Abstract
Western honey bees (Apis mellifera) far exceed the commonly observed 1–2 meiotic recombination events per chromosome and exhibit the highest Metazoan recombination rate (20 cM/Mb) described thus far. However, the reasons for this exceptional rate of recombination are not sufficiently understood. In a comparative study, we report on the newly constructed genomic linkage maps of Apis florea and Apis dorsata that represent the two honey bee lineages without recombination rate estimates so far. Each linkage map was generated de novo, based on SNP genotypes of haploid male offspring of a single female. The A. florea map spans 4,782 cM with 1,279 markers in 16 linkage groups. The A. dorsata map is 5,762 cM long and contains 1,189 markers in 16 linkage groups. Respectively, these map sizes result in average recombination rate estimates of 20.8 and 25.1 cM/Mb. Synteny analyses indicate that frequent intra-chromosomal rearrangements but no translocations among chromosomes accompany the high rates of recombination during the independent evolution of the three major honey bee lineages. Our results imply a common cause for the evolution of very high recombination rates in Apis. Our findings also suggest that frequent homologous recombination during meiosis might increase ectopic recombination and rearrangements within but not between chromosomes. It remains to be investigated whether the resulting inversions may have been important in the evolutionary differentiation between honey bee species.
Keywords: crossover, linkage map, social evolution, genome structure, recombination rate, chromosome evolution
Introduction
Crossovers leading to meiotic recombination are required for proper segregation of homologous chromosomes and also serve an important evolutionary role by generating new allelic combinations within chromosomes. Abundant evidence for substantial variation within genomes exists (Coop and Przeworski 2007; Comeron et al. 2012; Webster and Hurst 2012; Hunter et al. 2016). Inter-specifically, meiotic recombination rates largely vary as a function of genome size and chromosome number because crossovers occur only once or twice per chromosome in most Metazoan species (Lynch 2006). However, evidence for adaptive inter-specific variation is growing (Dumont and Payseur 2008). Social insects, particularly the Western honey bee (Apis mellifera L.), represent an important exception to this rule. While a large-scale analysis (Ross, Blackmon, et al. 2015) failed to support the theoretical notion that social insect evolution should be accompanied by an increase in chromosome number (Sherman 1979), the few existing estimates of genome-wide recombination rates indicate that highly social species have exceptionally high rates of recombination among the Metazoa (Beye et al. 2006; Wilfert et al. 2007).
Despite the wealth of genomic data, relatively few accurate measures of genome-wide recombination rates exist because complete linkage maps are still laborious and costly to produce. The assertion that social evolution is associated with high recombination is thus based on genome-wide estimates of recombination for the leaf cutter ant Acromyrmex echinatior (Sirviö et al. 2006), the harvester ant Pogonomyrmex rugosus (Sirviö, Pamilo, et al. 2011), the wasp Vespula vulgaris (Sirviö, Johnston, et al. 2011), the bumblebee Bombus terrestris (Stolle et al. 2011), and the two closely related honey bee species A. mellifera (Beye et al. 2006; Liu et al. 2015; Wallberg et al. 2015) and A. cerana (Shi et al. 2013). Hypotheses to explain the high recombination rates can principally be distinguished into two lines of arguments. The first invokes immediate, short-term benefits of increasing offspring (=colony) genetic diversity to enhance division of labor and disease resistance (Sirviö et al. 2006). However, the theoretical increase of intra-colonial genetic diversity due to recombination is small compared with the effect of multiple mating (Rueppell et al. 2012). Nevertheless, a negative correlation between the varying levels of polyandry in the genus Apis (Tarpy et al. 2004) and recombination rate may be predicted if the evolution of both was due to selection for increased intra-colonial genetic diversity: Genetic diversity could be increased by either mechanism without the need for two redundant mechanisms and polyandry is predicted to outweigh any effect of recombination (Rueppell et al. 2012). The second set of arguments to explain high recombination rates in social insects is based on the evolutionary history of strong, divergent selection within the genome or ongoing directional selection coupled with small effective population sizes in social insects (Beye et al. 2006; Sirviö et al. 2006). Selection in highly social insects may be particularly strong due to disease pressure (Wilfert et al. 2007) or functionally independent evolution among castes (Kent and Zayed 2013; Liu et al. 2015). This second hypothesis predicts that all Apis species share a similar recombination rate due to their shared queen-worker divergence and caste differences, similar mating biology, and similar social life style (Oldroyd and Wongsiri 2006). Preliminary evidence for a genus-wide high recombination in the honey bee genus Apis exists (Meznar et al. 2010), but more conclusive evidence is needed.
