Abstract
The advent of whole-genome sequencing has provided an unprecedented detail about the evolution and genetic significance of species-specific variations across the whole Mycobacterium tuberculosis Complex. However, little attention has been focused on understanding the functional roles of these variations in the protein coding sequences. In this work, we compare the coding sequences from 74 sequenced mycobacterial species including M. africanum, M. bovis, M. canettii, M. caprae, M. orygis, and M. tuberculosis. Results show that albeit protein variations affect all functional classes, those proteins involved in lipid and intermediary metabolism and respiration have accumulated mutations during evolution. To understand the impact of these mutations on protein functionality, we explored their implications on protein ductility/disorder, a yet unexplored feature of mycobacterial proteomes. In agreement with previous studies, we found that a Gly71Ile substitution in the PhoPR virulence system severely affects the ductility of its nearby region in M. africanum and animal-adapted species. In the same line of evidence, the SmtB transcriptional regulator shows amino acid variations specific to the Beijing lineage, which affects the flexibility of the N-terminal trans-activation domain. Furthermore, despite the fact that MTBC epitopes are evolutionary hyperconserved, we identify strain- and lineage-specific amino acid mutations affecting previously known T-cell epitopes such as EsxH and FbpA (Ag85A). Interestingly, in silico studies reveal that these variations result in differential interaction of epitopes with the main HLA haplogroups.
Keywords: Mycobacterium, lineages, coding sequences, epitope polymorphisms, epitope-HLA binding, protein ductility
Introduction
Tuberculosis (TB) is an ancient disease that has accompanied animals and humans for thousands of years. Globally, it is estimated that TB has killed 1 billion people in the past 200 years, which turns it as the biggest killer compared with other infectious diseases as plague, influenza, smallpox, malaria, cholera, or AIDS among others (Paulson 2013). TB is responsible for 1.8 million deaths each year (WHO 2016). The main pathogen causing TB in humans is Mycobacterium tuberculosis. Other species such as M. africanum and M. canettii are also able to infect humans with less pathogenic potential and they are phlylogeographically restricted to certain populations in West- and East-African countries, respectively (de Jong et al. 2010; Supply et al. 2013). On the other hand, animal-adapted ecotypes such as M. bovis and M. caprae infecting cattle and goats, respectively, represent an economic burden as well as a zoonotic risk for humans (Aranaz et al. 2003; Smith 2012; de la Fuente et al. 2015). Several studies have addressed the phylogenetic classification of TB-causing mycobacteria. Pioneering works based on comparative genomics demonstrated that Mycobacterium species evolved by irreversible loss of genomic regions, which cannot be compensated by horizontal gene transfer. This pattern of reductive evolution led to propose an evolutionary scenario for these species (Brosch et al. 2002). Subsequent studies based on whole-genome sequencing confirmed and refined the previous deletion-based phylogeny and proposed the existence of eight major lineages (L1–L7 and animal-adapted species), which include the human-adapted ecotypes M. tuberculosis (L1–L4 and L7), M. africanum (L5 and L6), and M. canettii and the animal-adapted ecotypes M. bovis, M. caprae, M. microti, M. pinnipedii, M. orygis and M. mungi (Comas et al. 2013). With the exception of M. canettii, considered an ancestral lineage from which the remaining species emerged, these lineages comprise the M. tuberculosis complex (MTBC).
Despite the MTBC is strictly clonal and it is characterized by less than 0.05% sequence divergence between lineages, for a 4 Mb genome, this value is readily translated into 2000 polymorphisms per genome (Brosch et al. 2002; Comas et al. 2013). Considering that MTBC species code for roughly 4000 genes, this means that half of the genes might be virtually affected by polymorphisms. Even if a number of these variations might represent silent mutations or affect non-coding sequences, it is tempting to speculate that some missense or nonsense mutations might determine lineage-specific phenotypes or even impact on the host range of human- and animal-adapted species.
It is well known that Beijing-L2 strains are associated with a massive spread of drug resistance in Europe and Asia and this phenotype appear to be related with positive selection of single nucleotide polymorphisms in selected genes (Merker et al. 2015). A comparative study of the mutation rate between some L2 and L4 strains suggest that acquisition of drug resistance in Beijing-L2 strains might be related to the higher basal mutation rate of this lineage (Ford et al. 2013). However, the specific polymorphism(s) underlying Beijing-L2 phenotypes are poorly understood. In this same line, it is still unknown why M. africanum L5 and L6 are geographically restricted to West African populations even if it has been recently sequenced the largest genome repertoire of this lineage (Winglee et al. 2016). This is also applied to M. canettii that is restricted to East African populations but the genetic determinants responsible for its phylogeographic preference start to be delineated (Supply et al. 2013; Blouin et al. 2014; Boritsch et al. 2016). Another attractive and yet unresolved question is the host-pathogen specificity of the MTBC. It is intriguing how so phylogenetically related bacteria infect a variety of hosts ranging from humans to ungulates, rodents and pinnipeds. Although the answer is hampered due to the relative scarcity of genome sequences from animal-adapted species, recent studies have started to address this issue (Bos et al. 2014; de la Fuente et al. 2015).
Previous genomic studies were predominantly focused on the human pathogen M. tuberculosis, mainly due to the scarcity of whole genome sequences from other members of the MTBC. Today, the advent of next generation sequencing has provided the first collection of genomic data from M. africanum and animal-adapted species (de la Fuente et al. 2015; Winglee et al. 2016). Thus, studies covering the whole MTBC are now possible. On the other hand, irrespective of this cumulative genomic data, studies regarding comparison of the coding sequences in the MTBC are more limited. In particular, investigations concerning the structural and functional implications of single amino acid mutations have been poorly reported. A recent study has proved the value of investigating amino acid polymorphisms to understand the evolution of virulence phenotypes in the MTBC. This work showed that a single amino acid substitution in the PhoPR virulence system from M. africanum-L6 and the animal-adapted ecotypes ablates PhoPR function. Consequently, L6 and animal-adapted strains lack acyltrehalose-derived lipids and other PhoPR-dependent phenotypes (Gonzalo-Asensio et al. 2014). Given the essential role of PhoPR in M. tuberculosis virulence (Broset et al. 2015), it is plausible to think that PhoPR polymorphisms have shaped the evolution of the MTBC. Thus, characterizing differences in protein coding sequences between mycobacterial species is expected to reveal yet unexplored virulence features. With this aim we have analyzed in this work 74 complete proteomes from six Mycobacterium species including M. africanum, M. bovis, M. canettii, M. caprae, M. orygis and M. tuberculosis (fig. 1). The effect of sequence variations on the structural and functional features of proteins will be discussed.
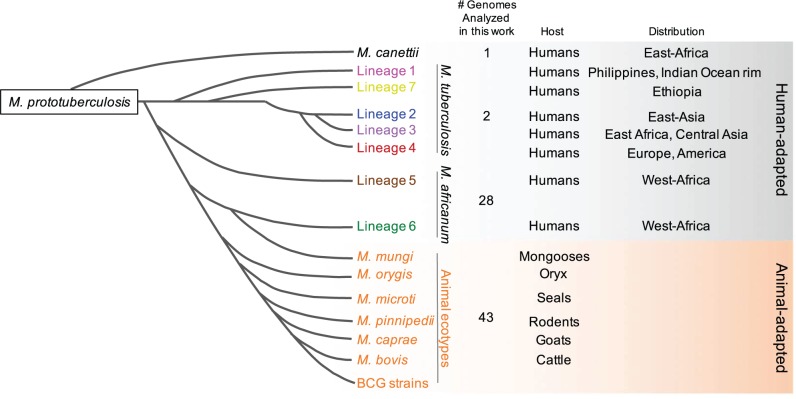
Fig. 1.—
Schematic phylogenetic relationships of the MTBC showing the number of genome sequences analyzed in this work. The figure also shows the phylogenetic distribution of M. canettii, the L1–L7 lineages and the animal-adapted species, as well as the geographic distribution of each lineage and the preferred host.
