Abstract
Understanding how mammalian genomes have been reshuffled through structural changes is fundamental to the dynamics of its composition, evolutionary relationships between species and, in the long run, speciation. In this work, we reveal the evolutionary genomic landscape in Rodentia, the most diverse and speciose mammalian order, by whole-genome comparisons of six rodent species and six representative outgroup mammalian species. The reconstruction of the evolutionary breakpoint regions across rodent phylogeny shows an increased rate of genome reshuffling that is approximately two orders of magnitude greater than in other mammalian species here considered. We identified novel lineage and clade-specific breakpoint regions within Rodentia and analyzed their gene content, recombination rates and their relationship with constitutive lamina genomic associated domains, DNase I hypersensitivity sites and chromatin modifications. We detected an accumulation of protein-coding genes in evolutionary breakpoint regions, especially genes implicated in reproduction and pheromone detection and mating. Moreover, we found an association of the evolutionary breakpoint regions with active chromatin state landscapes, most probably related to gene enrichment. Our results have two important implications for understanding the mechanisms that govern and constrain mammalian genome evolution. The first is that the presence of genes related to species-specific phenotypes in evolutionary breakpoint regions reinforces the adaptive value of genome reshuffling. Second, that chromatin conformation, an aspect that has been often overlooked in comparative genomic studies, might play a role in modeling the genomic distribution of evolutionary breakpoints.
Keywords: rodents, evolutionary breakpoints, recombination, lamina associated domains, KRAB genes, epigenome
Introduction
Unlocking the genetic basis of speciation is of crucial importance to explain species diversity and adaptation to a changing environment. Similarly, understanding the role that large-scale chromosomal rearrangements play in reproductive isolation has long been a focus of evolutionary biologists (White 1978; Ayala and Coluzzi 2005). Particularly, discussions have been focussed on whether genome reshuffling act as barriers to gene flow (Rieseberg 2001; Navarro and Barton 2003; Faria and Navarro 2010; Farré et al. 2013) or by modifying both the structure and regulation of genes located at, or near, the affected regions (Murphy et al. 2005; Larkin et al. 2009; Ullastres et al. 2014). The main motivation behind these studies has been to find evidence of the adaptive value of genome reshuffling and of the mechanisms of its formation during mammalian diversification [reviewed in Farré et al. (2015)].
A large body of studies has provided the basis for establishing models that can explain genome dynamics through comparative genomics of both closely and distantly related mammalian species (Murphy et al. 2005; Ruiz-Herrera et al. 2006; Larkin et al. 2009; Farré et al. 2011; Ruiz-Herrera et al. 2012). This allowed the delineation of genomic regions where the order of markers where conserved between species (so-called homologous synteny blocks, HSBs). Such reconstructions revealed that genomic regions implicated in structural evolutionary changes, disrupting the genomic synteny (evolutionary breakpoint regions, EBRs) are clustered in regions more prone to break and reorganize (Bourque et al. 2004; Murphy et al. 2005; Ruiz-Herrera et al. 2005, 2006; Larkin et al. 2009, Farré et al. 2011). Compelling evidence has shed light on genomic features that characterize EBRs. Repetitive elements including segmental duplications (Bailey and Eichler 2006; Kehrer-Sawatzki and Cooper 2007; Zhao and Bourque 2009), tandem repeats (Kehrer-Sawatzki et al. 2005; Ruiz-Herrera et al. 2006; Farré et al. 2011), and transposable elements (Carbone et al. 2009; Longo et al. 2009; Farré et al. 2011) have all been associated with their presence. However, given the diversity of repetitive elements found in EBRs it is likely that sequence composition is not alone in influencing genome instability during evolution. In fact, the genomic distribution of mammalian EBRs can be considered a multifactorial affair, involving repetitive elements, functional constrains and changes in the chromatin state (Farré et al. 2015). It was initially reported that EBRs are located in gene-rich regions (Murphy et al. 2005; Lemaitre et al. 2009), among others, those containing gene functional process networks, such as genes related to the immune system (Groenen et al. 2012; Ullastres et al. 2014). This suggests that changes in gene expression caused by genome reshuffling could reflect a selective advantage through the development of new adaptive characters specific to mammalian lineages (Larkin et al. 2009; Groenen et al. 2012; Ullastres et al. 2014). This view has been recently unified in the “integrative breakage model” (Farré et al. 2015), which postulates that the permissiveness of some genomic regions to undergo chromosomal breakage could be influenced by chromatin conformation. That is, certain properties of local DNA sequences together with the epigenetic state of the chromatin and the effect on gene expression are key elements in determining the genomic distribution of evolutionary breakpoints (Farré et al. 2015). But how universal this pattern is among mammals needs further validation.
Rodentia is the most diverse and species rich mammalian order with more than 2,000 defined species (Carleton and Musser 2005) that occupy a wide range of habitats and exhibit many adaptive features. Although the rodent phylogeny has been heavily contested due to its complexity, recent studies suggest recognizing three major clades (Huchon et al. 2002; Montgelard et al. 2008; Blanga-Kanfi et al. 2009; Churakov et al. 2010): (i) the mouse-related clade, (ii) the squirrel-related clade, and (iii) the clade Ctenohystrica (guinea pig and relatives). Rodentia are generally considered to present specific features such as higher rates of nucleotide substitution (Wu and Li 1985), lower recombination rates and higher genome reshuffling rates [although this is mainly based on Mus (Wu and Li 1985) when compared with other Laurasiatheria (Dumont and Payseur 2011; Segura et al. 2013). In fact, one of the most intriguing features that characterize rodents is the high chromosomal variability. This is exemplified by a wide range of diploid numbers ranging from 2n = 10 in Akodon spp. (Myodonta clade) to 2n = 102 in Tympanoctomys barerae (Ctenohystrica clade) (Silva and Yonenaga-Yassuda 1998; Gallardo et al. 2004). Previous comparative studies have provided relevant information on both ancestral karyotype reconstructions for the group (Bourque et al. 2004; Froenicke et al. 2006; Ma et al. 2006; Graphodatsky et al. 2008; Mlynarski et al. 2010; Romanenko et al. 2012) and specific large-scale chromosomal rearrangements (Pevzner and Tesler 2003; Zhao et al. 2004; Froenicke et al. 2006; Mlynarski et al. 2010). However, the reason(s) behind the extremely high rate of genomic reshuffling is far to be fully understood. Therefore, a more comprehensive picture of rodent genome evolution at the finer scale remains to be uncovered.
With the availability of fully sequenced genomes from several different rodent species we can now delineate the fine-scale evolutionary history of genomic reshuffling in rodents in order to better understand both the adaptive value of chromosomal rearrangements within the group and the mechanisms underlying this pattern. Here we present a refined analysis of the Rodentia evolutionary genome reshuffling by comparing the house mouse genome (Mus musculus) with those of five rodent species (Heterocephalus glaber, Jaculus jaculus, Spalax galilii, Microtus ochrogaster, and Rattus norvegicus) and six mammalian outgroup species (Homo sapiens, Macaca mulatta, Pongo pygmaeus, Bos taurus, Equus caballus, and Felis catus). This has permitted the delineation of two specific objectives: (i) the examination at the finest scale of EBRs across the Rodentia phylogeny and (ii) testing their association with gene content, recombination rates, lamina associated domains, DNase I hypersensitivity sites and a wide variety of chromatin modifications. Our results provide the first evidence for the presence of rodent specific genetic and epigenetic signatures, reinforcing the adaptive role of genomic reshuffling. Moreover, our results suggest that chromatin conformation might play a role in modeling the genomic distribution of evolutionary breakpoints, opening new avenues for our understanding of the mechanistic forces governing mammalian genome organization.