Moreover, the long-term consequences of high recombination rates for genome structure and chromosome integrity are not yet understood in general. On the one hand, crossovers are regarded as necessary to ensure proper chromosome segregation (Baker et al. 1976) and consequently more crossovers may stabilize the chromosome tetrads during meiosis. On the other hand, double-strand breaks that are required to initiate recombination events have been associated with chromosomal instability (van Gent et al. 2001). Thus, highly recombining genomes may be more or less prone to chromosomal rearrangements over evolutionary time and genome divergence in the genus Apis with a minimum age of 20 million years (Ramírez et al. 2010; Cardinal and Danforth 2013) presents a good opportunity to test for genome consequences of high recombination.
To investigate the rate of recombination throughout the genus Apis and study its consequences for chromosome evolution, we constructed two new linkage maps of the genomes of the giant honey bee (Apis dorsata), representing the subgenus Megapis, and the dwarf honey bee (Apis florea), representing the subgenus Micrapis, and compared the marker order of the linkage maps to A. mellifera to assess synteny.
Results
For A. florea, 2906 high-quality SNP and small InDel markers were selected. Due to missing data for individual genotypes, the average number of high quality markers per individual was 2,555 (range: 1,715–2,785) and markers contained data from 72 individuals on average (range: 53–82). For A. dorsata, 3,548 high-quality markers were obtained. Each A. dorsata drone was covered on average by 3,151 SNP loci (range: 2,760–3,380), and each worker by 2,074 loci (range: 2,021–2,132) after excluding the uninterpretable heterozygous genotypes. Together, markers in A. dorsata contained informative data from 77 individuals on average (range: 51–93).
After duplicate markers and markers with biased allele distribution or elevated counts of missing data were excluded, 1,297 markers remained in the A. florea dataset for linkage mapping. These markers were genotyped on average in 88.4% of the individuals and mapped to 16 final linkage groups (fig. 1) corresponding to the 16 honey bee chromosomes (Fahrenhorst 1977). After elimination of 135 double crossovers, the final map was determined to be 4781.9 cM long, with linkage groups ranging from 156.0 to 640.7 cM (table 1), resulting in a genome-wide recombination rate estimate of 20.8 cM/Mb.
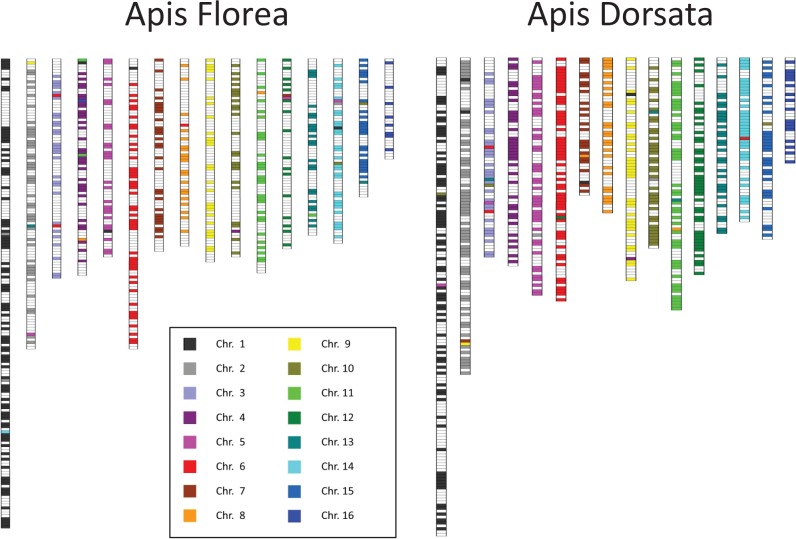
Fig. 1.—
Linkage maps of the dwarf (A. florea) and the giant honey bee (A. dorsata), with linkage maps ordered according to the homologous A. mellifera chromosomes. Each horizontal line indicates a SNP marker and their vertical position indicates recombination distances.
