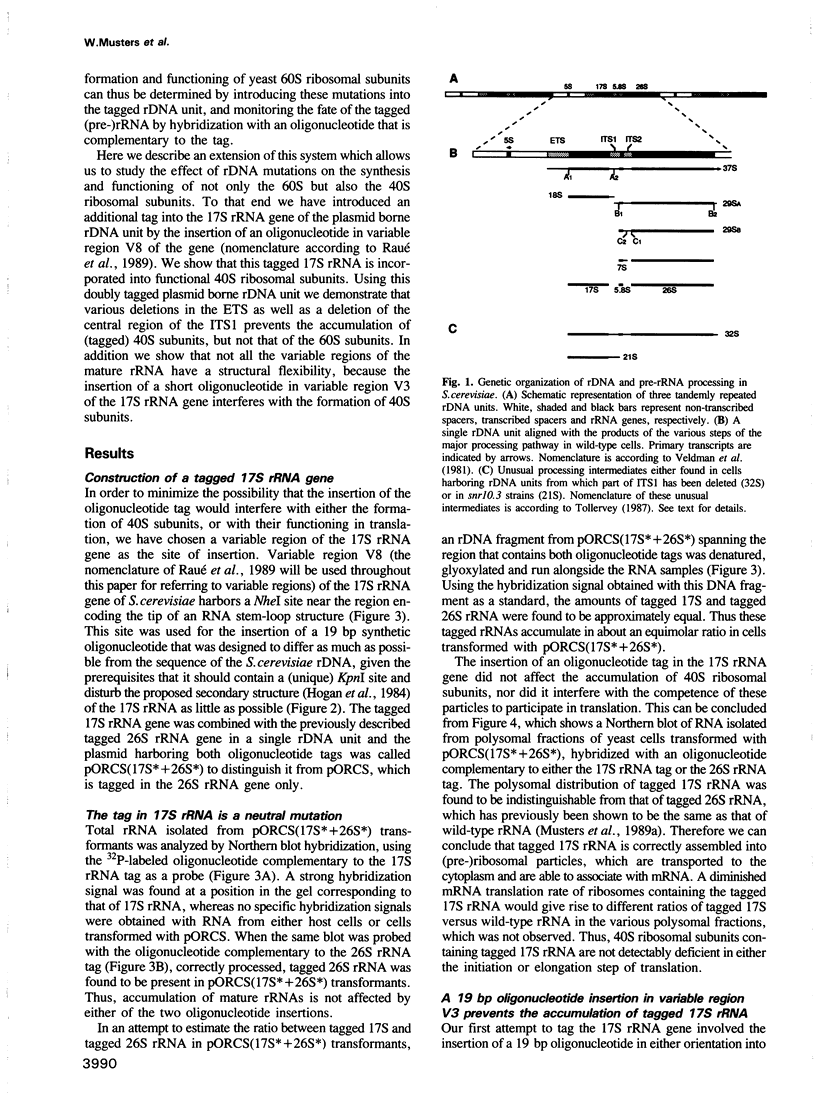
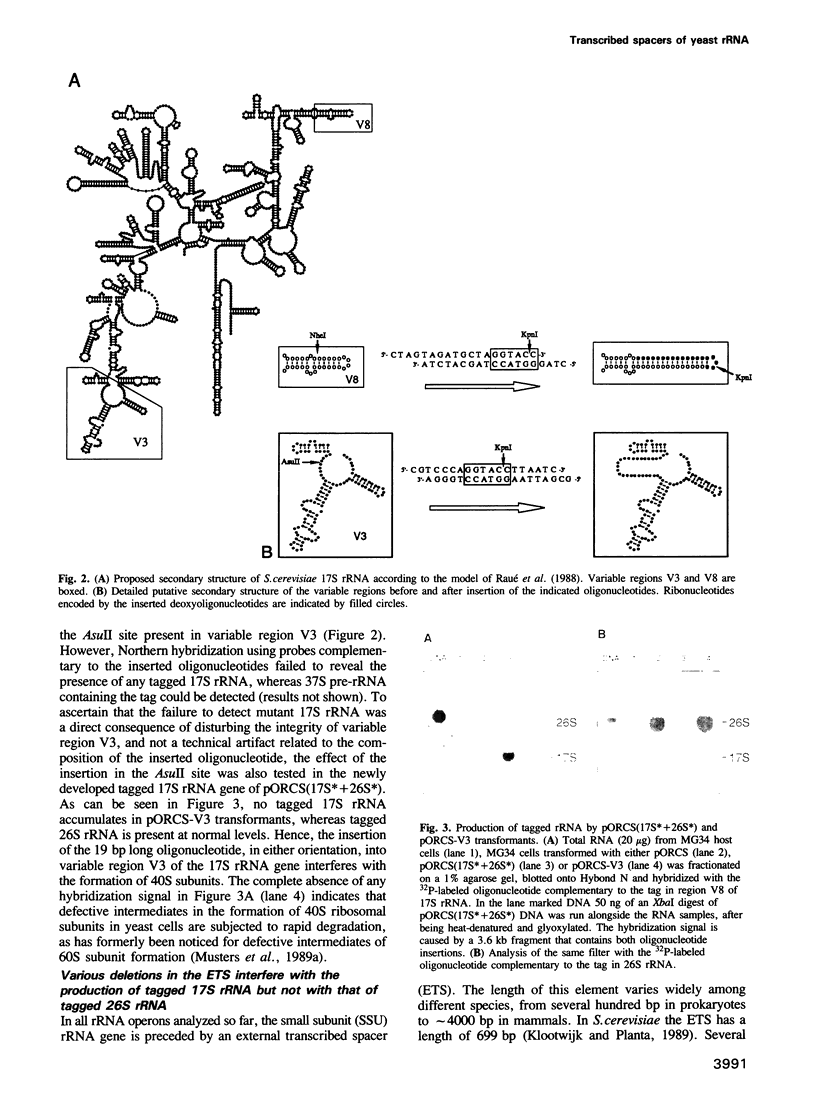
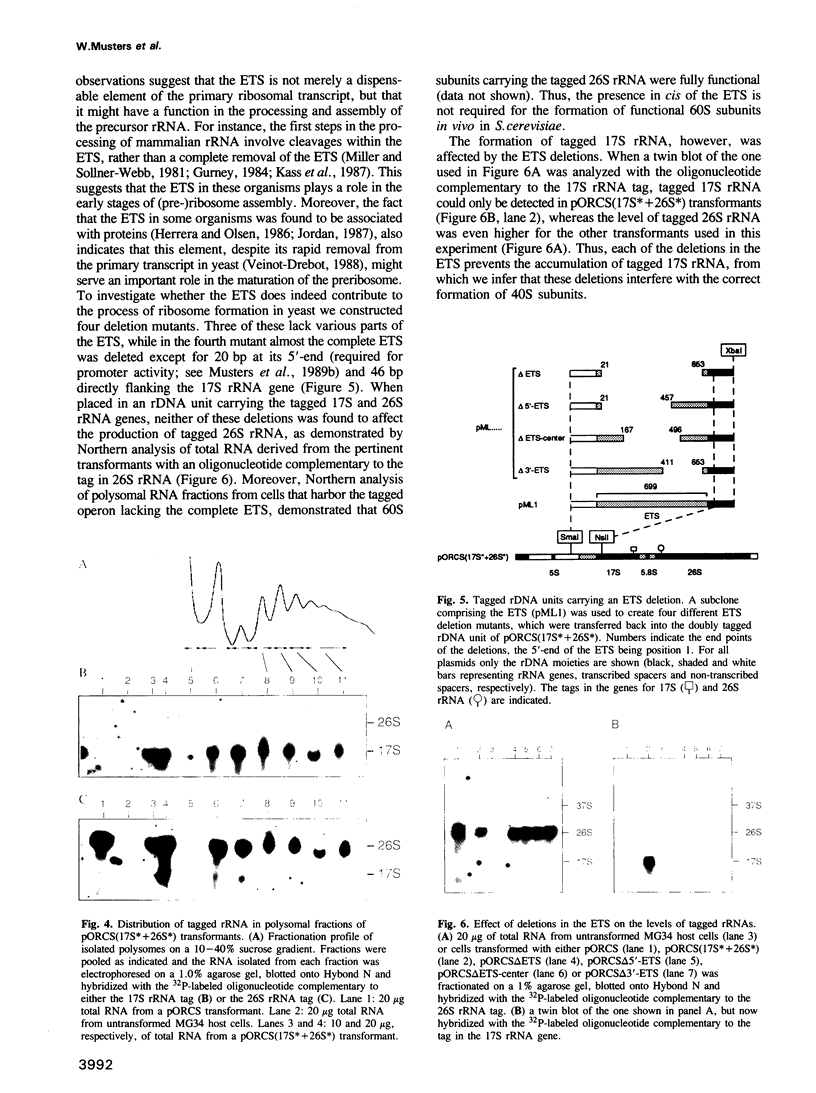
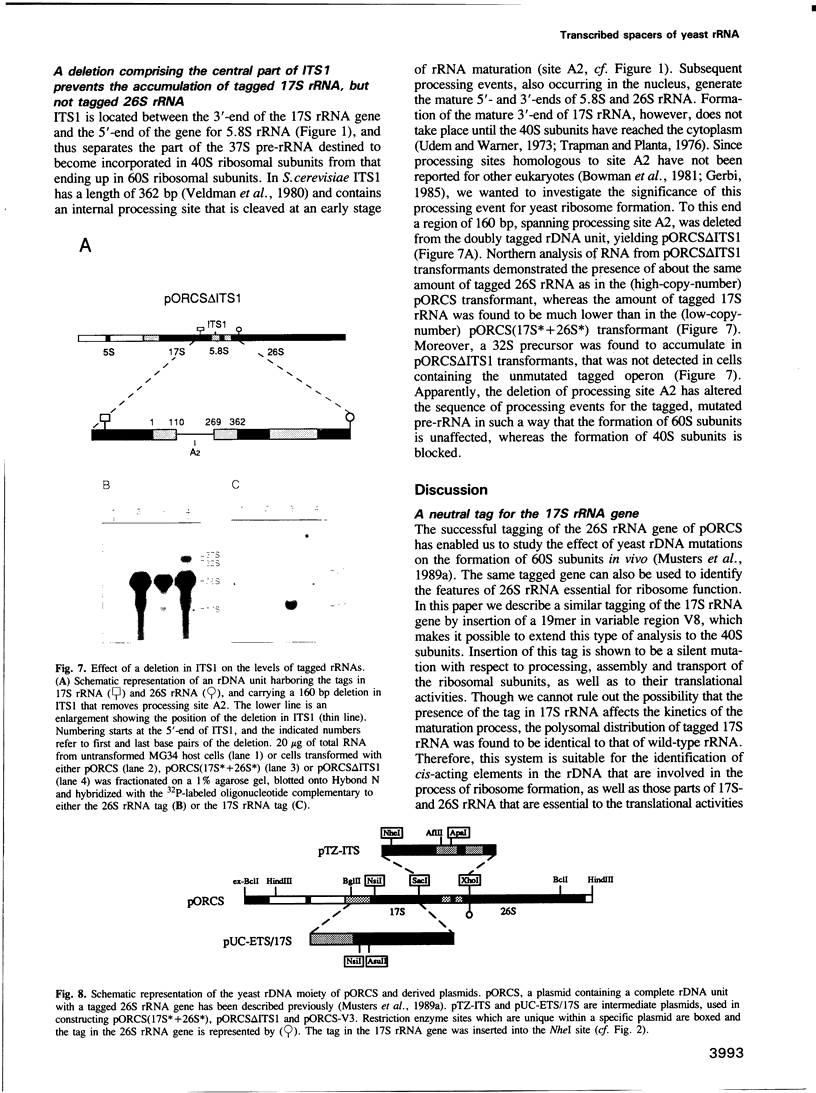
Making use of an rDNA unit, containing oligonucleotide tags in both the 17S and 26S rRNA gene, we have analyzed the effect of various deletions in the External Transcribed Spacer (ETS) and in one of the Internal Transcribed Spacers 1 (ITS1) on the process of ribosome formation in yeast. By following the fate of the tagged transcripts of this rDNA unit in vivo by Northern hybridization we found that deleting various parts of the ETS prevents the accumulation of tagged 17S rRNA and its assembly into 40S subunits, but not the formation of 60S subunits. Deleting the central region of ITS1, including a processing site that is used in an early stage of the maturation process, was also found to prevent the accumulation of functional 49 S subunits, whereas no effect on the formation of 60S subunits was detected. The implications of these findings for yeast pre-rRNA processing are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Boer P. H., Gray M. W. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell. 1988 Nov 4;55(3):399–411. doi: 10.1016/0092-8674(88)90026-8. [DOI] [PubMed] [Google Scholar]
- Bowman L. H., Rabin B., Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981 Oct 10;9(19):4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Crick F. H. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Dams E., Hendriks L., Van de Peer Y., Neefs J. M., Smits G., Vandenbempt I., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988;16 (Suppl):r87–173. doi: 10.1093/nar/16.suppl.r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. A. Molecular evolution. rDNA world falling to pieces. Nature. 