Materials and Methods
Whole-Genome Comparisons
Pair-wise alignments were established between the genomes of the mouse (NCBIm37 assembly) and 11 representative species of mammalian phylogeny by Satsuma Synteny (Grabherr et al. 2010) (supplementary table S1, Supplementary Material online). Based on the sequence alignments provided by Satsuma Synteny, the SyntenyTracker algorithm (Donthu et al. 2009) was used to establish regions of homology (syntenic regions) between the mouse genome (reference genome) and each of the mammalian species included in the analysis based on a minimum block size threshold. We differentiated two types of syntenic regions: (i) HSBs when pair-wise comparisons were established between genomes assembled into chromosomes, and (ii) Syntenic Fragments (SFs), for pair-wise comparisons between genomes only assembled at scaffold level (supplementary table S2, Supplementary Material online). For each pair-wise alignment, three different syntenic block sizes (including both HSBs and SFs) were defined (100, 300, and 500 kbp) (supplementary table S4 and fig. S1, Supplementary Material online). This allowed us to evaluate genome assembly reliability. When the number of HSBs or SFs was not proportional between the three resolutions, it was assumed that the genome contained assembly errors.
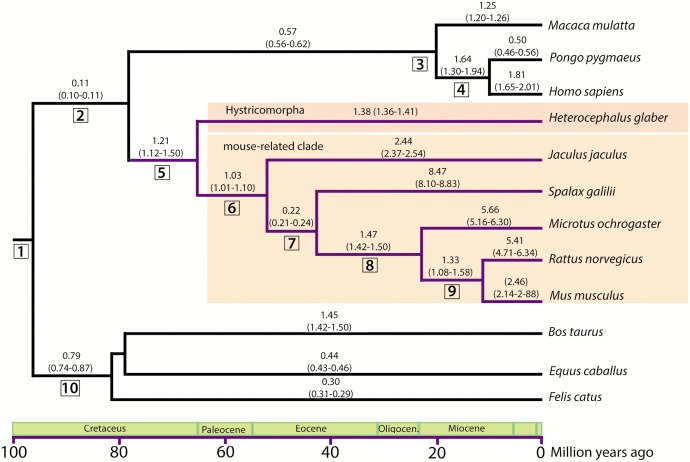
Once syntenic regions were established for all species, EBRs were defined and classified using the approach described elsewhere (Farré et al. 2016) using 300 kbp as the reference block size resolution. All EBRs were detected in each lineage included in the study and reliability scores for each classification were estimated. The main values are determined by the ratio of the scores and the percentage of species with breakpoints with respect to genomic gaps. By taking the total number of species used in our analysis into account and the percentage of species that presented the genome in scaffolds, the threshold was fixed at a ratio ≥34, and a percentage >60%. Then, two different groups of EBRs were established: (i) EBRs corresponding to any of the 11 species studied (hereafter, lineage-specific EBRs) and (ii) EBRs that appeared in any of the differentiation nodes of the phylogenetic tree (hereafter, clade-specific EBRs; fig. 1, supplementary table S3, Supplementary Material online). In fact, and based on the phylogenetic relationships among the species included in the analysis, 10 different nodes/clades were considered (fig. 1): Clade 1—Boreoeutheria, which included all mammalian species compared in our analysis; Clade 2—Euarchontoglires, including all rodent and primate species; Clade 3—Catarrhini, which included H. sapiens, M. mulatta, and P. pygmaeus; Clade 4—Hominoidea, with only H. sapiens and P. pygmaeus; Clade 5—Rodentia, which included all rodent species compared; Clade 6—Myodonta, all rodents species compared, except H. glaber; Clade 7—Muroidea, with S. galilii, M. ochrogaster, R. norvegicus and M. musculus; Clade 8—Cricetidae + Muridae, including M. ochrogaster, R. norvegicus and M. musculus; Clade 9—Muridae, with R. norvegicus and M. musculus; and Clade 10—Laurasiatheria, with B. taurus, E. caballus, and F. catus. In order to estimate the average rate of EBRs occurring for each phylogenetic branch (number of EBRs per million years—Myr), divergence times (autocorrelated rates and hard-bounded constraints) were extracted from Meredith et al. (2011) for each lineage and clade phylogenetic branches, with the exception of Muridae. In this latter instance, data retrieved from dos Reis et al. (2012) was used (supplementary table S5, Supplementary Material online).
Fig. 1.—
EBRs mapped in the time tree of the mammalian species included in the study. Time tree was based on divergence times (autocorrelated rates and hard-bounded constraints) described by Meredith et al. (2011), to the exception of two species (M. musculus and R. norvegicus) and one clade (Muridae) which were estimated from dos Reis et al. (2012) time tree. In the upper section of each branch, the mean rate of EBRs per Myr and the range (in brackets) is shown. Numbers framed in squares represent mammalian phylogenetic nodes: 1: Boreoeutheria; 2: Euarchontoglires; 3: Catarrhini; 4: Hominoidea; 5: Rodentia; 6: Myodonta; 7: Muroidea; 8: Cricetidae + Muridae; 9: Muridae; 10: Laurasiatheria.
Gene Content and Ontology
Sequence coordinates of all mouse genes were obtained from BioMart (RefSeq genes, NCBIm37). Genes were clustered into two groups: (i) total genes, which included protein-coding genes, novel genes with unknown function, pseudogenes and RNA genes; and (ii) protein-coding genes, which included only genes with known function. Genes were assigned either to HSBs or EBRs when coordinates fell within these regions. Gene density was analyzed by calculating the mean number of genes contained in nonoverlapping windows of 10 kbp across the mouse genome as previously described (Ullastres et al. 2014). Four different genomic regions were taken into account: (i) HSBs, (ii) EBRs, (iii) interphase regions (regions overlapping with the start or the end coordinates of any given EBRs), and (iv) 100 kbp regions upstream or downstream from the EBRs coordinates. Given the high incidence of assembly errors at the telomeres/subtelomeres and the centromeric/pericentromeric areas, a 3 Mbp section of each region was excluded from the analysis.
The functional annotation and clustering tool DAVID (Database for Annotation, Visualization, and Integrated Discovery, v6.7) (Huang et al. 2009) was used to identify overrepresented biological terms contained in EBRs. Functional annotation clustering allows for the biological interpretation at a “biological module” level and functional annotation charts identify the most relevant (over-represented) biological terms associated with a given gene list (Huang et al. 2009). We used the Benjamini’s test to control false positives. This compares the proportion of genes in the analyzed regions (i.e., EBRs) to the proportion of the genes of the rest of the genome (i.e., HSBs), and produces an EASE score. EASE scores ≤0.05 and containing a minimum of two gene ontology terms were considered significantly over-represented.
Recombination Rates
The mouse genetic map was extracted from Brunschwig et al. (2012). This contains high-resolution recombination rate estimates across the mouse genome (the autosomic chromosomes) based on 12 classically sequenced mouse strains (129S5/SvEvBrd, AKR/J, A/J, BALB/cJ, C3H/HeJ, C57BL/6NJ, CBA/J, DBA/2J, LP/J, NOD/ShiLtJ, NZO/HILtJ, and WSB/EiJ). From this map, we estimated recombination rates for nonoverlapping windows of 10 Kbp across the mouse genome as previously described (Farré et al. 2013). For each 10 kbp window, the recombination rate was calculated as the average of all recombination rates. These values were subsequently merged with the genomic positions from the four different genomic regions included in the gene density analysis using in-house Perl scripts. Centromeric and telomeric regions were not included in the analysis.
Constitutive Lamina Associated Domains
Genomic data for mouse Lamina Associated Domains (LADs) was extracted from Meuleman et al. (2013) available at the NCBI Gene Expression Omnibus (accession number GSE36132). LADs were obtained using DamID maps (Peric-Hupkes and van Steensel 2010) of lamina A in mouse astrocytes and neural precursor cells and Lamina B1 in wild type and Oct1 knockout mouse embryonic fibroblasts (MEFs and Oct1koMEFs, respectively). Constitutive LADs (cLADs) resulted from selecting lamina regions that were identified in all cell types analyzed. Once cLADs positions were obtained, their genomic distribution was analyzed in nonoverlapping windows of 10 kbp as described above. Each 10 kbp window was subsequently classified into different genomic regions as was done in the gene content and recombination analyses (EBRs, HSBs, interphases, and 100 kbp adjacent regions) described above.