Materials and Methods
Mycobacterium Proteome Analysis
Complete 74 proteomes of 6 Mycobacterial species: M. tuberculosis (2 strains), M. africanum (28 strains), M. canettii NC_015848 (1 strain), M. bovis (41 strains), M. caprae (1 strain), and M. orygis (1 strain) were retrieved from NCBI GenBank (http://www.ncbi.nlm.nih.gov/). The strains analyzed include M. tuberculosis reference strain H37Rv (NC_000962), M. tuberculosis strain Beijing (NC_021054), M. orygis (112400015), M. canettii (NC_015848), M. caprae (CDHG01), and the remaining 69 strains listed in supplementary table S1, Supplementary Material online.
The determination of the core genome, which encodes 3009 proteins, from these 74 strains was carried out using the bioinformatic tool GET_HOMOLOGUES (Contreras-Moreira and Vinuesa 2013) and taking all core genes identified by both the OMCL and the COGS algorithms. The M. tuberculosis H37Rv strain was used as reference. Each set of homologous sequences were aligned and mutations were identified using custom Perl scripts, including amino acid substitutions, deletions, and insertions.
Epitope and Functional Cluster Analysis
Epitopes were retrieved from (Comas et al. 2010) and following the methods described in Copin et al. (2014). Functional cluster classification was as in Tuberculist (http://tuberculist.epfl.ch). Mutations were found using custom Perl scripts.
dN/dS Analyses
Thirty-four non-redundant genomes from M. canettii (1), M. africanum (13), M. bovis (16), and M. tuberculosis (4) were analyzed. Translated CDS sequences of single-copy core clusters were aligned with Clustal-omega v1.2.1 (Sievers et al. 2011) and the resulting alignments translated back to codon alignments using the primers4clades suite (Contreras-Moreira et al. 2009). Each codon alignment was then passed to yn00_cds_prealigned, obtained from https://github.com/hyphaltip/subopt-kaks (Yang, 1997) to estimate the ratio of nonsynonymous substitutions per nonsynonymous site (dN) to the number of synonymous substitutions per synonymous site (dS) of all pairs of pre-aligned sequences. Pairs of identical sequences in each cluster were not considered for dN/dS calculations.
Predictions of Intrinsic Disorder and Molecular Recognition Motifs
DISOPRED v3.1 (Jones and Cozzetto 2015) predictions were performed for all protein sequences of the Mycobacterium core proteome. All input sequences, plus the reference database uniref90, were low-complexity filtered with PFILT and scanned with three iterations of BLASTPGP with an E-value cut-off of 0.001. Predictions in PhoR protein were also done using IUPred and ANCHOR (http://iupred.enzim.hu), MoRFPred (http://biomine-ws.ece.ualberta.ca/MoRFpred/index.html), and RONN (https://www.strubi.ox.ac.uk/RONN).
Prediction of Epitope Binding to HLA Molecules
CD8+ T and CD4+ T cell epitope prediction was performed with NetMHCpan 3.0 (Andreatta and Nielsen 2015) and NetMHCIIpan 3.1 (Andreatta et al. 2015), respectively, because these tools were considered the best overall predictors in independent benchmarks (Chaves et al. 2012; Zhang et al. 2012; Trolle et al. 2015). Overlapping peptides comprising the amino acid residues of interest were tested for their binding affinity to all available classical HLA class I alleles (HLA-A, HLA-B and HLA-C; n = 2,915) and DR locus alleles for class II (n = 660) (supplementary table S4, Supplementary Material online). These HLA haplogroups were chosen due to higher predictive reliability (Andreatta et al. 2015) and data availability in the Immune Epitope Database (IEDB) (Vita et al. 2015). In the case of deletions, we predicted all the possible overlapping peptides that were present in the larger and smaller versions of the epitopes. Eight to 14-mers peptides were used in HLA class I predictions and nine to 25-mers in HLA class II. The cut-off for high binding affinity was IC50 < 50 nM or rank score < 0.5%. The fraction of the human population with the ability to recognize the predicted epitopes (population coverage) was estimated using the implementation of a previously described algorithm (Bui et al. 2006) at IEDB (http://tools.immuneepitope.org/tools/population/iedb_input).
Results
Comparative Proteome Polymorphism Analysis
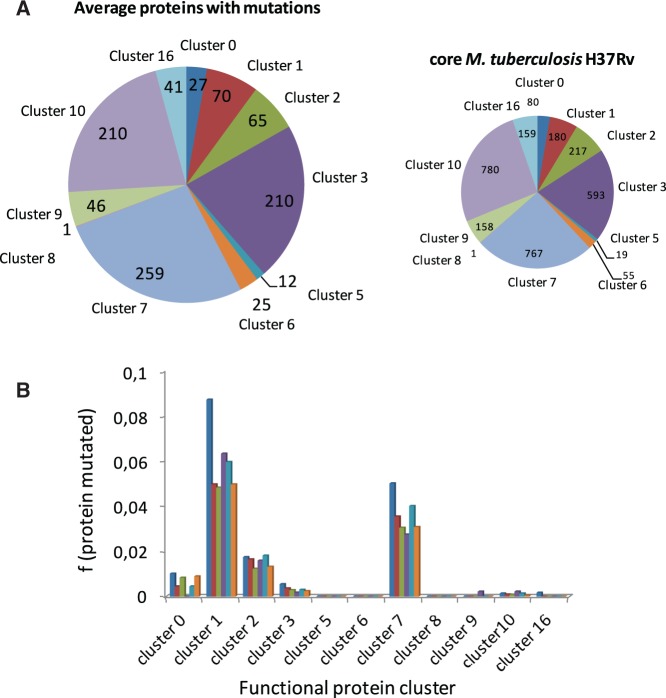
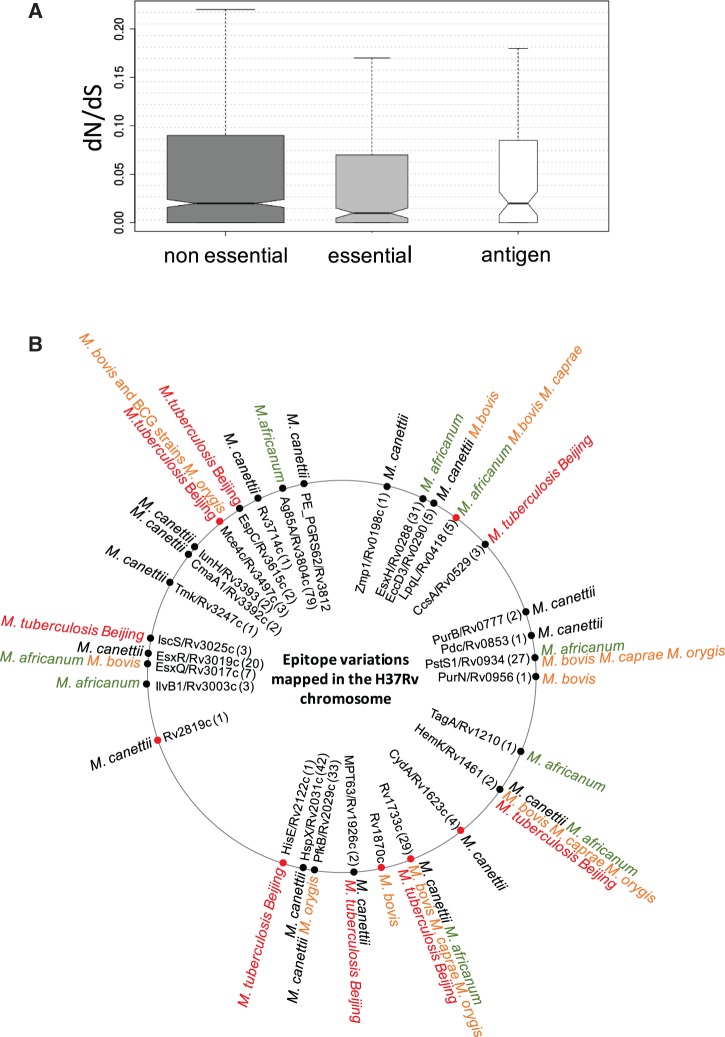
