Abstract
Transposable elements (TEs) have been recognized as potentially powerful drivers of genomic evolutionary change, but factors affecting their mobility and regulation remain poorly understood. Chaperones such as Hsp90 buffer environmental perturbations by regulating protein conformation, but are also part of the PIWI-interacting RNA pathway, which regulates genomic instability arising from mobile TEs in the germline. Stress-induced mutagenesis from TE movement could thus arise from functional trade-offs in the dual roles of Hsp90. We examined the functional constraints of Hsp90 and its role as a regulator of TE mobility by exposing nematodes (Caenorhabditis elegans and Caenorhabditis briggsae) to environmental stress, with and without RNAi-induced silencing of Hsp90. TE excision frequency increased with environmental stress intensity at multiple loci in several strains of each species. These effects were compounded by RNAi-induced knockdown of Hsp90. Mutation frequencies at the unc-22 marker gene in the offspring of animals exposed to environmental stress and Hsp90 RNAi mirrored excision frequency in response to these treatments. Our results support a role for Hsp90 in the suppression of TE mobility, and demonstrate that that the Hsp90 regulatory pathway can be overwhelmed with moderate environmental stress. By compromising genomic stability in germline cells, environmentally induced mutations arising from TE mobility and insertion can have permanent and heritable effects on both the phenotype and the genotype of subsequent generations.
Keywords: Hsp90, transposable elements, piRNAs, genome stability, stress-induced mutagenesis, evolution
Introduction
The capacity to buffer environmental and genetic perturbations is a fundamental property of all organisms (Waddington 1942; Pigliucci 1996). Stability in response to environmental and genetic disturbances requires numerous, highly responsive, and potentially overlapping mechanisms operating across multiple levels of biological organization (de Visser et al. 2003; Jarosz et al. 2010). At the cellular level, heat-shock proteins serve as molecular chaperones, helping fold other proteins into their appropriate conformations following environmental and physiological stress (Erlejman et al. 2014). Hsp90 is one such molecular chaperone, facilitating the proper functioning of many cellular proteins under normal conditions, as well as in response to hyperthermia, oxidative stress, and other environmental extremes (Feder and Hofmann 1999; Sangster et al. 2004). Hsp90 interacts with a broad, yet select set of client regulatory proteins, including various kinases and transcription factors (Mayer and Bukau 1999; Young et al. 2001), and experimentally induced perturbations of this chaperone function have shown that Hsp90 also acts as a buffer of pre-existing, cryptic genetic variability (Sangster et al. 2004; Gangaraju et al. 2011). Overload of the cellular stress response by either Hsp90 inhibition or acute environmental stress increases phenotypic variation in both plants and animals (Queitsch et al. 2002; Jarosz and Lindquist 2010). However, there is also evidence that Hsp90 contributes to the buffering of genetic change, both through the regulation of DNA repair complexes involved in maintaining genetic fidelity, and through the suppression of transposable element movements within the genome (Piacentini et al. 2014; Sekimoto et al. 2010).
Transposable elements (TEs) are mobile genetic elements found throughout most species’ genomes, and their movements within the host genome are often associated with induced mutations; they can alter the expression of genes by introducing alternative promoters, splice variants, frame shifts, and sequence deletions or duplications (Kazazian 2004; Cowley and Oakey 2013). Although TE-induced mutations can have deleterious effects, particularly in germline DNA, TEs have also been important in generating adaptive genomic variability across a wide range of taxa (Kazazian 2004; Biémont and Vieira 2006). TEs are often categorized into two classes, based on their mechanism of transposition (Kidwell and Lisch 2001; Sela et al. 2010). Mobility among Class I TEs or retrotransposons is restricted to “copy and paste” mechanisms via RNA intermediates, whereas Class II or DNA transposon mobility involves an autonomous “cut and paste” mechanism through a DNA intermediate (Bonchev and Parisod 2013).
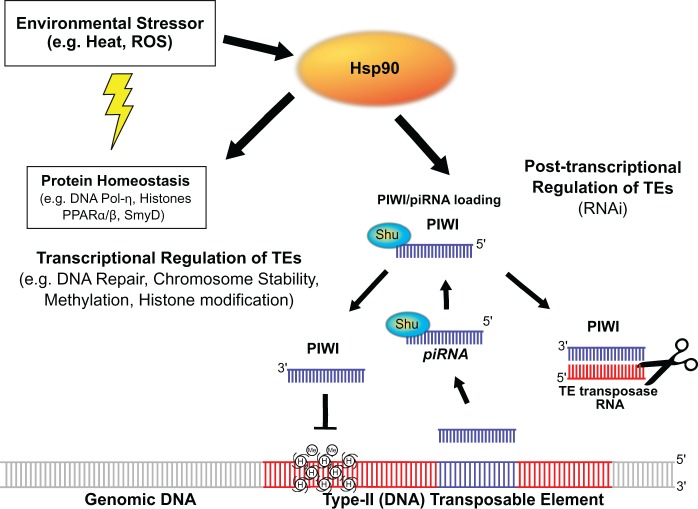
Transposon mobility can be regulated transcriptionally through histone modification and cytosine methylation (Yoder et al. 1997; Bestor 1998), as well as post-transcriptionally through the PIWI/piRNA pathway (Das et al. 2008). The PIWI/piRNA pathway involves a subfamily of Argonaute proteins, referred to as PIWI proteins, which act as catalysts in the gene silencing RNA-interference (RNAi) pathway, as well as a subclass of small RNAs that direct PIWI to its mRNA targets (PIWI-interacting RNAs or piRNAs). Along with a cochaperone known as Shu, the loading of pre-piRNAs onto the PIWI complex for TE suppression requires Hsp90 (Olivieri et al. 2012; Izumi et al. 2013) (fig. 1). In Drosophila melanogaster, transcription rates for several (Class I) retrotransposons were shown to increase in hsp83scratch flies hypomorphic for the Hsp90 analog, as well as in the testes of males treated with an Hsp90 inhibitor, geldanamycin (Specchia et al. 2010). What remains unclear is the extent to which Hsp90 suppresses TEs across taxa, whether Hsp90 suppresses DNA (Class II) in addition to Class I transposon excision and insertion frequency, and how environmental stress within a naturally occurring range might constrain the capacity of Hsp90 to buffer genomic instability.
Fig. 1.—
Pathway of the 90-kilodalton heat-shock protein (Hsp90) cellular stress response. Hsp90 expression increases in response to environmental stress, which perturbs protein form and function (protein homeostasis), including proteins involved in genomic DNA repair (e.g., DNA Pol-η), transcriptional activity (PPARα/β), chromosome integrity, and epigenetic regulation (e.g., histone acetyltransferase, SmyD). Hsp90, along with the cochaperone Shutdown (Shu) is also necessary for loading of piwi-interacting RNAs (piRNAs) onto the PIWI protein complex. The PIWI/piRNA pathway can silence transposable elements (TEs) transcriptionally (left), through histone methylation (Me) or other modifications to histones (H), or post-transcriptionally (right), through RNA-interference-mediated targeting of TE transposase RNAs. Genomic DNA (gray), TE DNA/RNA (red), and piRNA (purple) are also shown.