DNase I Hypersensitivity Sites and Chromatin Modifications
All available ChIP-seq and DNase-seq BED files based on M. musculus mm9 assembly were downloaded from Mouse ENCODE (The Mouse ENCODE Consortium). These included all available epigenetic marks from 58 different mouse cell lines, including the skeletal system, the muscular system, the circulatory system, the nervous system, the respiratory system, the digestive system, the excretory system, the endocrine system, the reproductive system, the lymphatic system, and stem cells.
Statistical Analysis
The genome-wide distribution of EBRs was estimated using an average frequency across the mouse genome and by assuming a homogeneous distribution of all detected EBRs. We used a χ2 test with a Bonferroni correction to assess any possible deviation from the homogeneous distribution. Mean comparison of gene density, recombination rates and cLADs with the genome wide division of 10 Kbp windows was performed with Kruskal–Wallis nonparametric test using JMP statistical package (release 7.1).
Genome wide association analysis between EBRs as well as control region datasets and different genomic features (gene content, cLADs, recombination rates, ChIP-seq, and DNase-seq data) were performed using RegioneR—a permutation-based approach implemented in the Bioconductor package regioneR (version 1.4.2) (Gel et al. 2016). RegioneR compares the number of observed overlaps between a query and a reference region-set to the distribution of the number of overlaps obtained by randomizing the regions-set over the genome for each chromosome. The tests were performed on canonical chromosomes with assembly gaps (AGAPS) and intra-contig ambiguities (AMB) masked using 10,000 permutations (min. P-value: 1e−04) and package-specific function overlapPermTest having nonoverlapping parameter set to false. If replicates were available for the same mark or tissue, P-values were combined using Fisher's method. For comparative analysis, two control region datasets were generated: (i) EBR-like—genomic regions with a gene density distribution similar to the EBRs, and (ii) genome-like—genomic regions with a gene density distribution similar to the whole mouse genome. For that, the mouse genome was divided in nonoverlapping windows of 100 kbp and their gene density was computed, excluding those windows overlapping EBRs, AGAPS, and AMB. Then, probability weights of observing gene densities in the EBRs and in the generated windows (whole genome) were calculated. According to probability weights, the EBR-like and the genome-like control region datasets with 200 randomly selected windows each were generated.
Results
The comparative genomic analysis performed in this study has permitted: (i) the delineation of genome reshuffling across Rodentia phylogeny and (ii) the study of genetic and epigenetic characteristics of EBRs in searching for the presence of specific evolutionary signatures that can account for genome reshuffling in rodents, such as gene content, recombination rates, and chromatic conformation.
Genome Reshuffling in Rodentia
Defining Syntenic Regions and Evolutionary Breakpoint Regions in Rodentia
In order to determine the evolutionary genomic landscape in Rodentia, we compared the mouse genome (M. musculus) to those of five rodent species: one representative of the Hystricognathi (H. glaber), group belonging to Ctenohystrica and four species of Myodonta (J. jaculus, S. galilii, M. ochrogaster, and R. norvegicus), group belonging to the mouse-related clade. In addition, the inclusion of six mammalian species from Primates (H. sapiens, M. mulatta, and P. pygmaeus), Cetartiodactyla (B. taurus), Carnivora (F. catus), and Perissodactyla (E. caballus) allowed us to refine the characterization of EBRs in a phylogenetic context (fig. 1).
We first determined the syntenic regions (HSBs and SFs) in the eleven species compared with the mouse genome (supplementary table S2, Supplementary Material online), identifying a total of 3,392 HSBs with a mean size ranging from to 5.56 Mbp in B. taurus to 13.22 Mbp in R. norvegicus (supplementary table S2, Supplementary Material online). We detected a total of 3,142 SFs, with a mean size ranging from 1.14 Mbp in S. galilii, to 5.14 Mbp in H. glaber (supplementary table S2, Supplementary Material online). The number of HSBs differed depending on species and ranged from 280 HSBs (representing the 95.60% of the mouse genome) between mouse and rat, to 521 HSBs (representing 91.11% of the mouse genome) between mouse and the cow (supplementary table S2, Supplementary Material online). In the case of scaffold-based genome comparisons, the number of SFs was slightly higher in J. jaculus (559, N50∼22 Mbp) and H. glaber (598, N50∼20 Mbp) and especially pronounced in S. galilii (1,985, N50∼4 Mbp). Because some of the SFs may merge when assembled into chromosomes to form HSBs, the syntenic regions detected in scaffold-based genomes may represent an overestimation. With this as caveat, the syntenic regions detected represented >80% of the mouse genome, reaching 95.6% in the mouse/rat comparison, and 93.5% for the mouse/horse comparison (supplementary table S2, Supplementary Material online). This is a reflection of the high conservation of their genomes.
Once the syntenic regions were determined for all species, we estimated the number and genomic distribution of EBRs in the mouse genome and classifed them in a phylogenetic context. We detected a total of 1,333 EBRs, the majority of which (1,179) were classified as unique EBRs (i.e., the occurrence of the same breakpoint in two species that do not share a recent common ancestor; see Murphy et al. 2005; Larkin et al. 2009) (fig. 1 and supplementary table S3, Supplementary Material online). The rest, representing 154 EBRs, were classified as reused (i.e., EBRs that are shared by a subset of species from the same clade). Of the unique EBRs detected, 1,049 were lineage-specific (i.e., specific for each of the species when compared with the mouse genome), and the remaining 130 EBRs were classified as clade-specific (Primate, Hominoidea, Laurasiatheria, Euarchontoglires, Rodentia, Myodonta, Muroidea, Cricetidae + Muridae, and Muridae) (supplementary table S3, Supplementary Material online). The number of lineage-specific EBRs was variable and ranged from 8 EBRs in P. pygmaeus to 360 EBRs in S. galilii. In the case of the clade-specific EBRs, the number of evolutionary breakpoint regions ranged from 2 EBRs in Euarchontoglires to 33 EBRs in Catarrhini (supplementary table S3, Supplementary Material online). Likewise, EBRs mean size varied in each pair-wise species comparison, ranging from 79.62 to 151.87 kbp and 55.58 to 135.32 kbp, respectively (supplementary table S3, Supplementary Material online). In order to corroborate the EBR estimations, we analyzed the number of syntenic blocks obtained at 100, 300, and 500 kbp resolutions for all pair-wise comparisons. Overall, the number of syntenic blocks was proportional between the three levels of resolution (e.g., between 1.29- and 1.70-fold increase between 100 kbp and 500 kbp resolutions, supplementary fig. S1 and table S4, Supplementary Material online) supporting the reliability of genome assemblies and EBR estimations. R. norvegicus was an exception to this pattern, showing between a 5.29-fold increase between 100 and 500 kbp resolutions.
To provide an estimation of the genome reshuffling rate (expressed as the number of EBRs detected in each phylogenetic branch per Myr) that occurred in Rodentia, we placed the total estimated EBRs in a phylogenetic context considering the species included in the study (fig. 1). We detected that the presence of EBRs in Rodentia was higher (1.21 EBRs/Myr) than in the rest of major mammalian clades (i.e., 0.79 EBRs/Myr for Laurasiatheria or 0.11 EBRs/Myr for Euarchontoglires) (fig. 1). This result corroborates initial observations that pose rodents as one of the mammalian orders with the highest genome reshuffling rates. There is, however, variability among Rodentia clades—the highest rate of the genome reshuffling was detected in the mouse-like group (Muridae, 1.47 EBRs/Myr) while a lower rate was detected in Muroidea (0.22 EBRs/Myr). In terms of the species-specific genome reshuffling rates, rodents in general showed higher rates than any other mammalian species included in the study (fig. 1). That was the case, for example, of J. jaculus (2.44 EBRs/Myr) and M. ochrogaster (5.66 EBRs/Myr). However, we need to be conservative in defining genome reshuffling rates in R. norvegicus because the number of HSBs detected was not proportional in the three different resolutions of Synteny Tracker (100, 300, and 500 kbp, supplementary fig. S1, Supplementary Material online).