Analysis of protein sequence variations was done in the group of proteins encoded by the core-genome of Mycobacterium species (M. africanum, M. bovis, M. canettii, M. caprae, M. orygis, and M. tuberculosis), which represents 3009/3645 proteins, ca. 82.5%. The Mycobacterium proteins encoded by the core-genome are distributed in nine functional classes as follows: virulence, detoxification and adaptation (80/3009 proteins, 2.6%), lipid metabolism (180/3009 proteins, 6.0%); information pathways (217/3009 proteins, 7.2%), cell wall and cell processes (593/3009 proteins, 19.7%), insertion sequences and phages (19/3009 proteins, 0.6%), proline–glutamate (PE), and proline–proline–glutamate (PPE) proteins (55/3009 proteins, 1.8%), intermediary metabolism and respiration (767/3009 proteins, 25.5%), unknown proteins (1/3009 proteins, 0.03%), regulatory proteins (158/3009 proteins, 5.2%), conserved hypothetical proteins (780/3009 proteins, 25.9%), and conserved hypothetical proteins with an ortholog in M. bovis (159/3009 proteins, 5.3%) (fig. 2A, right). This distribution is comparable to that of the complete M. tuberculosis H37Rv proteome. Analysis of polymorphisms revealed that they affect similarly all functional classes (fig. 2A, left). When the analysis is restricted to the subset of proteins with experimental evidence annotated in BioCyc (n = 323), the data reveal a similar trend, with the largest classes, lipid (19–30%) and intermediary metabolism and respiration (15%), harboring most mutations. Statistical analyses did not indicate deviations from the expected distribution of mutations along functional classes (fig. 2B;supplementary table S2, Supplementary Material online). These percentages were normalized taking into account the total proteins associated to each functional group and the sequence identity averages.
Fig. 2.—
(A) Overall distribution of proteins encoded by the core-genome of M. tuberculosis H37Rv (right panel) and the average mutated proteins found in 74 species of the MTBC analyzed in this work (left panel). Both diagrams are divided according to functional classes. (B) Distribution according to functional classes of mutated proteins with experimental evidence annotated in BioCyc of M. canettii (dark blue), M. africanum (red), M. bovis AF2122 (green), M. tuberculosis Beijiing (violet), M. orygis (ligth blue), and M. caprae (orange). Cluster 0, virulence, detoxification and adaptation; cluster 1, lipid metabolism; cluster 2, information pathways; cluster 3, cell wall and cell processes; cluster 5, insertion sequences and phages; cluster 6, proline–glutamate (PE) and proline–proline–glutamate (PPE) proteins; cluster 7, intermediary metabolism and respiration; cluster 8, unknown proteins; cluster 9, regulatory proteins; cluster 10, conserved hypothetical proteins; cluster 16, conserved hypothetical proteins with an orthologue in M. bovis. The percentages are normalized taking into account the total proteins associated to each functional group (A) and the sequence identity averages (B).
Polymorphisms in Protein Disordered/Ductile Regions
Research of the past decade and a half has provided valuable information about intrinsically disordered/ductile proteins (IDPs) and regions (IDRs) as well as their role in diseases (van der Lee et al. 2014; Yan et al. 2016). IDPs and IDRs are characterized by a number of specific features that distinguish them from those of ordered proteins and domains and make them predictable (He et al. 2009). Such particular structural and biochemical properties of disordered/ductile regions are ideal for proteins that mediate specific molecular recognition and interaction with partners or co-ordinate regulatory events (Uversky 2015). Accordingly, ductility and flexibility feature in proteins confers advantages for their functional versatility (Babu et al. 2011). But also variations in protein flexibility can modify protein function and phenotype.
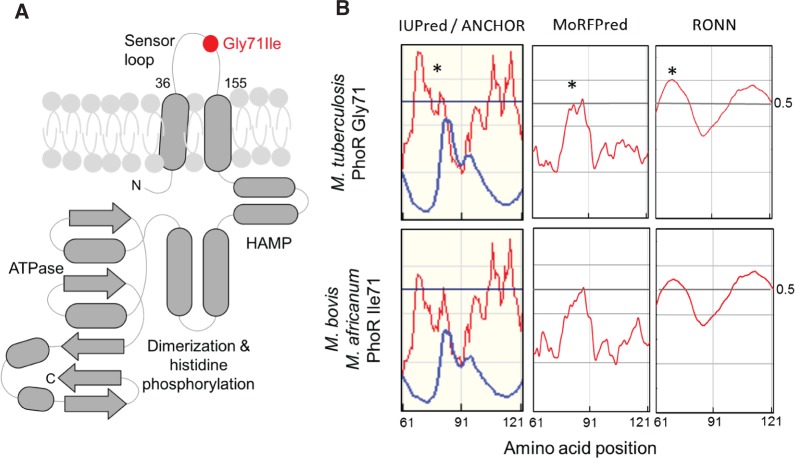
This is the case of the PhoPR two-component virulence system, where the single mutation Gly71Ile found in PhoR from M. bovis and M. africanum L6 cause strong phenotypic differences (Gonzalo-Asensio et al. 2014). The Gly71Ile mutation is located within the periplasmic loop of PhoR (Ser36 to Arg155) (fig. 3A). The intrinsically disorder predictions performed in this work show that such mutation reduce the flexibility of the Thr61–Gln91 amino acid segment. Thus, subtle variations in the flexibility of the periplasmic loop can cause a dramatic effect on the protein function and phenotype. The results obtained with IUPred (Dosztányi et al. 2005) and RONN (Yang et al. 2005) are shown in figure 3B. These are similar to those obtained with DISOPRED v3.1 (Jones and Cozzetto 2015) (supplementary fig. S1, Supplementary Material online). Moreover, ANCHOR (Dosztányi et al. 2009) and MorRFPred (Miri Disfani et al. 2012) point out that Thr61–Gln91 amino acid region in M. tuberculosis PhoR could be a molecular recognition motif. Besides, the glycine substitution by the branched-amino acid isoleucine probably impairs ordered-disordered transitions in such protein region of M. bovis/M. africanum modifying possible molecular interactions (fig. 3B).