Here, we examine the effects of several moderate environmental stresses on the capacity of Hsp90 to regulate the transcription and excision of two groups of Class II DNA transposons. We used the model nematode species Caenorhabditis elegans and C. briggsae, where the mechanisms for TE silencing have been extensively studied (Sijen and Plasterk 2003; Bagijn et al. 2012) and sensitive assays for measuring TE excision frequencies have been successfully applied (Brownlie and Whyard 2004; Brownlie et al. 2005). To broaden our comparative analysis, we included multiple strains of each species, and excision frequencies at several different loci for each strain. To test if Hsp90 faces functional constraints in buffering environmental and genetic instability, we subjected the nematodes to both heat and oxidative stresses, with and without RNAi-mediated knock-down of Hsp90 (daf-21). The stress-induced transposon excision and transposition events we observed in the progeny were exposure-dependent, and were amplified by RNAi-mediated Hsp90 knock-down. Our treatments, typically viewed as short-term physiological challenges, demonstrate that long-term, heritable genetic change may arise through TE-mediated activity following moderate environmental stress.
Materials and Methods
Nematode Strains and DNA Extractions
The C. elegans strains used in this study were N2 and AB2 (Egilmez et al. 1995) and the C. briggsae strains used in this study were transgenic lines of AF16 and DH1300 (supplementary fig. S1, Supplementary Material online). Genomic DNA was extracted from nematodes grown in liquid culture, at mixed developmental stages (Hope 1999), using the Genome 100 tip kit (Qiagen) according to the manufacturer’s instructions. DNA was extracted from single worms using a previously described method (Williams et al. 1992).
Development of sid-2 Transgenic Strains of C. briggsae
Transgenic lines of C. briggsae strains AF16 and DH1300 were produced using techniques established for C. elegans transformation (Evans 2006). The coding sequence of Cel-sid-2 (Genbank NC_003281) and its 5’ promoter and 3’ sequences were amplified by PCR from C. elegans genomic DNA using Phusion DNA polymerase and the primers Cel-sid2F (5’-CCAACTTCGTAGAGCTGAGCAGC-3’) and Cel-sid2R (5’-TTACTGCAAGCTGAGCTATTTTTTCG-3’). PCR products were gel-purified using the QIAquick Gel Extraction kit (Qiagen). The purified DNA (10 ng/µl) was independently injected into the gonad of adult worms, along with carrier pBS plasmid (150 ng/ml) and 10 ng/µl of the coinjection marker plasmid pWD47 (Jose et al. 2009), which carries the Pmyo-2::DsRed markers, to monitor progeny for visible phenotypes. Each injected animal was transferred singly to a culture plate. The DsRed phenotype was assayed in the F1 using a Leica MZ FL3 fluorescence stereomicroscope and those worms showing the phenotype were back-crossed to their respective parental stock for three generations before they were used for RNA interference assays. Individual worms (10) from each strain in the F4 were tested for the presence of the sid-2 gene by extracting their DNA and using a PCR screen using the primers described above.
RNA Interference Knockdown of Hsp90
RNAi experiments were performed by feeding dsRNA-expressing E. coli to the two strains of C. elegans and the two strains of transgenic C. briggsae carrying the Cel-sid-2 gene (supplementary fig. S1, Supplementary Material online), according to standard procedures (Kamath et al. 2003). The Hsp90 (daf-21; WBGene00000915) gene of C. elegans was amplified by PCR from cDNA derived from RNA extracted from multi-staged C. elegans N2 strain using standard techniques, with the primers CeHsp90F (5’-AGTTCTTTCGGTTTTCGGGT-3’) and CeHsp90R (5’-GCGAATGATTTCGAAACTAAAAA-3’). The primers used for PCR amplification of the Hsp90 gene in C. briggsae (Genbank XM_002637731) were as follows: CbrHsp90F, 5’-ATGTCCGAGAACGCTGAAAC-3’ and CbrHsp90R, 5’-ATGAGACCAACCTGGTAGCG-3’. The PCR products were cloned into the PCR cloning vector pDrive (Qiagen) and then subsequently excised using the restriction enzymes EcoRI/XhoI and ligated into a similarly digested dsRNA expression vector pL4440. The plasmids were sequenced prior transforming E. coli HT115 cells, which were grown in LB containing ampicillin and 1 mM IPTG to induce expression of the dsRNA within the bacteria. A pL4440 plasmid lacking any insert served as a negative control. After 4 h, the bacteria were plated on NGM agar plates containing 50 µg/ml ampicillin, 6 µg/ml tetracycline, 1 mM IPTG, and 5 µg/ml cholesterol. L1 larvae were grown on these plates at 20 °C and after 3 days, worms were collected for either RNA extractions to assess the extent of RNAi, or were subjected to different stresses, described below.
QRT-PCR Analysis of RNAi Treatments
To analyze the extent of daf-21 (Hsp90) transcript reduction in worms fed E. coli expressing dsRNA, quantitative reverse transcriptase PCR (QRT-PCR) experiments were performed. Worms were washed off their plates with M9 medium, and bacteria were removed by several washing steps. RNA was isolated from groups of 20 worms using an RNeasy Mini Kit (Qiagen). The RNA was subjected to reverse transcription with random hexamer primers using ALV reverse transcriptase (Promega). QRT-PCR was carried out according to the manufacturer's instructions, using SYBR Green MasterMix (BioRad) in an iQ5 Real-Time PCR Detection System (BioRad). Primer pairs were designed to amplify a 200–250 bp segment of the target gene, using one primer per pair that spanned an exon–exon junction. The primer pairs for C. elegans were for act-1 (Fwd: AATCCAAGA GAGGTATCCTTA, Rev: GATGGCGACATACATGGC T); pgk-1 (Fwd: TTTGATCCGTGTTGACTTCAAT, Rev: GAAGAGAACATCTTTTTTCAAGA); CeHsp90 (Fwd: AAGATGAGGAGGCTGTCGA, Rev: CATTGGACA AGCTCTTGTAGA). The primer pairs for C. briggsae were for Cbr-act-5 (Fwd: GCAACAGAGAAAAGATGACACAG, Rev: GATTTCTTCTTACCTCATAGATTGG); Cbr-pgk-1 (GAGTTGATGCTTCTGGATCCAAGG, Rev: CCGATCGCAATACCTTGCTGAAATAAG); and Cbr-Hsp90 (Fwd: GAGTTCCGTGCTCTTCTCTTCG, Rev: TTCCGAGCTTAAGGTTCTTTCCG). The log 2-fold changes of the genes of interest upon heat shock were normalized relative to the unc-60 (actin depolarizing factor) and pgk-1 (phosphoglycerate kinase 1) genes and averaged for three separate biological experiments.