Genome-Wide Distribution of Rodentia EBRs
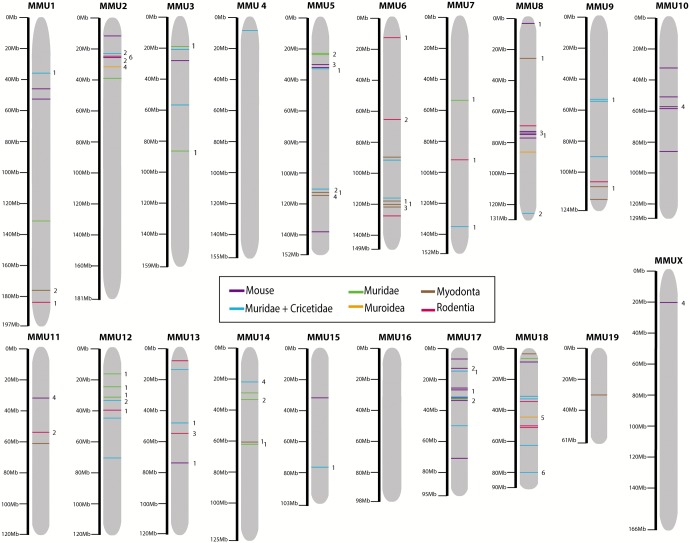
In order to define genome reshuffling in Rodentia, and more specifically, to determine the presence of genomic signatures that occurred during mouse evolution, we focused our efforts on analyzing the distribution of both Rodentia specific EBRs and mouse-specific EBRs across the mouse genome. Of the 891 EBRs detected in the rodent species analyzed, 105 (covering 0.31% of the mouse genome) appeared in the lineage leading to the Mus. These included 75 clade-specific EBRs: 15 EBRs defined Rodentia, 14 Myodonta, 3 Muroidea, 28 Cricetidae + Muridae, 15 Muridae, and 30 EBRs were specific to M. musculus (fig. 1 and supplementary table S3, Supplementary Material online). Assuming a homogeneous distribution across the genome, we observed that EBRs were not randomly distributed throughout the mouse genome (fig. 2 and supplementary fig. S2, Supplementary Material online). In fact, three chromosomes (chromosomes 8, 17, and 18) appeared to contain significantly more EBRs than expected under a random distribution (chromosome 17: χ2 = 13.57, P-value < 0.001 and chromosome 18: χ2 = 14.96, P-value < 0.001; supplementary fig. S2, Supplementary Material online). Additionally, three other chromosomes (chromosome 4, chromosome 16 and chromosome X) contained less EBRs than expected (chromosome 4: χ2 = 4.54, P-value < 0.05; chromosome 16: χ2 = 3.93, P-value <0.05; and chromosome X: χ2 = 4.81, P-value <0.05; supplementary fig. S2, Supplementary Material online). Moreover, EBRs appeared to be localized in clusters (i.e., genomic regions with a higher density of EBRs per Mbp), for example in chromosome 8 and chromosome 17 (fig. 2).
Fig. 2.—
EBRs mapped in the mouse genome. The positions of EBRs detected (lineage and clade-specific) are color-coded (see inset legend) along mouse (MMU, M. musculus) chromosomes. The number of protein-coding genes detected within each EBR is depicted on the right of each chromosome.
Rodentia EBRs Are Gene-Rich Regions
We further examined the genomic characteristics of EBRs searching for the presence of specific evolutionary signatures. To this end, we first analyzed the genome-wide distribution of genes, paying special attention to gene ontology. A total of 36,381 genes were identified and included in the analysis. These were divided into two groups: (i) all genes (n = 36,381) and (ii) protein-coding genes (n = 22,352). The mean distribution of genes (including protein-coding genes, noncoding RNA genes and pseudogenes) found in the mouse genome was 0.09 genes per 10 kbp, although these were nonhomogeneously distributed across chromosomes (Kruskal–Wallis test, P-value < 0.001). Mouse chromosomes 7, and 11 are gene-rich (0.14 genes per 10 kbp in both cases) whereas chromosomes 12, 18 and X (0.06 genes per 10 kbp in all cases) are low on genes.
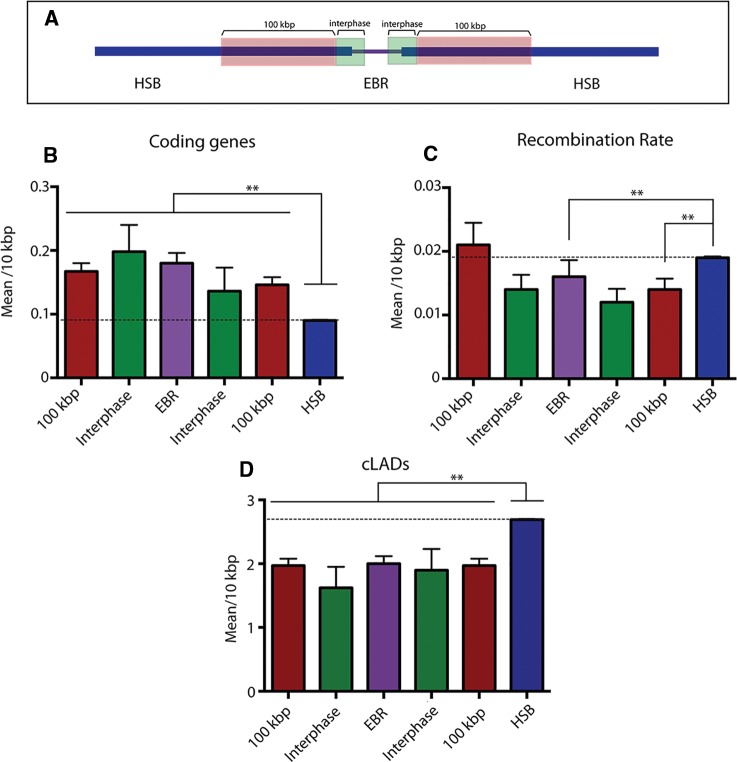
We then analyzed gene density for all Rodentia EBRs detected (including clade-specific and those that are mouse lineage-specific). Our results showed that EBRs are gene-rich regions with an average density of 0.18 genes per 10 kbp compared with the rest of the genome (0.09 genes per 10 kbp, Kruskal–Wallis test, P < 0.001). Density values were even higher (0.287 genes per 10 kbp) when considering only mouse lineage-specific EBRs. Gene enrichment was confirmed using a genome-wide permutation test (based on 10,000 permutations, P < 0.05) (table 1). When considering the gene density at the vicinity of EBRs (fig. 3a), we observed that these flanking regions have a high concentration of genes when compared with the rest of the genome (HSBs) (Kruskal–Wallis test, P-value < 0.001, fig. 3a), especially so in regions that are up-stream of EBRs. Additionally, we studied the presence of protein-coding genes (n = 22,352) overlapping either the start or the end coordinates of the analyzed EBRs (both clade- and mouse-specific). This allowed us to detect whether gene sequences were affected by the presence of the estimated EBRs coordinates. In total, we detected 63 protein-coding genes that were overlapping EBRs (35 genes at the start and 28 at the end of EBRs) representing all types of clade-specific and in mouse-specific EBRs (supplementary table S6, Supplementary Material online). Of these, 55 genes were overlapping in intronic regions (87.5%). In only 8 instances were EBR coordinates found to be positioned inside an exon (supplementary table S6, Supplementary Material online).
Table 1.
Gene Content in EBRs
| Protein-Coding Genes |
||
|---|---|---|
| EBR Type | P-Value | z-Score |
| Mouse specific | 0.029* | 2.53 |
| Muridae specific | 0.009** | 1.43 |
| Cricetidae + Muridae specific | 0.049* | 2.95 |
| Muroidea specific | 0.004** | 3.81 |
| Myodonta specific | 0.009** | 2.93 |
| Rodentia specific | 0.003** | 3.21 |
| All EBRs | 0.001** | 6.25 |
Note.—Analysis of 10,000 permutation test. P-values are represented for each type of EBR detected in the mouse genome. Significant P-values indicate an accumulation of genes for each EBR analyzed when compared with the rest of mouse genome.