Fig. 3.—
(A) Schematic representation of the topology of PhoR. Note the presence of two transmembrane helices spanning the periplasmic loop which contains the Gly71Ile mutation present in M. africanum L6 and animal-adapted species. (B) Disorder and molecular recognition motif predictions in the 61–121 amino acid segments of the periplasmic sensor loop of PhoR from M. tuberculosis and M. bovis/M.africanum L6. Y-axis values ≥ 0.5 means high probability of disorder. Note that the Gly71Ile mutation severely affects the ductility of the sensor domain. Predictions were carried out with IUPred and ANCHOR (left panel, red and blue lines, respectively), MoRFPred (middle panel, red line), and RONN (right panel, red line) (for details see “Materials and Methods” section).
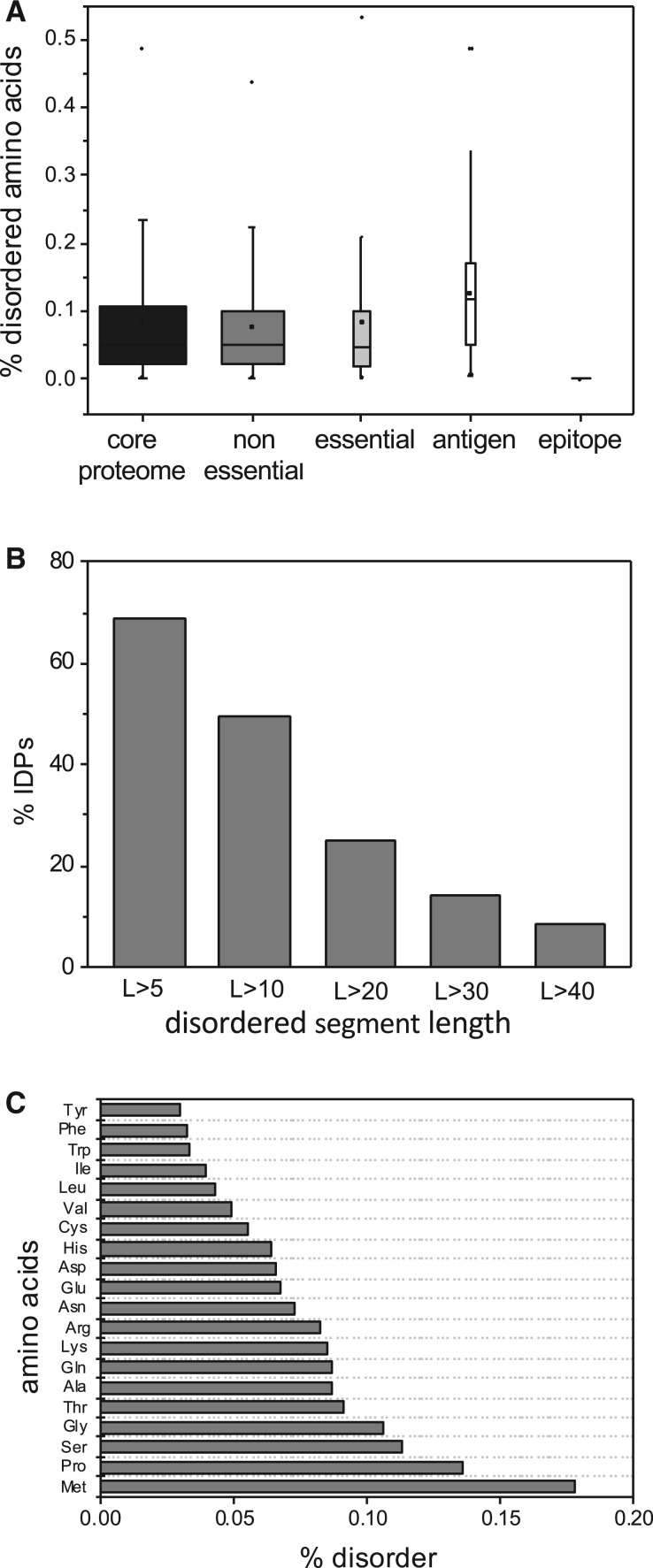
Taking into account the above results, further analyses were focused to investigate the presence of mutations in disordered/ductile regions (IDRs) of Mycobacterium proteomes in order to know their effect on structural and functional protein features. The results derived from DISOPRED v3.1 prediction indicated that disordered residues are present in all protein categories of M. tuberculosis being its content higher in antigens compared with essential and non-essential proteins (fig. 4A). Moreover, our results also show that epitopes are preferentially distributed in ordered regions in the MTBC and the higher frequency of disordered regions in antigenic proteins is restricted to non-epitope regions. This finding is agreement with previous investigations (Mitic et al. 2014; Pavlović et al. 2014). Furthermore, ca. 14% M. tuberculosis H37Rv proteins contain a long disordered segment (L > 30 aa), which is in agreement with other bacterial proteomes (fig. 4B). It is estimated that archaea and bacteria have 7–30% of such proteins (Pavlović-Lažetić et al. 2011). The amino acid composition of M. tuberculosis IDRs is enriched in methionine and proline (fig. 4C), which are characteristic of IDRs in different organisms (Dunker et al. 2008; Yruela and Contreras-Moreira 2012).
Fig. 4.—
(A) Average distribution of disordered residues in the proteins encoded by the core-genome of M. tuberculosis, essential proteins, nonessential proteins, antigens, and epitopes. (B) Percentage of intrinsically disordered/ductile proteins (IDPs) encoded by the core-genome of M. tuberculosis versus the length of disordered segments. (C) Frequency of amino acids within the disordered/ductile segments (L > 30).
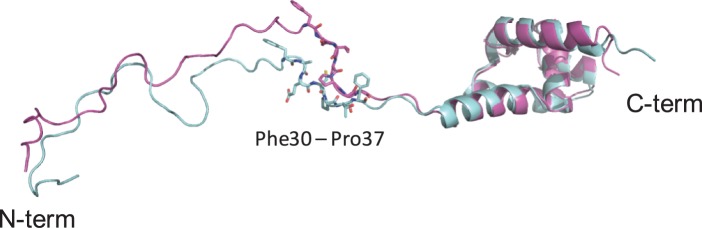
To know in detail the implications of mutations in ductile regions the attention was focused in well-annotated proteins with longer disordered segments (L > 30) because these regions are normally associated with high confidence to particular functions (Radivojac et al. 2007; Orosz and Ovádi, 2011; Yan et al. 2016). Twenty-one of these well-annotated proteins have mutations along their sequences. Among them only four proteins, Rv2215 (DlaT), Rv2358 (SmtB), Rv2495c (BkdC), and Rv3003c (IlvB1) have mutations inside IDRs (table 1). Interestingly, SmtB of M. tuberculosis Beiijing shows five amino acid substitutions in the Phe30–Pro37 motif of the N-terminal in comparison with M. tuberculosis H37Rv. The substitutions Ala31Ser, Glu32Thr, Cys33Ala, Thr35Gly, and Phe36Gly are predicted to induce a gain of flexibility in the sequence of M. tuberculosis Beijing strain compared with the H37Rv one (supplementary fig. S2, Supplementary Material online). ANCHOR (Dosztányi et al. 2009) predicted that this mutated region is more probably a molecular recognition motif (MoRF) in the Beijing strain. These variations might affect the function of the N-terminal of this protein.
Table 1.