Heat Shocks and Oxidative Stress Treatments
The nematodes were subjected to sub-lethal heat shocks or oxidative stresses to assess impacts on transposon activity (supplementary fig. S1, Supplementary Material online). For the oxidative stress treatments, adult worms were transferred to 96-well plates (20 worms/well) containing M9 buffer with 0 mM (negative controls), 0.25 mM (low oxidative stress; H2O2-Low), or 1.5 mM (high oxidative stress; H2O2-High) H2O2 for 8 h incubations at 20 °C. Thereafter, the worms were washed with 4 successive washes of M9 buffer before being transferred to E. coli-coated NGM agar plates for 24 h to recover. For the heat shock treatments, the nematodes, in 96-well plates containing M9 buffer were subjected to 35 °C for 1 h (low heat shock; Heat-Low), five serial exposures of 35 °C for 1 h, 30 min at 20 °C (serial heat shock, Heat-Serial), or 39 °C for 2 h (high heat shock; Heat-High). Negative control worms were held at 20 °C for 5 h (No Stress). After the heat stresses, the worms were transferred to agar plates for 24 h as previously described. Thereafter, the worms were pooled in groups of 20 and their DNA was extracted as described above, to assess transposon mobility.
Quantitative PCR Assays
Excision frequencies for three different Tc1 and three CemaT1 loci in C. elegans and three different Tcb1 and three CbmaT1 in C. briggsae were determined using a quantitative PCR (QPCR) approach (supplementary fig. S1, Supplementary Material online). For each locus examined, three primers were designed: 1) a forward primer that flanked the transposon approximately 30–50 bp from the transposon ITR; 2) a reverse primer that spanned the transposon ITR sequence and the flanking sequence; and 3) a reverse primer that spanned the site of a putative excision footprint. The first pair of primers detected nonexcised TEs, whereas the first and third primers detected transposon excisions. QPCR reactions were performed in triplicate with 50 ng of genomic DNA (obtained from 100 pooled worms subjected to the same treatment) using SYBR Green Supermix (BioRad) on a BioRad iQ5 Real-Time PCR Detection System. A melt-curve analysis was performed on all samples to assess the purity of amplified products. PCR primers (see supplementary table S1, Supplementary Material online) were designed such that the PCR products of the excised and nonexcised transposons were of similar size (∼80 bp) and GC% content. Efficiencies of the primer sets were calculated using the method of Pfaffl (2001), and were found to be essentially equivalent for all primers sets, ranging between 93% and 95%. The threshold cycle values for all excised and nonexcised transposon loci were normalized relative to the internal standard 26S rRNA gene. From these calculations, the percentage of loci containing a transposon excision footprint was determined, as previously described (Brownlie and Whyard 2004; Brownlie et al. 2005).
Assessment of Mutation Frequency
The frequency of spontaneous mutations at the unc-22 locus in one strain (N2) of C. elegans and two strains of (AF16 and DH1300) of C. briggsae was assessed using an assay similar to that described previously (Moerman and Baillie 1979) to screen for the generation of unc-22 mutants in C. elegans (supplementary fig. S1, Supplementary Material online). Briefly, single worms were placed on (100 mm) NGM plates and left until the plates were crowded (105 worms/plate). Worms were then washed off the plates in 5 ml of 1% nicotine solution (Sigma) and within 5 min. the animals were examined for the presence of any twitching individuals among the paralyzed worms. Although wild-type C. elegans worms become paralyzed when exposed to 1% nicotine, homozygous and heterozygous unc-22 mutants will twitch. Plates with one or more twitching worms were scored as positive and those on which only wild type worms were observed were scored as negative. Any twitching worms were recovered and their progeny were tested to ensure they produced unc-22 mutants. Three replicates of 20–30 plates for each combination of strain, stress treatment, and RNAi treatment were examined, with 2,717 plates examined for mutations overall. The proportion of plates not showing mutants (P0) was used to calculate the spontaneous mutant frequency using the Poisson distribution (P0 = e−aN, where a is the mutant frequency and N is the population size/plate).
Ten lineages of C. elegans and ten lineages of C. briggsae with heat or oxidative stress-induced unc-22 mutants were cultured and ∼50 progeny worms were pooled and their DNA was extracted, as described above (supplementary fig. S1, Supplementary Material online). The DNA was subjected to PCR using 10 different pairs of primers that spanned almost the entire length of unc-22, excluding large introns (see supplementary table S2, Supplementary Material online). The PCR products were resolved on 0.8% agarose gels and any PCR products that were between 0.9 and 1.6 kb larger than expected were sequenced to determine whether a transposon sequence had inserted within unc-22. All sequenced insertions appeared to be homozygous.
Statistical Analyses
Excision frequency was examined for CemaT1/CbmaT1 and Tc1/Tcb1 transposons at three loci each in two strains in C. elegans (N2 and AB2) and two strains in C. briggsae (AF16 and DH1300). TE mobility and insertion frequency was measured at the unc-22 gene for the N2 strain in C. elegans and both the AF16 and the DH1300 strains in C. briggsae. Excision frequency was analyzed using ordinary least squares regression, and was logged to meet model assumptions. The effects of treatment and Hsp90 RNAi on mutation frequency were analyzed using a general linear model with quasibinomial family to account for overdispersion. All models were initially run with strain, locus, treatment, RNAi, and all two-way interactions between these parameters. To avoid over-parameterization, three- and four-way interactions were not included. Nonsignificant two-way interactions were sequentially removed until only significant (P < 0.05) interactions remained. Nonsignificant main effects were also removed, unless they contributed to a significant two-way interaction. Effects of treatment and RNAi (fold increases) on excision or mutation frequencies were derived from corresponding regression or generalized linear model estimates, and are relative to nonstressed control animals in the absence of RNAi. The reference strain and locus varied by species, and final models including all parameters and estimates are provided (see supplementary tables S3–S5, Supplementary Material online). Estimates of treatment effects in the results are for the range of all treatments except Heat-Low, which was not significantly different for any analysis.
Results
Knockdown of Hsp90 in C. briggsae Transformed with Cel-Sid-2
When certain strains of C. elegans are fed dsRNA, either as free oligonucleotides or expressed within transformed E. coli, RNAi is induced throughout most of the animal’s tissues. This systemic RNAi is mediated by SID-2, a transmembrane protein in the worm intestine, and by SID-1, a related protein in most other tissues (Winston et al. 2002), both of which facilitate cellular uptake of the dsRNA. C. briggsae has a variant of sid-2 that appears unable to take up dsRNAs. Winston et al. (2007) demonstrated that C. briggsae could be induced to take up ingested dsRNAs by expressing the C. elegans sid-2 (Cel-sid-2) gene. Using standard transformation techniques, we similarly prepared two different strains of C. briggsae (AF16 and DH1300) that expressed the Cel-sid-2 gene. Representative F4 worms from each lineage were PCR-screened for the presence of the transgene, and all individuals carried the Cel-sid-2 gene. The worms were fed E. coli containing Cbr-daf-21/L4440, which expressed dsRNA that targets the Hsp90-encoding gene, and qRT-PCR of the F3 worms confirmed that the gene transcript levels were reduced >80%, relative to negative control worms, in both strains (table 1).