P-value < 0.05.
P-value < 0.01.
Fig. 3.—
Genome wide analysis of gene content and recombination rates. (A) Schematic representation of the genomic regions considered for the analysis (see “Materials and methods” section for details). (B) Distribution of protein-coding genes. The X-axis represents the genomic regions analyzed, whereas the Y-axis display the mean number of genes detected per 10 kbp. (C) Distribution of recombination rates. The X-axis represents the genomic regions analyzed, whereas the Y-axis displays the mean recombination rate detected per 10 kbp. (D) Distribution of constitutive Lamina Associated Domains (cLADs). The X-axis represents de genomic regions analyzed, whereas the y-axis display the mean number of cLADs identified per each 10 kbp windows. Standard error bars are represented. Punctuated lines represent genome-wide means. Asterisk indicates statistical significance (Kruskal–Wallis test, **P-value < 0.001).
Because chromosomal rearrangements can potentially affect the structure and regulation of genes in or nearby the affected regions, we focused on the putative adaptive role of EBRs by analyzing gene ontology of the 107 protein-coding genes detected within Rodentia-specific and one mouse-specific EBRs in the mouse genome. We found two gene families localized within individual EBRs. Moreover, there was one enrichment cluster in EBRs that presented the highest statistical support when compared with the rest of the genome (n = 3; EASE ≤ 0.05) (table 2 and supplementary table S7, Supplementary Material online). The first gene family included the Calycin superfamily and more specifically the Lipocalins (Lcn) that were localized within two nearby EBRs (one Rodentia-specific and one mouse-specific EBR) in mouse chromosome 2. In particular, we detected Lipocalin genes that were involved in the transportation of lipophilic molecules (Lcn4), sperm maturation (Lcn5), male fertility (Lcn13), retinoid carrier proteins within the epididymis (Lcn5 and Lcn13) and odorant binding proteins (Lcn14). The second gene family found was localized in mouse chromosome 11 and included four genes belonging to the hemoglobin family (involved in binding and/or transporting oxygen). All four genes were hemoglobin subunits and localized in a mouse-specific EBR which included Hemoglobin (Hb) X, hemoglobin alfa (Hba-alfa, chains 1, and 2), and hemoglobin theta A and B (Hb-Theta, 1B, and 1A). Moreover, our analysis revealed genes from the Lipocalin family in the oldest Rodentia EBRs (Rodentia-specific), whereas, both the hemoglobin family and the transcription regulation gene enrichment cluster were localized in the EBRs leading to the mouse lineage (transcription regulation gene cluster; n = 8 genes, enrichment score = 2.39; Benjamini test, P-value = 0.18).
Table 2.
Gene Clusters Found Enriched within EBRs
| Chr | EBR Analysis |
Gene Analysis |
||||
|---|---|---|---|---|---|---|
| Start (bp) | End (bp) | EBR Type | Gene Family | ID | Distance EBR Start (kbp) | |
| 2 | 25,510,722 | 25,615,814 | Rodentia specific | Calycin | Lcn5: Lipocalin 5 | −2.8 |
| Lcn6: Lipocain 6 | −21.6 | |||||
| Lcn10: Lipocain 10 | −27.5 | |||||
| Lcn13: Lipocalin 13 | −44.8 | |||||
| Lcn14: Lipocalin 14 | −81.8 | |||||
| 26,481,623 | 26,536,687 | Mouse specific | Lcn4: Lipocalin 4 | −41.6 | ||
| 11 | 32,168,628 | 32,232,893 | Mouse specific | Hemoglobin | Hba-X: Hemoglobin X | −7.7 |
| Hba-a1 and Hba-a2: Hemoglobin alpha-like embryonic chain in Hba complex | −14.9 | |||||
| Hbq1b: Hemoglobin, theta 1B | −18.3 | |||||
| Hbq1a: Hemoglobin, theta 1A | −31.4 | |||||
| 13 | 48,534,105 | 48,607,849 | Muridae specific | Krueppel associated box | Zfp169: zinc finger protein 169 | −50.4 |
| 17 | 15,680,043 | 15,701,318 | Muridae specific | Prdm9: PR domain containing 9 | −11.3 | |
| X | 20,596,836 | 20,735,882 | Mouse specific | Zfp182: zinc finger protein 182 | −9.2 | |
| 20,596,836 | 20,735,882 | Mouse specific | Zfp300: zinc finger protein 300 | −59.4 | ||
| 20,596,836 | 20,735,882 | Mouse specific | Ssxa1: Synovial sarcoma, X member A, breakpoint 1 | −96.1 | ||
Note.—For each EBR included in the table we have specified the mouse chromosome (chr), the start and end position (in bp), the corresponding gene enrichment cluster or gene family name, the ID and the distance of the gene start from the up-stream region of the EBR (in kbp).
Finally, and most intriguing, the only statistically significant enrichment cluster found in our analysis (Benjamini test, P-value = 0.02; table 2 and supplementary table S7) included five genes clustered as a Krueppel-associated box (KRAB) that were localized in three EBRs (classified as mouse- and Muridae-specific) and distributed in three different mouse chromosomes (table 2). KRAB proteins are transcription factors with zinc finger binding domains (Knight and Shimeld 2001) that are mainly expressed during meiosis (Baudat et al. 2010; Parvanov et al. 2010) and include, among others, Prdm9, the only known speciation-associated gene described for mammals (Mihola et al. 2009; Capilla et al. 2014).
Rodentia EBRs Correspond to Regions of Low Recombination Rates
It is known that genome reshuffling affects recombination (Rieseberg 2001; Navarro and Barton 2003), but data on the interplay between EBRs and recombination in mammals is restricted to few studies (Navarro et al. 1997; Larkin et al. 2009; Farré et al. 2013; Ullastres et al. 2014). To address this, we analyzed the genome-wide distribution of recombination rates in the mouse genome and tested whether there was a correlation with EBRs. We found that recombination rates were not homogeneously distributed across the mouse genome. Chromosomes 17 and 19 had the highest recombination rates (0.019 4Ner/kbp in both cases) while the chromosome 8 showed the lowest rate (0.003 4Ner/kbp). The mean genome-wide recombination rate was 0.015 4Ner/kbp. These observations corroborate previous observations in mammals that showed smaller chromosomes tends to have higher recombination rates than large chromosomes thereby ensuring their correct segregation during meiosis (Sun et al. 2005; Farré et al. 2013). Moreover, our analysis indicated that Rodentia EBRs presented a significantly lower mean recombination rate (0.016 4Ner/kbp) compared with the rest of the genome (0.019 4Ner/kbp, Kruskal–Wallis test, P < 0.001). To further explore these observations, we estimated the mean recombination rates for clade-specific and mouse-specific EBRs and found a significantly lower recombination rate in the mouse-specific and Muridae-specific EBRs (0.013 and 0.006 4Ner/kbp, respectively, Kruskal–Wallis test, P < 0.001). We also analyzed mean recombination rates around EBRs (fig. 3c). This analyses suggested a tendency of low recombination rates in EBRs flanking regions (0.014 and 0.012 4Ner/kbp) and then an increment in the following 100 kbp surrounding EBRs (0.021 and 0.019 4Ner/kbp) that tend to reach the values observed for HSBs (fig. 3c).