1988 Dec 15;336(6200):623–624. doi: 10.1038/336623a0. [DOI] [PubMed] [Google Scholar]
- Gurney T., Jr Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 +/- 100 nucleotides from the 5' end and the second 500 +/- 100 nucleotides from the 3' end of a 13.9 kb precursor. Nucleic Acids Res. 1985 Jul 11;13(13):4905–4919. doi: 10.1093/nar/13.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera A. H., Olson M. O. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry. 1986 Oct 7;25(20):6258–6264. doi: 10.1021/bi00368a063. [DOI] [PubMed] [Google Scholar]
- Hogan J. J., Gutell R. R., Noller H. F. Probing the conformation of 18S rRNA in yeast 40S ribosomal subunits with kethoxal. Biochemistry. 1984 Jul 3;23(14):3322–3330. doi: 10.1021/bi00309a032. [DOI] [PubMed] [Google Scholar]
- Jordan G. At the heart of the nucleolus. Nature. 1987 Oct 8;329(6139):489–490. doi: 10.1038/329489a0. [DOI] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987 Aug;7(8):2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., Planta R. J. Isolation and characterization of yeast ribosomal RNA precursors and preribosomes. Methods Enzymol. 1989;180:96–109. doi: 10.1016/0076-6879(89)80095-3. [DOI] [PubMed] [Google Scholar]
- Kraig E., Haber J. E., Rosbash M. Sporulation and rna2 lower ribosomal protein mRNA levels by different mechanisms in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1199–1204. doi: 10.1128/mcb.2.