Table 1.
Hsp90 Transcript Knockdown Levels in C. briggsae Carrying Cel-sid-2 and fed E. coli Expressing daf-21 dsRNAa
| Strain | Transgenic F1 | Transgenic F2 | % RNAi Knockdown in F4 Transformantsa |
|---|---|---|---|
| AF16 | 18/22 | 10/22 | 88 ± 4 |
| DH1300 | 7/25 | 4/25 | 84 ± 3 |
The extent of RNAi-mediated knockdown of daf-21 transcripts was calculated relative to the expression levels of worms fed E. coli carrying the empty vector pL4440. The values represent the means and standard errors of 3 replicates of 10 pooled worms. The number of worms depicted are those expressing DsRed in the F1 and F2 worms, relative to the number of surviving injectants.
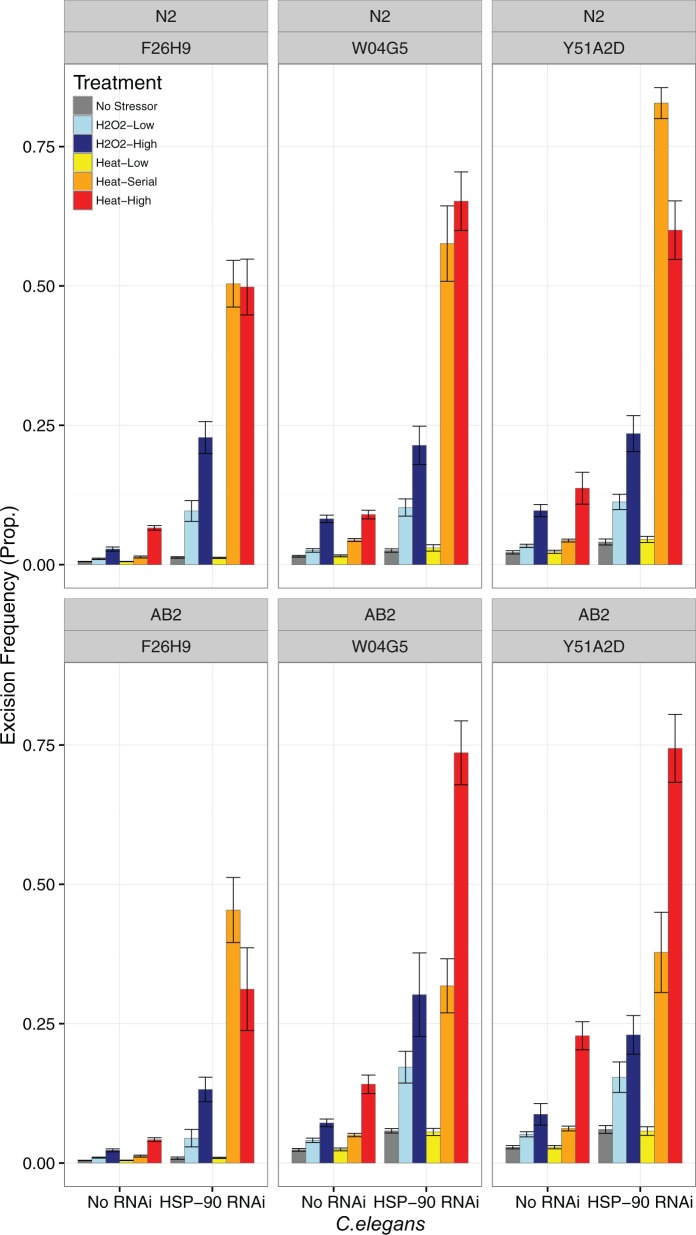
CemaT1 and Tc1 excision frequency in C. elegans
The excision frequencies of two different transposons, Tc1 and CemaT1, were examined in C. elegans exposed to different oxidative and heat stresses using a PCR-based assay developed previously (Brownlie and Whyard 2004). The assay compares the relative abundance of nonexcised transposons to excision footprints at a given locus. Three loci of each transposon were examined in two separate C. elegans strains, and the excision frequencies of these transposons in nonstressed, control worms were comparable to those observed in a previous study (Brownlie and Whyard 2004). For both CemaT1 and Tc1, certain loci showed significantly higher excision frequency than others (F2,358 = 208.5, P < 0.001 and F2,345 = 771.5, P < 0.001, respectively; see supplementary table S3, Supplementary Material online), suggesting that individual transposon activity is influenced by the local genomic environment. For Tc1, excision frequencies were also significantly lower in the AB2 strain compared with the N2 strain (F1,345 = 48.2, P < 0.001). Despite loci- and strain-specific differences in transposon activity, stress treatment had an overall significant effect on both CemaT1 and Tc1 excision frequencies (F5,358 = 303.3, P < 0.001 and F5,345 = 361.5, P < 0.001, respectively; fig. 2, supplementary fig. S2, Supplementary Material online. The two oxidative stress treatments and more extreme heat stresses (a series of 1 h 37 °C shocks or a 2h 39 °C shock) were associated with 2- to 7-fold increases in excision frequencies for both CemaT1 and Tc1 (P < 0.001 for all). Low-heat stress (a brief 1 h heat shock at 37 °C) did not significantly affect excision frequencies for either CemaT1 or Tc1 (P > 0.65 for both; see supplementary table S3, Supplementary Material online). QRT-PCR analyses confirmed that all stress treatments caused significant (P < 0.05), approximately 2-fold increases in levels of Hsp90 mRNA in both C. elegans strains (N2, AB2) relative to no treatment controls (see supplementary table S6, Supplementary Material online).
Fig. 2.—
Excision frequency for CemaT1 transposons at three loci (F26H9, W04G5, Y51A2D) for two strains of C. elegans (N2, AB2) in response to five different conditions of environmental stress, plus no stress controls. Excision frequency given as the proportion of excision footprints to the number of nonexcised transposons at a given locus. Bars show proportions of excision for Low- and High-oxidative stress, Low (35 °C for 1 h), Serial (five serial exposures of 35 °C for 1 h, 30 min at 20 °C) and High (39 °C for 2 h) heat exposures, and No Stress controls.