EBRs Are Associated with Open Chromatin States
We further investigated whether the distribution of EBRs in the mouse lineage was influenced by the spatial organization of chromatin inside the nucleus. We analyzed the distribution of constitutive lamina associated domains (cLADs) and found that the total 715,804 cLADs described in the mouse were not homogenously distributed across the genome, but were inversely correlated with gene distribution (supplementary fig. S3a) thus mirroring similar studies on human cells (Guelen et al. 2008). The X chromosome had the highest cLADs density (3.75 cLADs/10 kbp), whereas chromosomes 11 and 19 had the lowest (1.80 and 1.72 cLADs/10 kbp, respectively) (Kurskal–Wallis test, P < 0.001). Gene density was inversely correlated to cLADs density per chromosome, the only exceptions being chromosomes 4, 15, and 16 (supplementary fig. S3a, Supplementary Material online). When looking at the genome-wide distribution of cLADs in each chromosome, the same pattern was observed; cLADs tend to occur in genomic regions devoid in protein-coding genes (supplementary fig. S3b, Supplementary Material online). We subsequently analyzed the relationship between EBRs (both Rodentia and mouse lineage-specific EBRs) and cLADs. Our results indicated a significant decrease in cLADs density in all EBRs (2 cLADs/10 kbp) as well as in interphase regions (1.62 and 1.90 cLADs/10 kbp) when compared with the rest of the genome (2.68 cLADs/10 kbp; Kruskal–Wallis test, P < 0.001; fig. 3d). This pattern was corroborated by permutation tests (based on 10,000 permutations, z-score = −2.46; P < 0.05). Finally, the relationships between the three genomic characteristics studied in this work (gene content, recombination rate and cLADs) was examined using pair-wise correlations between all three variables. This indicated a significant negative correlation between the number of cLADs and the number of coding genes (Spearman correlation test, P = −0.093; P-value < 0.001) and less but also significant between cLADs and the recombination rates (Spearman correlation test, P = −0.015; P-value < 0.001).
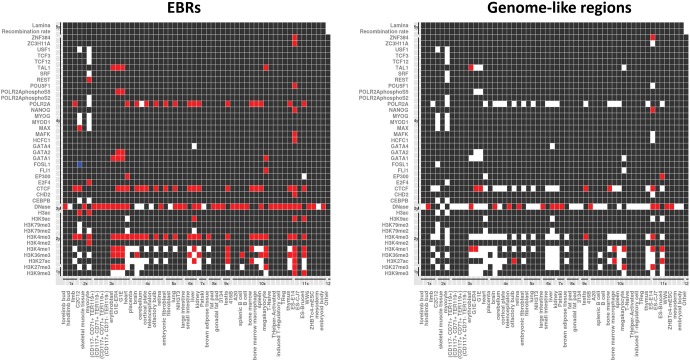
When considering DNAse-seq and ChIP-seq data available from ENCODE for a variety of mouse cell lines and tissues, we observed an association (based on 10,000 permutations, P < 0.05) with EBRs and different genomic features, representing 160 out of 244 mark-cell line combinations included in the analysis. The genomic features found to be statistically associated with EBRs included RNA pol II sites (normally associated with gene transcription), CCCTC-binding factor (CTCF) sites, DNase I hypersensitive sites (markers of regulatory and nuclease binding sites) and active chromatin marks, such as H3K4me3 (fig. 4). In order to test whether these associations were due to the high gene content observed in EBRs, two control region datasets were generated: (i) EBR-like regions, where the gene density is analogous to EBRs (0.29 genes per 10 kbp), and (ii) genome-like regions with the gene density distribution similar to the whole mouse genome (0.09 genes per 10 kbp). The observed associations with genomic features related to active chromatin marks were also present in the EBR-like regions (224 out of 244 mark-cell line combinations, representing 92% of the data set, were significantly enriched). However, a general depletion in the enrichment of these DNAse-seq and ChIP-seq marks was shown in the genome-like regions (31 out of 244 mark-cell line combinations, around a ∼13%, were significant with enrichment). These results suggest that these associations found between active chromatin markers and insulators with EBRs are likely due to the gene enrichment found in evolutionary regions in the mouse genome.
Fig. 4.—
Heat maps representing significant association found when comparing Rodentia EBRs (left panel) and control genome-like regions (right panel) with epigenetic modifications in 58 different mouse cell lines based on 10,000 permutation test with randomization (P-value < 0.05). Red squares indicate positive association (enrichment with P-value < = 0.05); white squares indicate no statistical association (P-value > 0.05), whereas blue squares indicate depletion (P-value < = 0.05). Black squares reflect no data available. The x-axis represents: (1x) Skeletal system, (2x) Muscular system, (3x) Circulatory system, (4x) Nervous system, (5x) Respiratory system, (6x) Digestive system, (7x) Excretory system, (8x) Endocrine system, (9x) Reproductive system, (10x) Lymphatic system, (11x) Stem cells, and (12x) Other. The y-axis shows: (1y) Histone modifications leading to “close” chromatin, (2y) Histone modifications associated with “open” chromatin, (3y) DNase-seq, (4y) Transcription factors, (5y) Other.
Discussion
The genome comparative analysis of six rodent species representative of two of the three major Rodentia clades (Ctenohystrica and mouse-related clade) together with six outgroup mammalian representative species has allowed us to reconstruct the most detailed comprehensive picture of the evolutionary rodent genome reshuffling. We have been able to identify lineage and clade-specific EBRs among the Rodentia species analyzed and to compare their rate of chromosome breakage (number of EBRs/Myr) as an estimate of genome reshuffling, with respect to other mammalian outgroups such as Primates, Perissodactyla, Cetartiodactila, and Carnivora. Our results are in agreement with previous studies that reflected a high genome reshuffling rate within Rodentia differentiation (either in the clades and species differentiation) (Murphy et al. 2005; Larkin et al. 2009). In fact, when considering the main mammalian diversification nodes, Rodentia presented approximately two orders of magnitude increase in EBRs per million years, than either Euarchontoglires or Laurasiathera. However, more intriguingly, this rate increased when analyzing lineage-specific EBRs. Previous cytogenetic studies indicated that the myomorph rodents showed more highly reorganized patterns [reviewed in Romanenko et al. (2012)], whereas the comparative genome analysis performed here showed the Muroidea species (S. galilii, M. ochrogaster, R. norvegicus, and M. musculus) were the ones with the highest rates of genome reshuffling (a 2- to 5-fold increase when compared with other eutherian mammals). Both differences in distinct levels of resolutions and sampling (i.e., species studied) can account for the discrepancies found between previous cytogenetic studies and the genome analysis herein presented.
In searching for signatures that characterize evolutionary genome reshuffling in rodents we detected a significantly higher gene density in EBRs when compared with the rest of the mouse genome. Although previous studies have detected this trend in other mammalian species (Murphy et al. 2005; Larkin et al. 2009; Lemaitre et al. 2009; Groenen et al. 2012), the reasons behind this pattern have remained unclear. Our results offer a substantial advance showing that both the state of the chromatin and the adaptive role of evolutionary breakpoints are most probably affecting the genomic distribution of EBRs in the mouse genome and it seems likely that this will hold for other mammalian orders.
EBRs Can Represent Opportunities for the Development of Novel Functions Involved in Adaptation in Rodents
Despite the possibility that genome reshuffling would disrupt genes essential for survival, and therefore be subject to purifying selection, EBRs can represent opportunities for the development of novel functions that may promote the adaptation of species. This is consistent with the idea that there is a connection between mammalian EBRs and the development of new adaptive gene functions, such as in the immune system or olfactory receptors (Larkin et al. 2009; Groenen et al. 2012; Ullastres et al. 2014). In this context, rodents are a particularly useful model because they are the largest mammalian order, whose species show an enormous array of evolutionary adaptations. We detected the presence of two gene families in our rodent data (lipocalins and hemoglobins) and one functional enrichment cluster (KRAB genes) within clade- and lineage-specific EBRs in the Rodentia phylogeny that might support the adaptive hypothesis of genome reshuffling.
The lipocalins found within rodent EBRs belong to two main functional groups: (i) odor-binding proteins involved in chemical communication (Snyder et al. 1989), and (ii) epididymal retinoic acid binding proteins, which are specifically expressed in the epididimys and, therefore, relevant for assuring fertility through sperm maturation acquire (Suzuki et al. 2007). Given that chemical communication in rodents is extremely important for sexual reproduction driving mate choice between individuals (Hurst and Beynon 2004), the original function of lipocalins may have been favored by natural selection during the evolution of the chemical communication in mice (Stopková et al. 2009). In addition to this observation, the impairment of antioxidative mechanisms in rodents have been also described to be adaptive under uncertain conditions, such as altitude or extreme thermal conditions, among others (Storz et al. 2007, 2009). In this context, developing new variants of hemoglobin can provide selective advantage, exemplified by the high levels of hemoglobin polymorphisms described in rodent species (Natarajan et al. 2013; Kotlík et al. 2014).