Proteins of MTBC Species with Mutations in Disordered/Ductile Segments L > 30 with Respect to M. tuberculosis H37Rv
| gene | protein | FNC | Genome with Mutation | IDR L > 30 | IDR L < 30 | Mutation Position | MoRF (ANCHOR) | MoRF (Disopred3.1) | Phospho site1 |
|---|---|---|---|---|---|---|---|---|---|
| Rv3003c | IlvB1 | 7 | M. canetii (1) | 1–37 | 86–92 | 35 (L/P)* | 32–45 | 1–3 | nd |
| 211–223 | 51–59 | 514–519 | |||||||
| 431 | 608–618 | ||||||||
| 515–519 | |||||||||
| 608–618 | |||||||||
| Rv2358 | SmtB | 9 | M. tuberculosis str. Beijing/(5) | 1–36 | nd | 31 (A/S)* | 1–22 | 1–9 | nd |
| 32 (E/T)* | |||||||||
| 33 (C/A)* | |||||||||
| 35 (T/G)* | |||||||||
| 36 (F/G)* | |||||||||
| Rv2215 | DlaT | 7 | M. canettii (2, insert) | 80–119 | 157–159 | 139 (I/T), 210 (insert | 1–11 | nd | nd |
| 200–239 | 287–288 | APKP)*, 265 (G/E) | 31–41 | ||||||
| 48–91 | |||||||||
| M. bovis BCG Pasteur (1) | 517 (L/V) | 117–-160 | |||||||
| 169–204 | |||||||||
| 236–298 | |||||||||
| 307–319 | |||||||||
| Rv2495c | BkdC | 7 | M. africanum (1) | 87–116 | 1–4 | 107 (T/A)* | 23–31 | 1–4 | Ser551 |
| M. canettii (2) | 67 (A/V), 107(T/A)* | ||||||||
| M. bovis (2) | 107 (T/A)*, 208 (R/W) | ||||||||
| M. tuberculosis str. Beijing/NITR203 (2) | 103 (Y/D)*, 107 (T/A)* | ||||||||
| M. orygis (2) | 107 (T/A)*, 158 (R/G) |
Mutations within disordered segments (L > 30).
SmtB is a Zn-binding transcription regulator, a member of the ArsR family (Canneva et al. 2005), which allows the organism to respond to a wide range of changes in its immediate microenvironment. The SmtB presents homology with the human p53 transcription factor. Particularly, the alignment shows similarity with the TransActivation Domain 2 (TAD2) (residues 40–60), and a proline-rich region, PR (residues 64–92) of its N-terminal (supplementary fig. S3, Supplementary Material online). It is well known that the N-terminal of p53 is involved in a multitude of interactions with a wide spectrum of partners (Xue et al. 2013). Considering these observations, the amino acid substitutions in the N-terminal of SmtB of M. tuberculosis Beijing might perturb the interaction with their partners or to increase their number. The tridimensional structure of SmtB protein is still unknown and there is not information about the possible molecular interactions with their partners. However, structural homologues could provide information about these features and help us to understand the effect of such variations. In this sense, structural models of the SmtB from both M. tuberculosis H37Rv and Beijing (E-value = 2.2e−14) were produced using the NMR tridimensional structure (pdb 2lkp) of the NmtR transcription factor of M. tuberculosis (Rv3744) as template, which is also a member of the ArsR family. The comparison of both SmtB structure models shows relative conformational variations in the N-terminal (fig. 5).
Fig. 5.—
Structural models of SmtB transcription factor of M. tuberculosis H37Rv (magenta) and M. tuberculosis Beijing (light blue) using the NMR structure of NmtR transcription factor of M. tuberculosis (pdb 2lkp) as template. The residues of the non-conserved motif Phe30, Ala31, Glu32, Cys33, Thr35, Phe36, Pro37 in M. tuberculosis H37Rv and Phe30, Ser31, Thr32, Ala33, Gly35, Gly36, Pro37 in M. tuberculosis Beijing are shown in sticks. The three-dimensional cartoons were drawn using PyMol 1.4.1 (Schrodinger LLC).
Furthermore, Rv2495c, a component of branched-chain alpha-ketoacid dehydrogenase complex (BkdC, UniProt O06159) shows mutations in Tyr103Asp and Thr107Ala in M. tuberculosis Beijing compared with M. tuberculosis H37Rv which increase the flexibility in this region as indicate IUPred/ANCHOR predictions (supplementary fig. S3, Supplementary Material online). The functional implications for this higher flexibility remain to be elucidated.
Polymorphism Analysis in Human Antigens and T-Cell Epitopes
It is generally well assumed that human antigens containing T-cell epitopes in M. tuberculosis are evolutionarily hyperconseved. Accordingly, dN/dS of antigens and epitope regions are lower than dN/dS of essential and non-essential proteins (Comas et al. 2010). However, to the best of our knowledge, the hyperconservation of T-cell epitopes has not been interrogated in other species different from M. tuberculosis and BCG vaccines (Copin et al. 2014; Coscolla et al. 2015; Stucki et al. 2016). The overall ratio of nonsynonymous/synonymous substitutions (dN/dS) was calculated for the aligned CDS sequences from MTBC species listed above based on the number of nonredundant synonymous and nonsynonymous changes. Note that for these calculations a subset of 34 nonredundant genomes was analyzed, leaving out those with average peptide identity over 95.3%. The calculated dN/dS values indicated that antigens are evolutionarily conserved to the same extent than essential and nonessential genes (fig. 6A). This result allows expanding the general assumption about hyperconservation of human T-cell epitopes to the whole MTBC.
Fig. 6.—
(A) dN/dS in MTBC species calculated for nonessential genes, essential genes and antigens. Note that antigens present dN/dS ratios comparable to essential and nonessential genes, indicative of antigen hyperconservation in the whole MTBC. (B) Localization in the M. tuberculosis H37Rv chromosome of T-cell antigens showing one (black dots) or more than one (red dots) polymorphism in the MTBC species analyzed in this work. The inner circle indicates the protein name and gene number according to H37Rv nomenclature. The number of epitopes contained in every antigen is provided in parentheses. The outer circumference contains information about those MTBC species carrying epitope mutations. For detailed information about every polymorphism see supplementary table S3, Supplementary Material online.
However, it should be noted that interrogating the whole MTBC implies a higher degree of antigenic variation than comparing M. tuberculosis species alone (Comas et al. 2010). In this context, it has been also demonstrated that epitope variability is higher in widely distributed L4 sublineages relative to those geographically restricted (Stucki et al. 2016). Specifically, despite of antigen conservation, we identified single mutations in 51/852 epitopes of 27 antigenic proteins encoded by the core-genome of the MTBC species analyzed. In addition, double and even higher number of mutations were found in 9/852 epitopes of seven antigenic proteins. These polymorphisms are detailed in fig. 6B and supplementary table S3, Supplementary Material online.
Some polymorphisms are specific of one specie as the case of Ala71Ser in EsxH and Ala177Val in IlvB1, which are highly represented in M. africanum, and Thr46Pro in Mce4C of M. bovis. Others are conserved in different species such as Val352Ala in PstS1 and Gln68His in Rv1733c which are represented in M. africanum, M. bovis, M. orygis, M. caprae, and M. tuberculosis Beijing. The effect of some of these polymorphisms in antigenic proteins was partially investigated.
This is the case of EsxH, a short protein of 96 amino acids in length, which is a member of the Esx family (Uniprot P9WNK3; Ilghari et al. 2011). M. tuberculosis encodes 23 Esx proteins (EsxA–W), which are generally short in length (∼100 residues) and are organized in pairs within the genome. Of these, the EsxA/EsxB (ESAT-6/CFP10) pair is the best known mycobacterial virulence factor and promotes escape of M. tuberculosis from the phagosome to the cytosol (van der Wel et al. 2007; Houben et al. 2012) The EsxG/EsxH pair is co-ordinately regulated forming a small operon. Both proteins interact with each other to form a tight 1:1 complex (Lightbody et al. 2008). EsxG·EsxH complex contains a specific Zn(2+) binding site in the N-terminal.