RNAi-mediated knockdown of Hsp90 increased excision frequencies of the CemaT1 transposon overall (F1,358 = 856.2, P < 0.001; fig. 2), and amplified the effect of stress treatment for all (RNAi × Treatment: P < 0.001 for all) except the Heat-Low treatment (Heat-Low: P = 0.931; see supplementary table S3, Supplementary Material online). Hsp90 RNAi also increased the excision frequency for Tc1 overall (F1,345 = 35.95, P < 0.001; supplementary fig. S2, Supplementary Material online), but only amplified the effects of stress treatment on excision frequency for H2O2-High and Heat-High treatments (P < 0.05 for both). For Tc1, RNAi-induced knockdown of Hsp90 had no effect on excision frequency for No Stress controls (P = 0.303; see supplementary table S3, Supplementary Material online), indicating that environmental stress is required to induce the effect of RNAi. QRT-PCR analyses confirmed that RNAi was effective, even under stress conditions; control, unstressed worms and stress-treated worms alike showed more than 70% knockdown of Hsp90 transcripts (see supplementary table S7, Supplementary Material online). The combined effect of the environmental stress treatment and RNAi-mediated knockdown of Hsp90 increased excision frequency between 2- and 9-fold for Tc1 and between 7- and 41-fold for CemaT1.
CbmaT1 and Tcb1 Excision Frequencies in C. briggsae
The genome of C. briggsae has autonomous transposons related to Tc1 and CemaT1, Tcb1, and CbmaT1, which have been previously shown to be active (Brownlie et al. 2005). As in C. elegans, there were significant differences in excision frequency for CbmaT1 and Tcb1 between loci (F2,272 = 541.37, P < 0.001 and F2,271 = 1095.25, P < 0.001, respectively; see supplementary table S4, Supplementary Material online). Excision frequencies at CB015K23 and CB046A04 were lower than at the CBRG21D19 locus for CbmaT1 (P < 0.02 for both), and C007801047.Contig1 and C012001013.Contig3 were significantly lower than the C009001188.Contig2 locus for Tcb1 (P < 0.001 for both). Compared with the AF16 strain, there was an increase in excision frequency for the DH1300 strain for CbmaT1 (F1,272 = 51.52, P < 0.001), and significant locus by strain interactions for both CbmaT1 and Tcb1 (F2,272 = 300.54, P < 0.001 and F2,271 = 33.80, P < 0.001, respectively; see supplementary table S4, Supplementary Material online). Stress treatment in general affected excision frequencies for both CbmaT1 and Tcb1 (F2,272 = 305.10, P < 0.001 and F2,271 = 118.05, P < 0.001, respectively; supplementary figs. S3 and S4, Supplementary Material online). More specifically, all stress treatments except the Heat-Low treatment increased excision frequency of CbmaT1 and Tcb1 (P < 0.001 for all except Heat-Low; see supplementary table S4, Supplementary Material online). QRT-PCR analyses confirmed that all stress treatments caused significant (P < 0.05), approximately 2-fold increases in levels of Hsp90 mRNA in both C. briggsae strains (AF16, DH1300) relative to no treatment controls (see supplementary table S6, Supplementary Material online).
RNA-mediated knockdown of Hsp90 increased overall excision frequencies of CbmaT1 (F1,272 = 63.84, P < 0.001; supplementary fig. S3, Supplementary Material online) and Tcb1 (F1,272 = 26.80, P < 0.001; supplementary fig. S4, Supplementary Material online). However, as with the Tc1 transposon, Hsp90 RNAi had no independent effect on excision frequency for No Stress controls for either CbmaT1 or Tcb1 (P = 0.290 and P = 0.437, respectively; see supplementary table S4, Supplementary Material online), indicating that environmental stress is necessary to induce the effect of RNAi. For both C. briggsae transposons, RNAi amplified the effect of stress treatment overall (RNAi × Treatment: F5,272 = 6.65, P < 0.001 and F5,271 = 2.65, P = 0.023). However, this effect was limited to Heat-High and H2O2-High treatments, where excision frequencies were significantly higher for both CbmaT1 and Tcb1 when combined with RNAi-induced knockdown of Hsp90 than for these treatments alone (P < 0.02 for all; see supplementary table S4, Supplementary Material online). QRT-PCR analyses confirmed that RNAi was effective, even under stress conditions; control, unstressed worms and stress-treated worms alike showed more than 70% knockdown of Hsp90 transcripts (see supplementary table S7, Supplementary Material online). The combined effects of stress treatment and RNAi increased excision frequencies from 2- to 5-fold in Tcb1, and 2- to 9-fold in CbmaT1.
unc-22 Mutation Frequency
Spontaneous unc-22 mutations in C. elegans and C. briggsae appear to be rare events, based on the frequencies observed in nonstressed, non-RNAi treated worms, but the frequencies observed in this study are comparable to those reported in other studies (Moerman and Waterston 1984; Brownlie et al. 2005). In C. briggsae, where we examined two strains, we observed mutation frequencies that mirrored excision frequency, with higher overall mutation frequency in the DH1300 compared with AF16 strain ( = 20.87, P < 0.001; fig. 3). Similar to the observed excision frequencies in C. elegans and C. briggsae, mutation frequency in unc-22 generally increased in response to RNAi ( = 143.23, P < 0.001 and = 156.70, P < 0.001), and stress treatment ( = 497.35, P < 0.001 and = 836.28, P < 0.001, respectively; fig. 3). In the absence of RNAi, mutation frequency increased for all but the Heat-Low and H2O2 treatments in C. elegans (P < 0.002 for Heat-High, Heat-Serial, and H2O2-High), whereas in C. briggsae all but the Heat-Low treatment increased mutation frequency (P < 0.006 for all except Heat-Low).
Fig. 3.—
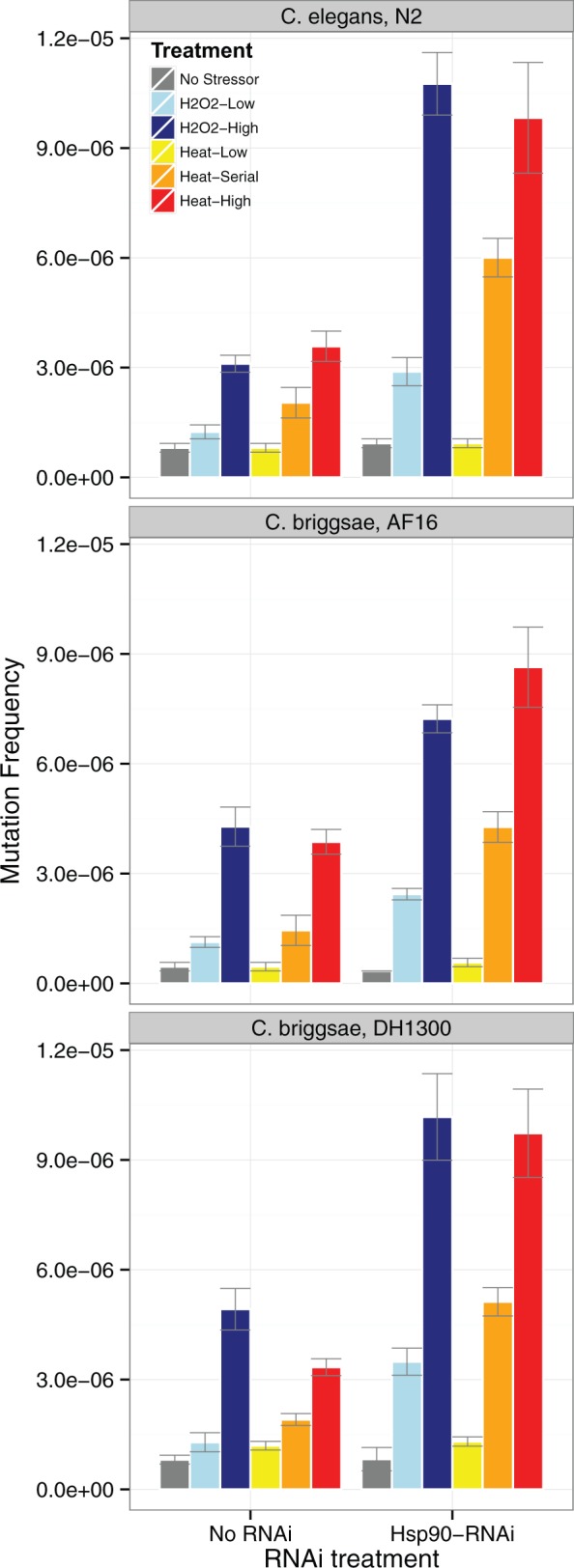
Proportion of plates with one or more mutants at the unc-22 gene following exposure to one of the five environmental stresses or No Stress controls, with and without RNAi-induced knockdown of Hsp90. Experiments were carried out with one strain of C. elegans (N2) and two strains of C. briggsae (AF16 and DH1300). Mutants were detected by observing the “twitching” phenotype in response exposure to 1% nicotine solution, with proportions based on the average of three replicates comparing between 20 and 30 plates per replicate (2717 plates in total).