However, perhaps the most relevant result was the presence of an enrichment cluster in rodent EBRs that included KRAB genes, a group of transcription factors with zinc finger (ZNF) domains. Most of the KRAB-ZNF proteins, with the exception of Prdm9, are not functionally fully characterized, but are known to be organized in clusters (Huntley et al. 2006; Ding et al. 2009) and are thought to play a role in speciation given their role in reproductive isolation (Turner et al. 2014; Nowick et al. 2013). In fact, studies in mouse have shown that the PRDM9 protein, a meiotic-specific histone methyltransferase, determines the position where recombination occurs (Brick et al. 2012) as well as determining recombination rates in mice natural populations (Capilla et al. 2014). KRAB-ZNF genes are, indeed, fast evolving [for a review see Nowick et al. (2013)] and, in the case of Prdm9, a large diversity in the number and sequence of zinc fingers have been reported (Oliver and Greene 2009; Steiner and Ryder 2013; Buard et al. 2014; Capilla et al. 2014). Strikingly, we found Prdm9 together with poorly characterized KRAB genes, such as Zfp169, Zfp182 and Zfp300 in different Rodentia EBRs. It may be possible that the rapid evolution characterizing this gene family might be related to the instability created by genome reshuffling within these regions which could alter both sequence composition and expression patterns of the genes located within EBRs.
Considering the results obtained, can evolutionary breakpoint regions be considered ‘genomic islands of speciation’ (as referred by Turner et al. 2005)? Previous studies found that EBRs tend to show higher divergence rates than other regions in the genome (Navarro et al. 1997; Marques-Bonet and Navarro 2005) and lower recombination rates (Farré et al. 2013). Mirroring these results, we detected a significant reduction on recombination rates within EBRs when compared with the rest of the mouse genome. This reduction was only maintained in EBRs corresponding to the mouse lineage and the Muridae clade, in consonance with the short effect of chromosomal rearrangements on recombination rates along the species evolution (Coop and Myers 2007). However, one may ask whether the presence of speciation genes within EBRs (here exemplified by Prdm9) combined with low recombination rates might give rise to linkage disequilibrium that facilitates selection. Genes involved in reproductive isolation are expected to be found in regions of low recombination (Rieseberg 2001; Noor 2002; Navarro and Barton 2003). In fact, gene incompatibilities, reduced introgression and higher differentiation are associated with genomic regions with reduced recombination (Geraldes et al. 2011; Seehausen et al. 2014; Janoušek et al. 2015). Therefore, low recombination rates in EBRs could lead to a high genomic differentiation and the fixation of new mutations in genes related to the species-specific phenotypes (such as genes involved in mating and individual recognition, reproductive isolation and oxidative stress), thereby reinforcing the adaptive value of genome reshuffling.
Active Chromatin Regions as Facilitators of Genome Reorganization?
We also detected an association between genome distribution of EBRs and genome organization. Several lines of evidence have suggested that factors independent of the DNA sequence are probably affecting genome plasticity, such as changes in chromatin conformation [see Farré et al. (2015) for a review]. We first observed that rodent EBRs were depleted in cLADs and that these structural genomic regions negatively correlated with gene content. Nuclear lamina anchor chromosomal domains in mammalian chromatin by interacting with constitutive LADs (cLADs). Previously it was thought that cLADs interact with the nuclear lamina independently of cell type and are conserved in human and mouse (Meuleman et al. 2013). The pattern that we observed is most probably related with the fact that the chromatin status in cLADs is mostly transcriptionally inactive and silenced (Reddy et al. 2008; Kind and van Steensel 2010; Peric-Hupkes et al. 2010; Kohwi et al. 2013). Therefore, genomic regions outside cLADs are expected to be more exposed to the transcription machinery. As a consequence of this spatial chromatin organization and according to the new Integrative Breakage Model proposed for genome evolution (Farré et al. 2015) gene-rich regions would be more susceptible to the occurrence of large-scale chromosomal reorganizations, due to their accessibility. In fact, we detected an association with EBRs and RNA pol II sites (normally associated with gene transcription), CCCTC-binding factor (CTCF) sites, DNase I hypersensitive sites (markers of regulatory and nuclease binding sites), and histone marks typically associated with open chromatin, such as H3K4me3. Our observation of a depletion of cLADs in rodent EBRs, in conjunction with a high-density of protein-coding genes, supports this view. That is, “open” chromatin configurations in regions with high transcriptional activity are gene-rich and may drive genome reshuffling. Therefore, certain properties of local DNA sequences together with the epigenetic state of the chromatin could promote the change of chromatin to an open configuration and this can contribute to genome reshuffling.
Conclusions
The present study represents the first attempt at reconstructing the evolutionary breakpoint regions across rodent phylogeny at the genomic level. Our results in rodents suggest that the presence of genes related to species-specific phenotypes in evolutionary breakpoint regions would reinforce the adaptive value of genome reshuffling. Moreover, we found association of the evolutionary breakpoint regions with active chromatin state landscapes, most probably related to gene enrichment. Overall, we postulate that chromatin conformation, an aspect that has been often overlooked in comparative genomic studies, might play a role in modeling the genomic distribution of evolutionary breakpoints. In order to fully understand the mechanism(s) shaping mammalian genomes and driving speciation, it will be necessary to take not only the functional constrains that would accompany genome reshuffling, but also the analysis of the structural organization of genomes into consideration.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
LC was the beneficiary of a FPI predoctoral fellowship (BES-2011-047722). RASG was the recipient a postdoctoral grant from “Alianza 4 Universidades”. AP is supported by a “Doctorats Industrials” predoctoral fellowship (Agència de Gestió d'Ajuts Universitaris i de Recerca). This study was supported by research grants from the Spanish Ministerio de Economia y Competitividad (CGL-2010-20170, CGL-2014-54317-P and BFU2015-71786-REDT) to ARH. The authors thank J. Alföldi and K Lindblad-Toh from Vertebrate Genome Biology Group at the Broad Institute for allowing us to use J. jaculus and M. ochrogaster genomes. The authors are grateful to V. Olmos for her contribution during the development of the work. T.J. Robinson is also acknowledged for insightful comments on early versions of the manuscript.