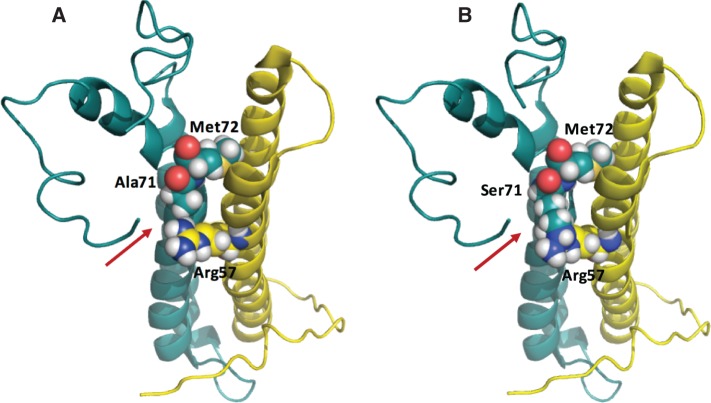
The tridimensional structure of EsxG·EsxH complex has been resolved by NMR spectroscopy (Ilghari et al. 2011). The contact surface between EsxG and EsxH is essentially hydrophobic and the Ala71 residue is located in the intermolecular interface of EsxG·EsxH complex close to Met72, which together Met18 form the base of the cleft. The conserved substitution Ala71Ser in M. africanum probably destabilizes this hydrophobic interaction. The comparison of the native complex structure and that of the mutant Ala71Ser shows that the polar Ser71 residue perturbs the interface close to EsxG–Arg57 and EsxH–Met72 (fig. 7). The adjacent conserved Ala71/Ser71 replacement could also perturb the metal binding. Thus, besides the potential role of these epitope polymorphisms in altering binding to MHC molecules (as will be studied below), mutations in these antigens might also impact on protein functionality.
Fig. 7.—
(A) Native and (B) mutated structures of the EsxG/EsxH (Rv0287/Rv0288) antigens (pdb 2kg7; Ilghari et al. 2011). The EsxG (yellow) and EsxH (blue) proteins are shown in ribbon. The amino acids Arg 57 of EsxG and Ala71/Ser71 and Met72 of EsxH are shown as spheres. The three-dimensional cartoons were drawn using PyMol 1.4.1 (Schrodinger LLC).
Other antigen carrying amino acid mutations is Rv0934 (Uniprot P9WGU1), which corresponds with the phosphate-binding protein PstS1 and functions in inorganic phosphate uptake (Peirs et al. 2005). The tridimensional 2.16 Å structure of phosphate-bound PstS-1 has been resolved by X-ray crystallography (Vyas et al. 2003). The residues Asn66 and Val352 are localized in the surface of the N-terminus (residues 42–128) and the C-terminus (residues 314–374) of the protein, respectively, which are exposed to the solvent (supplementary fig. S5, Supplementary Material online). The substitution Asn66Ser can modify slightly the electrostatic surface of the protein as shown in supplementary figure S5, Supplementary Material online. This polymorphism might perturb the electrostatic balance and the interaction with the outer cell wall membrane and/or the capture of ligands as phosphate in the medium. The Val352Ala polymorphism has been reported earlier in M. bovis and BGC by Liu et al. (2013). It is worth noting that our analysis indicates that this substitution is also highly represented in other species such as M. africanum, M. orygis, and M. caprae. Because the mycobacterial antigen PstS1 is a highly immunogenic and immunostimulatory component of the mycobacterial cell membrane such mutation might be an evolutionary adaptation and have caused immune evasion (Liu et al. 2013).
Additionally, the 1.5 Å resolution structure of MPT63 (Rv1926c, Uniprot P9WIP1, pdb 1lmi) shows that the substitution Val93Ile found in M. canettii is localized in the only helical secondary structure, a 310 helix of three residues at the start of β-strand 6 (supplementary fig. S6, Supplementary Material online). This position is in the vicinity of a negatively charged cavity and a positively charged channel, features probably with implications in the MPT63 function (Goulding et al. 2002).
Effect of Epitope Polymorphisms on HLA Binding Calculated by Predictive Algorithms
Binding of Mycobacterium epitopes to MHC class II molecules (named HLA in humans) plays a central role to mount an efficient adaptive immunity mediated by T-cells (O’Garra et al. 2013). Accordingly, polymorphisms in epitopes might alter the binding affinities of these molecules to the HLA cleft and the subsequent immune responses (Ivanyi 2014). Because HLA genes harbor a high degree of polymorphisms, testing the binding affinity of individual epitopes is challenging. Hence, in silico algorithms are useful prediction tools to compare the binding of epitope variants across representative HLA haplotypes. Using such immunoinformatics approach, we demonstrate that some epitope polymorphisms greatly impact the number of HLAs able to bind epitopes with high affinity. Consequently, because HLA molecules have distinct frequencies across the world, the fraction of the human population that will theoretically respond to the described epitopes (population coverage) can be affected (Bui et al. 2006). In our data, population coverage across several world regions was increased for Val34Met in Rv0288 (EsxH), Leu262Arg in Rv0290 (EccD3), Ile212Thr in Rv0853c (Pdc), Ala162Thr in Rv1733c, Val93Ile in Rv1926c (MPT63), Pro194Ser in Rv3714c, and Thr145Ala in Rv3804c (FbpA/Ag85A). On the other hand, a decrease was observed for Pro9Leu and Ala71Ser in Rv0288 (EsxH), diverse mutations in Rv2819c, Leu270Val in Rv3025c (IscS) and Leu76Arg in Rv3615c (EspC) (fig. 8;supplementary table S5, Supplementary Material online). Our results also suggest that T-cell vaccine design might benefit from taking into account MTBC strain variation and global HLA haplogroup distributions. Indeed, a polymorphism in Rv3804c (FbpA/Ag85A) found in a specific M. africanum strain results in stronger interaction of this epitope variant by most HLA haplogroups and specially in central American people (fig. 8). Conversely, some epitope mutations cause little or no alteration in population coverage (supplementary table S5, Supplementary Material online). Predictions for HLA class I binding was also performed (supplementary table S5 and fig. S7, Supplementary Material online).
Fig. 8.—
In silico prediction of the interaction of epitopes with the main HLA class II haplotypes. Each graph represents the population coverage of HLA class II alleles predicted to bind specific epitope variants (Y-axis) across 15 geographic regions (X-axis). Population coverages for the M. tuberculosis H37Rv epitope are indicated by orange lines and population coverages for the mutant variants in other MTBC species are shown by blue lines.
Discussion
According to the clonal origin of MTBC species, it is observed that mutations in coding sequences are uniformly distributed across the MTBC. The M. canettii outgroup has ca. 1.7 more mutations per protein than the average and this observation can be associated to its greater genetic diversity. Irrespective of this uniform distribution, proteins involved in lipid and respiration pathways have accumulated mutations during the MTBC evolution. During infection M. tuberculosis uses lipids (fatty acids and cholesterol) from the host macrophages as the main energy source (Schnappinger et al. 2003; Rohde et al. 2012). In this context, it is tempting to hypothesize that polymorphisms in the lipid metabolism might provide selective benefits to survive in the intracellular fatty acid environment. Those strains with efficient metabolic flux through lipid metabolism routes would be associated with higher virulence and vice versa. Concerning the higher frequency of polymorphisms of proteins involved in intermediary metabolism and respiration, TB-causing bacteria should be able to shift from an aerated environment during transmission to a hypoxic environment during infection. Accordingly, we can postulate that polymorphisms in respiration pathways might influence the infectious cycle. A recent study has linked the hypoxic response with lipid metabolism in M. tuberculosis (Galagan et al. 2013) and consequently it is possible that both metabolic pathways have co-evolved in different Mycobacterium species.