In the absence of stress treatment, RNAi-induced knockdown of Hsp90 did not significantly increase mutation frequency in either C. elegans or C. briggsae (P = 0.604 and P = 0.721 for No Stress controls). However, for both species, the effect of treatment was amplified by RNAi-induced knockdown of Hsp90 (RNAi × Treatment interaction: = 33.52, P < 0.001 and = 33.18, P < 0.001 for C. elegans and C. briggsae, respectively). RNAi increased mutation frequency in response to stress treatments for all except the Heat-Low treatment in both C. elegans (P < 0.005 for all except H202-Low, where P = 0.050), and C. briggsae (P < 0.003 for all). The combined effect of stress treatment and RNAi-induced knockdown of Hsp90 increased unc-22 mutation frequency by 3- to 20-fold for C. elegans, and 5- to 23-fold for C. briggsae. Incidence rate estimates for all parameters are provided in supplementary table S7, Supplementary Material online.
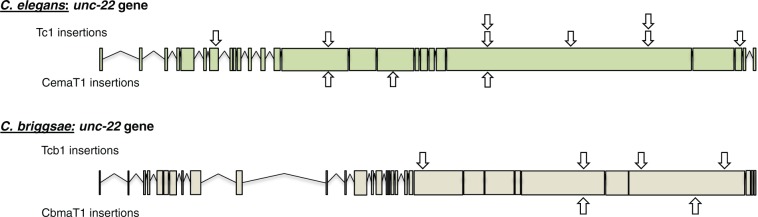
The mutant phenotypes observed in the worms were indistinguishable from C. elegans unc-22 mutants, with uncoordinated movements and persistent body twitching when exposed to nicotine. A minimum of five putative unc-22 worms were isolated from any positive (mutant) plate, and all showed segregation of mutant phenotypes in their progeny, indicating that the mutations were heritable. A total of 8 Tc1 and 3 CemaT1 insertions were detected in the induced putative unc-22 mutant progeny of the oxidative and heat stressed worms (fig. 4). In C. briggsae, 4 Tcb1, and 2 CbmaT1 insertions were detected (fig. 4). Both transposons target TA dinucleotides to insert, and consequently, there were some loci that independently had either transposon integrated. There were not enough insertions observed to confirm that some of these insertion sites are transposon insertion hotspots, although several insertion sites also showed insertion of the same transposon in independently derived worms, which suggests that some TA loci may be favored more than others.
Fig. 4.—
Transposable element (TE) insertion sites within the unc-22 gene in Caenorhabditis elegans (top) and C. briggsae (bottom) subjected to environmental stress. Arrows above the gene indicate insertions of Tc1 (C. elegans) or Tcb1 (C. briggsae) TEs; arrows below indicate CemaT1 and CbmaT1 insertions. Locations with stacked arrows are loci where multiple independent TE insertions were detected.
Discussion
In this study, we demonstrate that moderate environmental stress can increase the excision and insertion frequency of two Class II DNA transposons, and that these effects can be amplified through RNAi-mediated knockdown of Hsp90. Our findings support a role for Hsp90 in the suppression of transposon mobility, and suggest a functional constraint on Hsp90 owing to its dual role in promoting protein homeostasis and genetic stability. Under conditions of stress, Hsp90 may prioritize cellular physiological functioning and protein homeostasis, but at the cost of failing to protect the genome from the mutagenic activity of transposable elements. The heritable nature of the TE-induced genetic changes that we observed provides additional support – and a molecular mechanism – for the suggestion that Hsp90 may serve as a capacitor for evolutionary change (Queitsch et al. 2002; Rutherford 2003; Rohner et al. 2013; Peuß et al. 2015).
Each of the environmental stress treatments used in this study—except the low, short-duration heat exposure—increased transposon excision frequency in an exposure-dependent manner. Excision frequency increased in response to the stress treatments, despite concurrent up-regulation of daf-21 (Hsp90 gene) transcription. The level of daf-21 transcripts was enhanced in all stress treatments, including the low, short duration heat exposure, but the level of these transcripts did not increase significantly with the more intense stresses. As the number of cellular targets of Hsp90 increases during stress (Sangster et al. 2004; Jarosz and Lindquist 2010; Hartson and Matts 2012), the pool of available and newly synthesized Hsp90 was likely insufficient to suppress TE activity adequately. RNAi-mediated knock-down of daf-21 transcripts also increased TE excision frequency in some, but not all, nonstressed nematodes, suggesting that Hsp90 may in some cases be limiting in both the presence and absence of stress.
The effects of heat/oxidative stress treatment and RNAi-mediated knockdown of Hsp90 on TE mobility could occur through several pathways, including either the disruption of the proteins associated with DNA maintenance, repair, or chromatin packaging and/or activation of the PIWI/piRNA pathway. We observed differences in TE excision frequency in response to stress treatment and RNAi-mediated knockdown of Hsp90 between both species and strains. More interestingly, we also observed differences in excision frequency across loci within each species and strain. These genomically localized differences in TE mobility suggest that suppression of TE activity is dependent on many factors, including DNA secondary structure, and the availability of Hsp90 and the proteins with which it interacts.