Literature Cited
- Ayala FJ, Coluzzi M. 2005. Chromosome speciation: humans, Drosophila, and mosquitoes. Proc Natl Acad Sci U S A. 102(Suppl):6535–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. 2006. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 7:552–564. [DOI] [PubMed] [Google Scholar]
- Baudat F, et al. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanga-Kanfi S, et al. 2009. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol Biol. 9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA, Tesler G. 2004. Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res. 14:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunschwig H, et al. 2012. Fine-scale maps of recombination rates and hotspots in the mouse genome. Genetics 191:757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, et al. 2014. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS One 9:e85021.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capilla L, et al. 2014. Genetic recombination variation in wild Robertsonian mice: on the role of chromosomal fusions and Prdm9 allelic background. Proc Biol Sci. 281:pii: 20140297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, et al. 2009. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 5:e1000538.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton MD, Musser GG. Order Rodentia 2005. Mammal species of the world. Baltimore (MD: ): The Johns Hopkins University Press; p. 745–752. [Google Scholar]
- Churakov G, et al. 2010. Rodent evolution: back to the root. Mol Biol Evol. 27:1315–1326. [DOI] [PubMed] [Google Scholar]
- Coop G, Myers SR. 2007. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 3:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Lorenz P, Kreutzer M, Li Y, Thiesen H-J. 2009. SysZNF: the C2H2 zinc finger gene database. Nucleic Acids Res. 37:D267–D273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donthu R, Lewin HA, Larkin DM. 2009. SyntenyTracker: a tool for defining homologous synteny blocks using radiation hybrid maps and whole-genome sequence. BMC Res Notes 2:148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, et al. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc Biol Sci. 279:3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2011. Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet. 7:e1002116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria R, Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 25:660–669. [DOI] [PubMed] [Google Scholar]
- Farré M, Bosch M, López-Giráldez F, Ponsà M, Ruiz-Herrera A. 2011. Assessing the role of tandem repeats in shaping the genomic architecture of great apes. PLoS One 6:e27239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, Micheletti D, Ruiz-Herrera A. 2013. Recombination rates and genomic shuffling in human and chimpanzee—a new twist in the chromosomal speciation theory. Mol Biol Evol. 30:853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, et al. 2016. Novel insights into chromosome evolution in birds, archosaurs, and reptiles. Genome Biol Evol. 8:2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, Robinson TJ, Ruiz-Herrera A. 2015. An Integrative Breakage Model of genome architecture, reshuffling and evolution: the Integrative Breakage Model of genome evolution, a novel multidisciplinary hypothesis for the study of genome plasticity. Bioessays 37:479–488. [DOI] [PubMed] [Google Scholar]
- Froenicke L, et al. 2006. Are molecular cytogenetics and bioinformatics suggesting diverging models of ancestral mammalian genomes?. Genome Res. 16:306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo MH, Garrido O, Bahamonde R, González M. 2004. Gametogenesis and nucleotypic effects in the tetraploid red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae). Biol Res. 37:767–775. [DOI] [PubMed] [Google Scholar]
- Gel B, et al. 2016. RegioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32:289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Smith KL, Nachman MW. 2011. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol Ecol. 20:4722–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2010. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics 26:1145–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graphodatsky AS, et al. 2008. Tracking genome organization in rodents by Zoo-FISH. Chromosome Res. 16:261–274. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491:393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453:948–951. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huchon D, et al. 2002. Rodent phylogeny and a timescale for the evolution of glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol. 19:1053–1065. [DOI] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, et al. 2006. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 16:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst JL, Beynon RJ. 2004. Scent wars: the chemobiology of competitive signalling in mice. Bioessays 26:1288–1298. [DOI] [PubMed] [Google Scholar]
- Janoušek V, Munclinger P, Wang L, Teeter KC, Tucker PK. 2015. Functional organization of the genome may shape the species boundary in the house mouse. Mol Biol Evol. 32:1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN. 2007. Understanding the recent evolution of the human genome : insights from human–chimpanzee genome comparisons. Hum Mutat. 28:99–130. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, et al. 2005. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes). Hum Mutat. 25:45–55. [DOI] [PubMed] [Google Scholar]
- Kind J, van Steensel B. 2010. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol. 22:320–325. [DOI] [PubMed] [Google Scholar]
- Knight RD, Shimeld SM. 2001. Identification of conserved C2H2 zinc-finger gene families in the Bilateria. Genome Biol. 2:RESEARCH0016.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai S-L, Miller MR, Doe CQ. 2013. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlík P, Marková S, Vojtek L, Stratil A, Slechta V, Hyršl P, et al. 2014. Adaptive phylogeography: functional divergence between haemoglobins derived from different glacial refugia in the bank vole. Proc Biol Sci. 281:pii:20140021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin DM, et al. 2009. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 19:770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre C, et al. 2009. Analysis of fine-scale mammalian evolutionary breakpoints provides new insight into their relation to genome organisation. BMC Genomics 10:335.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo MS, Carone DM, Green ED, O’Neill MJ, O’Neill RJ. 2009. Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genomics 10:334.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, et al. 2006. Reconstructing contiguous regions of an ancestral genome. Genome Res. 16:1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Navarro A. 2005. Chromosomal rearrangements are associated with higher rates of molecular evolution in mammals. Gene 353:147–154. [DOI] [PubMed] [Google Scholar]
- Meredith RW, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334:521–524. [DOI] [PubMed] [Google Scholar]
- Meuleman W, et al. 2013. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323:373–375. [DOI] [PubMed] [Google Scholar]
- Mlynarski EE, Obergfell CJ, O’Neill MJ, O’Neill RJ. 2010. Divergent patterns of breakpoint reuse in Muroid rodents. Mamm Genome 21:77–87. [DOI] [PubMed] [Google Scholar]
- Montgelard C, Forty E, Arnal V, Matthee CA. 2008. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol Biol. 8:321.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, et al. 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309:613–617. [DOI] [PubMed] [Google Scholar]
- Natarajan C, et al. 2013. Epistasis among adaptive mutations in deer mouse hemoglobin. Science 340:1324–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. 2003. Chromosomal speciation and molecular divergence-accelerated evolution in rearranged chromosomes. Science 300:321–324. [DOI] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, Ruiz A. 1997. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 146:695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. 2002. Is the biological species concept showing its age? Trends Ecol Evol. 17:153–154. [Google Scholar]
- Nowick K, Carneiro M, Faria R. 2013. A prominent role of KRAB-ZNF transcription factors in mammalian speciation? Trends Genet. 29:130–139. [DOI] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. 2009. Transposable elements: powerful facilitators of evolution. Bioessays 31:703–714. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327:835.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, van Steensel B. 2010. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb Symp Quant Biol. 75:517–524. [DOI] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. 2003. Genome rearrangements in mammalian evolution: lessons from human and mouse genomes. Genome Res. 13:37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452:243–247. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16:351–358. [DOI] [PubMed] [Google Scholar]
- Romanenko SA, Perelman PL, Trifonov VA, Graphodatsky AS. 2012. Chromosomal evolution in Rodentia. Heredity 108:4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, et al. 2005. Evolutionary conserved chromosomal segments in the human karyotype are bounded by unstable chromosome bands. Cytogenet Genome Res. 108:161–174. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Castresana J, Robinson TJ. 2006. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 7:R115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Farré M, Robinson TJ. 2012. Molecular cytogenetic and genomic insights into chromosomal evolution. Heredity 108:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, et al. 2014. Genomics and the origin of species. Nat Rev Genet. 15:176–192. [DOI] [PubMed] [Google Scholar]
- Segura J, et al. 2013. Evolution of recombination in eutherian mammals: insights into mechanisms that affect recombination rates and crossover interference. Proc Biol Sci. 280:20131945.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MJ, Yonenaga-Yassuda Y. 1998. Karyotype and chromosomal polymorphism of an undescribed Akodon from Central Brazil, a species with the lowest known diploid chromosome number in rodents. Cytogenet Cell Genet. 81:46–50. [DOI] [PubMed] [Google Scholar]
- Snyder SH, Sklar PB, Hwang PM, Pevsner J. 1989. Molecular mechanisms of olfaction. Trends Neurosci. 12:35–38. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Ryder OA. 2013. Characterization of Prdm9 in equids and sterility in mules. PLoS One 8:e61746.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopková R, Hladovcová D, Kokavec J, Vyoral D. 2009. Multiple roles of secretory lipocalins (Mup, Obp) in mice. Folia Zool. 58:29–40. [Google Scholar]
- Storz JF, et al. 2009. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 106:14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, et al. 2007. The molecular basis of high-altitude adaptation in deer mice. PLoS Genet. 3:e45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Trpkov K, Rademaker A, Ko E, Martin RH. 2005. Variation in meiotic recombination frequencies among human males. Hum Genet. 116:172–178. [DOI] [PubMed] [Google Scholar]
- Suzuki K, et al. 2007. Epididymis-specific lipocalin promoters. Asian J Androl. 9:515–521. [DOI] [PubMed] [Google Scholar]
- Turner LM, White MA, Tautz D, Payseur BA. 2014. Genomic networks of hybrid sterility. PLoS Genet. 10:e1004162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3:e285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullastres A, Farré M, Capilla L, Ruiz-Herrera A. 2014. Unraveling the effect of genomic structural changes in the rhesus macaque—implications for the adaptive role of inversions. BMC Genomics 15:530.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. 1978. Modes of speciation. San Francisco: W. H. Freeman and Company. [Google Scholar]
- Wu CI, Li WH. 1985. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci U S A. 82:1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Bourque G. 2009. Recovering genome rearrangements in the mammalian phylogeny. Genome Res. 19:934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, et al. 2004. Human, mouse, and rat genome large-scale rearrangements: stability versus speciation. Genome Res. 14:1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.