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak W., Denman R., Nurse K., Hellmann W., Boublik M., Gehrke C. W., Agris P. F., Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987 Apr 21;26(8):2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie. 1987 Jan;69(1):11–23. doi: 10.1016/0300-9084(87)90267-7. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Musters W., Knol J., Maas P., Dekker A. F., van Heerikhuizen H., Planta R. J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989 Dec 11;17(23):9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Venema J., van der Linden G., van Heerikhuizen H., Klootwijk J., Planta R. J. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989 Feb;9(2):551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raué H. A., Klootwijk J., Musters W. Evolutionary conservation of structure and function of high molecular weight ribosomal RNA. Prog Biophys Mol Biol. 1988;51(2):77–129. doi: 10.1016/0079-6107(88)90011-9. [DOI] [PubMed] [Google Scholar]
- Raziuddin, Little R. D., Labella T., Schlessinger D. Transcription and processing of RNA from mouse ribosomal DNA transfected into hamster cells. Mol Cell Biol. 1989 Apr;9(4):1667–1671. doi: 10.1128/mcb.9.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTER M. RIBOSOMAL RNA ON THE SURFACE OF RIBOSOMES. Science. 1963 Sep 13;141(3585):1049–1050. doi: 10.1126/science.141.3585.1049. [DOI] [PubMed] [Google Scholar]