The suppression of TEs through the PIWI/piRNA pathway has been established in a number of species, including C. elegans (Ketting et al. 1999; Tabara et al. 1999; Das et al. 2008). Mutations in proteins in the PIWI/piRNA pathway have been associated with increases in TE transcription, although the specific mechanisms for TE suppression appear to differ across taxa (Mani and Juliano 2013). In contrast to Drosophila, where piRNA biogenesis is thought to involve “ping–pong” amplification of piRNAs that then target TE transcripts for destruction, TE silencing in C. elegans seems to be mediated through piRNAs that facilitate the production of small-interfering RNAs that then target TE transcripts (Das et al. 2008). In Drosophila, the formation of the PIWI/piRNA complex appears to require the cochaperone Shu, as well as Hsp90 (Olivieri et al. 2012; Izumi et al. 2013; Izumi and Tomari 2014). Despite the differences in how piRNAs are produced and how they may mediate TE suppression in flies and nematodes (Gillan and Devaney 2014), our study provides evidence that Hsp90 may indeed share a similar role in nematodes to reduce TE activity. In Drosophila, where the PIWI/piRNA pathway in TE suppression has been investigated, hsp83scratch mutants with reduced Hsp90 function exhibited an impaired ability to produce several piRNAs essential for the suppression of repetitive Stellate sequences and nearby transposons (Tritto et al. 2003; Specchia et al. 2010). The majority of these transposons were of the Class I (copy and paste) variety, including one I element-like insertion responsible for a mutation in the noc gene (Specchia et al. 2010).
In our study, we observed that RNAi-mediated knockdown of Hsp90 enhanced Class II (cut and paste) transposon activity. This increased TE activity was associated with increased TE-induced mutagenesis at the unc-22 reporter gene. Our findings thus extend the work of Specchia et al. (2010) to the phylum Nematoda and to Class II (cut and paste) transposons, and demonstrate a direct link between Hsp90 and both TE excision and insertion frequencies. Importantly, we have shown that moderate environmental stress can have similar effects on transposon insertion frequency and mutagenesis as Hsp90 suppression using pharmacological means (i.e., geldanamycin) or using mutant strains. This supports the hypothesis that naturally occurring, stress-induced TE mobility and insertion mutagenesis could serve to radically reorganize host—or germline—genomes at precisely the time when environmental conditions threaten to overwhelm host’s capacity to buffer them (McClintock 1984). Such rapid and widespread genomic reorganization in response to environmental or physiological extremes could have important implications for evolution and speciation (Zeh et al. 2009; Piacentini et al. 2014), particularly in light of the fact that all of the mutations observed in our stressed nematodes were heritable.
In nematodes the impacts of TE-activity can be heritable (Bégin and Schoen 2007), and tied to an increase in frequency of mildly deleterious mutations with effects on fecundity, longevity, and body size (Bégin and Schoen 2006). Poor genetic quality has also been associated with declines in phenotypic quality in Drosophila (Sharp and Agrawal 2012, 2016), which itself contributes to increased mutation rates over time (so-called “mutational meltdown”; Sharp and Agrawal 2012). Stress arising from mismatches with novel environments could similarly affect phenotypic quality and mutation rate, increasing standing genetic variation and providing the substrate for accelerated adaptation. Thus, depending on the relationship between genetic load, mutation rate, and fitness, stress-induced TE-activity could lead to either rapid extinction or adaptation to novel environments (Shaw and Baer 2011). Our study provides insights into several mechanisms by which stress could impact genetic change and population evolution. In addition to its role in protein homeostasis and the regulation of TE mobility, Hsp90 is considered an important facilitator of DNA repair, and numerous proteins associated with DNA double strand break repair machinery are client proteins of Hsp90 (Pennisi et al. 2015). Under conditions of stress, it is conceivable that Hsp90’s role in regulating genomic stability could be compromised at both the level of TE regulation and in managing general DNA repair. Both of the above pathways point to Hsp90 as a nexus for genetic change arising from physiological and environmental stress.
Bégin and Schoen (2006) observed that while transposition frequencies can vary in different strains of C. elegans, most transposon activity does not produce readily detectable phenotypes. The unc-22 gene is one of the rare sites where a transposition-induced phenotype is easily observable, and our finding that multiple TEs inserted in the same locations within the unc-22 gene in both nematode species suggests that transposon insertions are not entirely random and may be indicative of TE insertion “hot-spots” (Hackett et al. 2007). Although our experimental design limits our ability to comment on TE insertion patterns or frequency elsewhere in the genome, the number of such insertions no doubt far exceeds those detected in the unc-22 reporter gene, providing a potentially important source of genetic variation. Moreover, a number of factors are thought to contribute to biases in TE insertion sites, including local DNA sequence and structure (Yant et al. 2005). Intriguingly, regions that are relatively highly expressed may also be biased towards TE or retroviral integration (Wu et al. 2003; Campos-Sanchez et al. 2014). Therefore, to the extent that expression levels increase TE insertion probability, actively expressed genes responding to an environmental stressor may also provide biased targets for mutational change. Whether selection has favored TE insertion biases in transcriptionally active regions—or whether they arise incidental to the biochemical underpinnings of TE mobility, or from functional constraints such as those we have described for Hsp90—is not yet clear (Capy et al. 2000). However, Hsp90 expression levels appear to be restricted by the requirement for cellular buffering and protein homeostasis on one hand, and cytotoxic effects of Hsp90 accumulation on the other (Krebs and Loeschcke 1994; Feder and Hofmann 1999; Casanueva et al. 2012). By limiting peak Hsp90 expression levels, Hsp90 cytotoxicity would limit the buffering capacity of Hsp90 within the cell, giving rise to the functional constraints between protein homeostasis and genetic buffering proposed here.
We have shown that moderate environmental stress—and the reduction of Hsp90 using RNAi—has the capacity to increase the excision and insertion frequencies of two Class II DNA transposons at multiple loci in multiple strains of two different species of nematode. Differences in excision frequency between loci suggest a role for Hsp90 independent of protein buffering of DNA regulatory machinery. More empirical studies examining the role of Hsp90 and the PIWI/piRNA pathway in TE suppression are crucial to understanding the dynamic regulation of transposable elements as a dangerous, but potentially powerful force for evolutionary change.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors also wish to thank S. Halliburton and L. Crompton for technical assistance. This work was supported by a Natural Sciences and Engineering Research Council of Canada grant to SW (NSERC #312690). Nematode strains used in this study were kind gifts of the CGC at the University of Minnesota, USA.