The comparative analyses of protein variations carried out in this work point out that changes in flexibility/ductility of MTBC protein regions can explain phenotype differences. It is known that IDRs in proteins are relevant in molecular recognition and protein-protein interactions (Vacic et al. 2007; Yan et al. 2016). This fact makes that these regions can have an important role in bacterial pathogenesis and virulence. A previous study demonstrated that a Gly71Ile substitution severely impacts on the functionality of PhoPR. The periplasmic loop of PhoR is involved in the recognition of a yet unknown activating stimulus. Accordingly, changes in ductility of the PhoR sensor domain might impair recognition of the activating stimulus which otherwise would result in lack of PhoP transcriptional activation of virulence genes (Broset et al. 2015). It is important to remember that the Gly71Ile substitution is exclusive of M. africanum and animal-adapted Mycobacterium species. These lineages are associated with slower progression to active disease and lower pathogenicity in humans than the human-adapted pathogen M. tuberculosis (Gonzalo-Asensio et al. 2014). In line with this observation, A.K. Dunker and co-workers have reported that evolution of disordered regions can be related to pathogenic microbes in comparison with non-pathogenic ones (Mohan et al. 2008). Altogether, it is worth noting that this structural analysis provides the first relationship between such protein structural variation and reported phenotype differences.
Here, we also demonstrate that hyperconservation of T-cell epitopes can be extrapolated to the whole MTBC. Accordingly, we can assume that human-adapted lineages (M. tuberculosis L1–L4 and L7 and M. africanum L5 and L6) equally benefit from being recognized by the human immune system. This finding could be also applied to the zoonotic transmission of M. bovis from cattle to humans, because our results demonstrate hyperconservation of human epitopes in the cow pathogen M. bovis. However, because we do not know the epitope repertoire in mammalian hosts infected by animal-adapted species, we cannot confirm whether animal T-cell epitopes are also evolutionarily hyperconserved.
Interestingly, despite epitopes are hyperconserved some of them show mutations. These variations could affect the structure and function of their corresponding antigens such is the case of EsxH. A recent study has demonstrated that EsxH is required for proper iron acquisition in M. tuberculosis and deletion of esxH results in decreased virulence in mice (Tufariello et al. 2016). Because a similar mutation has been reported in the non MTBC species M. marinum and M. ulcerans, it might represent an evolutionary adaptation of these species to modulate iron availability and/or virulence to specific hosts.
It is also worth of mentioning that mutations found in epitopes are predicted to modify the binding affinity to HLA proteins. A recent study expanded the search for antigenic variations by examining 1226 epitopes in 216 diverse strains of M. tuberculosis. Seven antigenic proteins showed amino acid changes between M. tuberculosis lineages. Epitopes of these proteins were immunogenic in whole blood assays and importantly, amino acid mutations in these epitopes produced differential immune stimulation (Coscolla et al. 2015). Another recent study demonstrated differences in dN/dS of epitopes from L4 sublineages. Those sublineages considered generalists associated with worldwide distribution present higher epitope variation than geographically restricted specialist sublineages (Stucki et al. 2016).These pioneering studies have paved the way to study rare epitope variants and our findings could help to translate this knowledge to other MTBC species that are also an important cause of mortality in animals and restricted human populations. In this regard, a Leu262Arg in EccD3 of M. bovis found in this work greatly increases the population coverage of HLAs class II that theoretically respond to this epitope. It remains to be explored whether this polymorphism is associated to more efficacious zoonotic transmission of certain M. bovis strains. Different polymorphisms in Rv0288 (EsxH) from M. africanum are associated with higher (Val34Met) and lower (Pro9Leu and Ala71Ser) human population coverages of HLAs class II responding to these variants. These might be sites involved in the adaption of the different species to humans or to specific subsets of the human population.
Finally, even if immunogenicity does not necessarily correlate with protection, our results could have extraordinary implications in vaccine development. Future vaccine candidates might greatly benefit from containing epitope variants with increased potential to stimulate immunity. This could be applied to either live attenuated vaccines constructed in genetically immunodominant backgrounds or to subunit vaccines containing antigens with immunodominant epitopes. One of the most advanced vaccine candidates named MVA85A consist on the Modified Vaccinia Ankara virus expressing FbpA/Ag85A. This vaccine was designed to boost the immunity provided by BCG. Although MVA85A was safe and immunogenic it failed to confer significant protection compared with BCG (Tameris et al. 2013). Our predictions indicate that FbpA/Ag85A variant found in M. africanum binds with more affinity to most HLA haplogroups. If this FbpA/Ag85A variant proves to be differentially immunogenic comparatively to the wild type, new vaccines based on modified MVA85A could be efficacious strategies to combat TB. Currently, 4 of 8 subunit vaccines (Ad5Ag85A, ChAdOx185A, MVA85A, and TB/FLU-04L) contain Ag85A expressed from a viral vector. Other subunit vaccines use FbpB/Ag85B, a related member to the Fbp family (Fletcher and Schrager 2016). Because there is a dominance of vaccines that express or contains Fbp proteins, these strategies could greatly benefit from comparative and immunological studies to select the best antigenic variants.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Gobierno de Aragón (DGA-GC B18 and B25), the Spanish Ministry of Science and Competitiveness (BIO2014-52580P, CSIC13-4E-2490), Instituto de Salud Carlos III (PI12/01970) and the European Commission Horizon 2020 (H2020-PHC-643381). Some of these grants were partially financed by the EU FEDER Program. This work was also supported by Fundação para a Ciência e Tecnologia, Portugal (IF/00474/2014) and cofunded by Programa Operacional Regional do Norte (ON.2—O Novo Norte), Quadro de Referência Estratégico Nacional (QREN), through the Fundo Europeu de Desenvolvimento Regional (FEDER).