- Sweeney R., Yao M. C. Identifying functional regions of rRNA by insertion mutagenesis and complete gene replacement in Tetrahymena thermophila. EMBO J. 1989 Mar;8(3):933–938. doi: 10.1002/j.1460-2075.1989.tb03454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapprich W. E., Hill W. E. Involvement of bases 787-795 of Escherichia coli 16S ribosomal RNA in ribosomal subunit association. Proc Natl Acad Sci U S A. 1986 Feb;83(3):556–560. doi: 10.1073/pnas.83.3.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapman J., Planta R. J. Maturation of ribosomes in yeast. I Kinetic analysis by labelling of high molecular weight rRNA species. Biochim Biophys Acta. 1976 Sep 6;442(3):265–274. doi: 10.1016/0005-2787(76)90301-4. [DOI] [PubMed] [Google Scholar]
- Udem S. A., Warner J. R. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem. 1973 Feb 25;248(4):1412–1416. [PubMed] [Google Scholar]
- Veinot-Drebot L. M., Singer R. A., Johnston G. C. Rapid initial cleavage of nascent pre-rRNA transcripts in yeast. J Mol Biol. 1988 Jan 5;199(1):107–113. doi: 10.1016/0022-2836(88)90382-8. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., van Heerikhuizen H., Planta R. J. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon. Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res. 1981 Oct 10;9(19):4847–4862. doi: 10.1093/nar/9.19.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware V. C., Renkawitz R., Gerbi S. A. rRNA processing: removal of only nineteen bases at the gap between 28S alpha and 28S beta rRNAs in Sciara coprophila. Nucleic Acids Res. 1985 May 24;13(10):3581–3597. doi: 10.1093/nar/13.10.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M. C., Yao C. H. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol Cell Biol. 1989 Mar;9(3):1092–1099. doi: 10.1128/mcb.9.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]