Literature Cited
- Bagijn MP, et al. 2012. Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science 337:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bégin M, Schoen DJ. 2006. Low impact of germline transposition on the rate of mildly deleterious mutation in Caenorhabditis elegans. Genetics 174:2129–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bégin M, Schoen DJ. 2007. Transposable elements, mutational correlations, and population divergence in Caenorhabditis elegans. Evolution 61:1062–1070. [DOI] [PubMed] [Google Scholar]
- Bestor TH. 1998. Gene silencing: methylation meets acetylation. Nature 393:311–312. [DOI] [PubMed] [Google Scholar]
- Biémont C, Vieira C. 2006. Genetics: junk DNA as an evolutionary force. Nature 443:521–524. [DOI] [PubMed] [Google Scholar]
- Bonchev G, Parisod C. 2013. Transposable elements and microevolutionary changes in natural populations. Mol Ecol Resour. 13:765–775. [DOI] [PubMed] [Google Scholar]
- Brownlie JC, Johnson NM, Whyard S. 2005. The Caenorhabditis briggsae genome contains active CbmaT1 and Tcb1 transposons. Mol Genet Genomics. 273:92–101. [DOI] [PubMed] [Google Scholar]
- Brownlie J, Whyard S. 2004. CemaT1 is an active transposon within the Caenorhabditis elegans genome. Gene 338:55–64. [DOI] [PubMed] [Google Scholar]
- Campos-Sanchez R, Kapusta A, Feschotte C, Chiaromonte F, Makova KD. 2014. Genomic landscape of human, bat, and ex vivo DNA transposon integrations. Mol Biol Evol. 31:1816–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Gasperi G, Biémont C, Bazin C. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85:101–106. [DOI] [PubMed] [Google Scholar]
- Casanueva MO, Burga A, Lehner B. 2012. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science 335:82–85. [DOI] [PubMed] [Google Scholar]
- Cowley M, Oakey RJ. 2013. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 9:e1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 31:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Visser JAGM, et al. 2003. Perspective: evolution and detection of genetic robustness. Evolution 57:1959–1972. [DOI] [PubMed] [Google Scholar]
- Egilmez NK, Ii RHE, Reis RJS. 1995. Strain evolution in Caenorhabditis elegans: transposable elements as markers of interstrain evolutionary history. J Mol Evol. 40:372–381. [DOI] [PubMed] [Google Scholar]
- Erlejman AG, Lagadari M, Toneatto J, Piwien-Pilipuk G, Galigniana MD. 2014. Regulatory role of the 90-kDa-heat-shock protein (Hsp90) and associated factors on gene expression. BBA-Gene Regul Mech. 1839:71–87. [DOI] [PubMed] [Google Scholar]
- Evans T. 2006. Transformation and microinjection.In: The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.108.1. http://www.wormbook.org. [Google Scholar]
- Feder ME, Hofmann GE. 1999. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 61:243–282. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, et al. 2011. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 43:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillan V, Devaney E. 2014. Nematode Hsp90: highly conserved but functionally diverse. Parasitology 141:1203–1215. [DOI] [PubMed] [Google Scholar]
- Hackett CS, Geurts AM, Hackett PB. 2007. Predicting preferential DNA vector insertion sites: implications for functional genomics and gene therapy. Genome Biol. 8:S12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartson SD, Matts RL. 2012. Approaches for defining the Hsp90-dependent proteome. BBA-Mol Cell Res. 1823:656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA. editor. 1999. C. elegans: A Practical Approach. Oxford: Oxford University Press. [Google Scholar]
- Izumi N, et al. 2013. Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins. Rna 19:896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Tomari Y. 2014. Diversity of the piRNA pathway for nonself silencing: worm-specific piRNA biogenesis factors. Genes Dev. 28:665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. 2010. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 330:1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Taipale M, Lindquist S. 2010. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu Rev Genet. 44:189–216. [DOI] [PubMed] [Google Scholar]
- Jose AM, Smith JJ, Hunter CP. 2009. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. PNAS 106:2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–237. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. 2004. Mobile elements: drivers of genome evolution. Science 303:1626–1632. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp THA, van Luenen HGAM, Plasterk RHA. 1999. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99:133–141. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1–24. [DOI] [PubMed] [Google Scholar]
- Krebs RA, Loeschcke V. 1994. Costs and benefits of activation of the heat-shock response in Drosophila melanogaster. Funct Ecol. 8:730. [Google Scholar]
- Mani SR, Juliano CE. 2013. Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol Reprod Dev. 80:632–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. 1999. Molecular chaperones: the busy life of Hsp90. Curr Biol. 9:R322–R325. [DOI] [PubMed] [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 226:792–801. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Baillie DL. 1979. Genetic organization in Caenorhabditis elegans: fine-structure analysis of the unc-22 gene. Genetics 91:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Waterston RH. 1984. Spontaneous unstable unc-22 IV mutations in C. elegans var. Bergerac. Genetics 108:859–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti K-A, Subramanian S, Sachidanandam R, Brennecke J. 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell. 47:954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi R, Ascenzi P, di Masi A. 2015. Hsp90: A New Player in DNA Repair? Biomolecules 5:2589–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuß R, Eggert H, Armitage SAO, Kurtz J. 2015. Downregulation of the evolutionary capacitor Hsp90 is mediated by social cues. Proc R Soc B. 282:20152041.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucl Acids Res. 29:e45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Specchia V, Bozzetti MP, Berloco M, Palumbo G, Pimpinelli S. 2014. Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma. 123(4):345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. 1996. How organisms respond to environmental changes: from phenotypes to molecules (and vice versa). Trends Ecol Evol. 11:168–173. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624. [DOI] [PubMed] [Google Scholar]
- Rohner N, et al. 2013. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in Cavefish. Science 342:1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL. 2003. Between genotype and phenotype: protein chaperones and evolvability. Nat Rev Genet. 4:263–274. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Lindquist S, Queitsch C. 2004. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays 26:348–362. [DOI] [PubMed] [Google Scholar]
- Sekimoto T, et al. 2010. The molecular chaperone Hsp90 regulates accumulation of DNA polymerase η at replication stalling sites in UV-irradiated cells. Mol Cell. 37:79–89. [DOI] [PubMed] [Google Scholar]
- Sela N, Kim E, Ast G. 2010. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 11(6):R59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp NP, Agrawal AF. 2012. Evidence for elevated mutation rates in low-quality genotypes. Proc Natl Acad Sci. 109:6142–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp NP, Agrawal AF. 2016. Low genetic quality alters key dimensions of the mutational spectrum. PLOS Biol. 14:e1002419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Plasterk RHA. 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426:310–314. [DOI] [PubMed] [Google Scholar]
- Specchia V, et al. 2010. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463:662–665. [DOI] [PubMed] [Google Scholar]
- Shaw FH, Baer CF. 2011. Fitness-dependent mutation rates in finite populations. J Evol Biol. 24:1677–1684. [DOI] [PubMed] [Google Scholar]
- Tabara H, et al. 1999. The rde-1 Gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123–132. [DOI] [PubMed] [Google Scholar]
- Tritto P, et al. 2003. Structure, regulation and evolution of the crystal–stellate system of Drosophila. Genetica 117:247–257. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150:563–565. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131:609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456–2459. [DOI] [PubMed] [Google Scholar]
- Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Natl Acad Sci. 104:10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749–1751. [DOI] [PubMed] [Google Scholar]
- Yant SR, et al. 2005. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 25:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335–340. [DOI] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU. 2001. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 154:267.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeh DW, Zeh JA, Ishida Y. 2009. Transposable elements and an epigenetic basis for punctuated equilibria. BioEssays 31:715–726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.