Literature Cited
- Andreatta M, et al. 2015. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 67:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M, Nielsen M. 2015. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranaz A, Cousins D, Mateos A, Dominguez L. 2003. Elevation of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to species rank as Mycobacterium caprae comb. nov., sp. nov. Int J Syst Evol Microbiol. 53:1785–1789. [DOI] [PubMed] [Google Scholar]
- Babu MM, van der Lee R, de Groot NS, Gsponer J. 2011. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 21:432–440. [DOI] [PubMed] [Google Scholar]
- Blouin Y, et al. 2014. Progenitor “Mycobacterium canettii” clone responsible for lymph node tuberculosis epidemic, Djibouti. Emerg Infect Dis. 20:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boritsch EC, et al. 2016. pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat Microbiol. 1:19. [DOI] [PubMed] [Google Scholar]
- Bos KI, et al. 2014. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514:494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 99:3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broset E, Martin C, Gonzalo-Asensio J. 2015. Evolutionary Landscape of the Mycobacterium tuberculosis complex from the viewpoint of PhoPR: implications for virulence regulation and application to vaccine development. MBio 6:e01289–e01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H-H, et al. 2006. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 7:153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canneva F, Branzoni M, Riccardi G, Provvedi R, Milano A. 2005. Rv2358 and FurB: two transcriptional regulators from Mycobacterium tuberculosis which respond to zinc. J Bacteriol. 187:5837–5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves FA, Lee AH, Nayak JL, Richards KA, Sant AJ. 2012. The utility and limitations of current Web-available algorithms to predict peptides recognized by CD4 T cells in response to pathogen infection. J Immunol. 188:4235–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, et al. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 42:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, et al. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 45:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin R, Coscolla M, Efstathiadis E, Gagneux S, Ernst JD. 2014. Impact of in vitro evolution on antigenic diversity of Mycobacterium bovis bacillus Calmette-Guerin (BCG). Vaccine 32:5998–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscolla M, et al. 2015. M. tuberculosis T cell epitope analysis reveals paucity of antigenic variation and identifies rare variable TB antigens. Cell Host Microbe 18:538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Moreira B, Sachman-Ruiz B, Figueroa-Palacios I, Vinuesa P. 2009. primers4clades: a web server that uses phylogenetic trees to design lineage-specific PCR primers for metagenomic and diversity studies. Nucleic Acids Res. 37(Web Server issue): W95–W100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 79:7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J, et al. 2015. Comparative genomics of field isolates of Mycobacterium bovis and M. caprae provides evidence for possible correlates with bacterial viability and virulence. PLoS Negl Trop Dis. 9:e0004232.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong BC, Antonio M, Gagneux S. 2010. Mycobacterium africanum—review of an important cause of human tuberculosis in West Africa. PLoS Negl Trop Dis. 4:e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmók V, Tompa P, Simon I. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. [DOI] [PubMed] [Google Scholar]
- Dosztányi Z, Mészáros B, Simon I. 2009. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 25:2745–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 18:756–764. [DOI] [PubMed] [Google Scholar]
- Fletcher HA, Schrager L. 2016. TB vaccine development and the End TB Strategy: importance and current status. Trans R Soc Trop Med Hyg. 110:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CB, et al. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 45:784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, et al. 2013. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499:178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo-Asensio J, et al. 2014. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator. Proc Natl Acad Sci U S A. 111:11491–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding CW, et al. 2002. Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at 1.5-A resolution. Protein Sci. 11:2887–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, et al. 2009. Predicting intrinsic disorder in proteins: an overview. Cell Res. 19:929–949. [DOI] [PubMed] [Google Scholar]
- Houben D, et al. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 14:1287–1298. [DOI] [PubMed] [Google Scholar]
- Ilghari D, et al. 2011. Solution structure of the Mycobacterium tuberculosis EsxG·EsxH complex: functional implications and comparisons with other M. tuberculosis Esx family complexes. J Biol Chem. 286:29993–30002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi J. 2014. Function and potentials of M. tuberculosis epitopes. Front Immunol. 5:107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Cozzetto D. 2015. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31:857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightbody KL, et al. 2008. Molecular features governing the stability and specificity of functional complex formation by Mycobacterium tuberculosis CFP-10/ESAT-6 family proteins. J Biol Chem. 283:17681–17690. [DOI] [PubMed] [Google Scholar]
- Liu H, et al. 2013. pstS1 polymorphisms of Mycobacterium tuberculosis strains may reflect ongoing immune evasion. Tuberculosis (Edinb) 93:475–481. [DOI] [PubMed] [Google Scholar]
- Merker M, et al. 2015. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 47:242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri Disfani F, et al. 2012. MoRFpred, a computational tool for sequence-based prediction and characterization of disorder-to-order transitioning binding sites in proteins. Bioinformatics 28:i75–i83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitic NS, Pavlovic MD, Jandrlic DR, 2014. Epitope distribution in ordered and disordered protein regions—part A. T-cell epitope frequency, affinity and hydropathy. J Immunol Methods 4:83–103. [DOI] [PubMed] [Google Scholar]
- Mohan A, Sullivan WJ, Jr, Radivojac P, Dunker AK, Uversky VN. 2008. Intrinsic disorder in pathogenic and non-pathogenic microbes: discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol Biosyst. 4:328–340. [DOI] [PubMed] [Google Scholar]
- O'Garra A, et al. 2013. The immune response in tuberculosis. Annu Rev Immunol. 31:475–527. [DOI] [PubMed] [Google Scholar]
- Orosz F, Ovádi J. 2011. Proteins without 3D structure: definition, detection and beyond. Bioinformatics 27:1449–1454. [DOI] [PubMed] [Google Scholar]
- Paulson T. 2013. Epidemiology: a mortal foe. Nature 502:S2–S3. [DOI] [PubMed] [Google Scholar]
- Pavlović MD, Jandrlic DR, Mitic NS. 2014. Epitope distribution in ordered and disordered protein regions. Part B—Ordered regions and disordered binding sites are targets of T- and B-cell immunity. J Immunol Methods 407:90–107. [DOI] [PubMed] [Google Scholar]
- Pavlović-Lažetić GM, et al. 2011. Bioinformatics analysis of disordered proteins in prokaryotes. BMC Bioinformatics 12:66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirs P, et al. 2005. Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect Immun. 73:1898–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic s, et al. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci U S A. 107(16):7521–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, et al. 2007. Intrinsic disorder and functional proteomics. Biophys J. 92:1439–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde KH, Veiga DF, Caldwell S, Balazsi G, Russell DG. 2012. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 8:e1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 198:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NH. 2012. The global distribution and phylogeography of Mycobacterium bovis clonal complexes. Infect Genet Evol. 12:857–865. [DOI] [PubMed] [Google Scholar]
- Stucki D, et al. 2016. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. doi: 10.1038/ng.3704. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Marceau M, Mangenot S. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 45:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameris MD, et al. 2013. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 381:1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolle T, et al. 2015. Automated benchmarking of peptide-MHC class I binding predictions. Bioinformatics 31:2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufariello JM, et al. 2016. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci U S A. 113:E348–E357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. 2015. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 282:1182–1189. [DOI] [PubMed] [Google Scholar]
- Vacic V, et al. 2007. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 6:2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, et al. 2014. Classification of intrinsically disordered regions and proteins. Chem Rev. 114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N, et al. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129:1287–1298. [DOI] [PubMed] [Google Scholar]
- Vita R, et al. 2015. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 43:D405–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. 2003. Crystal structure of M tuberculosis ABC phosphate transport receptor: specificity and charge compensation dominated by ion-dipole interactions. Structure 11:765–774. [DOI] [PubMed] [Google Scholar]
- WHO. 2016. Global Tuberculosis Report 2016. Geneva: WHO.
- Winglee K, et al. 2016. Whole genome sequencing of Mycobacterium africanum strains from mali provides insights into the mechanisms of geographic restriction. PLoS Negl Trop Dis. 10:e0004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Brown CJ, Dunker AK, Uversky VN. 2013. Intrinsically disordered regions of p53 family are highly diversified in evolution. Biochim Biophys Acta. 1834:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Dunker AK, Uversky VN, Kurgan L. 2016. Molecular recognition features (MoRFs) in three domains of life. Mol Biosyst. 12:697–710. [DOI] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13:555–556. [DOI] [PubMed] [Google Scholar]
- Yang ZR, Thomson R, McNeil P, Esnouf RM. 2005. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 15:3369–3376. [DOI] [PubMed] [Google Scholar]
- Yruela I, Contreras-Moreira B. 2012. Protein disorder in plants: a view from the chloroplast. BMC Plant Biol. 12:165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Udaka K, Mamitsuka H, Zhu S. 2012. Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools. Brief Bioinform. 13:350–364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.