Table 1.
Chromosome Characteristics of the Linkage Maps of the Dwarf and Giant Honey Bees
| Dwarf Honey Bee: Apis florea |
Giant Honey Bee: Apis dorsata |
|||
|---|---|---|---|---|
| Linkage Group | Length (cM) | Marker Number* | Length (cM) | Marker Number* |
| 1 | 640.7 | 173 (87) | 830.0 | 163 (78) |
| 2 | 374.9 | 107 (48) | 399.1 | 108 (66) |
| 3 | 288.3 | 81 (31) | 408.3 | 68 (37) |
| 4 | 310.4 | 80 (37) | 377.8 | 71 (35) |
| 5 | 332.2 | 73 (26) | 382.0 | 81 (43) |
| 6 | 366.8 | 107 (45) | 324.3 | 83 (54) |
| 7 | 288.3 | 71 (28) | 331.6 | 47 (24) |
| 8 | 266.7 | 69 (21) | 321.6 | 53 (27) |
| 9 | 267.6 | 75 (28) | 353.1 | 76 (35) |
| 10 | 267.7 | 73 (25) | 341.9 | 65 (37) |
| 11 | 296.1 | 79 (31) | 317.2 | 86 (50) |
| 12 | 262.5 | 70 (18) | 318.6 | 74 (42) |
| 13 | 222.5 | 65 (23) | 303.8 | 60 (29) |
| 14 | 241.9 | 68 (32) | 276.1 | 56 (36) |
| 15 | 200.5 | 51 (25) | 292.4 | 62 (30) |
| 16 | 156.0 | 37 (8) | 184.2 | 36 (18) |
Markers with homologous matches in the A. mellifera genome are shown in brackets.
In A. dorsata, workers were excluded after an initial analysis revealed map expansion when workers and drones were analyzed together. A final set of 1,201 non-redundant markers with the least missing data and most even allele distribution were used for linkage mapping. The resulting map contained 16 major linkage groups (fig. 1) and two much smaller groups (8 and 4 markers), which were excluded from the analysis. After elimination of 141 double crossovers, the overall length of the final map was determined to be 5,761.8 cM long, with chromosomes ranging from 184.2 to 830.0 cM (table 1), resulting in a genome-wide recombination rate estimate of 25.1 cM/Mb.
Blast-searches of the A. mellifera genome with the sequences of the RAD-tag markers identified homologous sequences for 513 of the 1,279 mapped A. florea markers and 641 of the 1,189 mapped A. dorsata markers (supplementary table S1, Supplementary Material online). Only 5.3% of these homologous sequences for A. florea and 3.6% for A. dorsata were located on A. mellifera chromosomes that did not match the chromosome that corresponded to the respective linkage group (fig. 2). Almost all of these “misplaced” markers matched multiple locations in the A. mellifera genome with similar identity scores. Map saturation was confirmed by constructing linkage maps with the subset of markers with homologous sequences on respective chromosomes in the A. mellifera genome: These linkage maps were 4,657.7 and 6,811.8 cM long for A. florea and A. dorsata, containing subsets of 476 and 608 markers, respectively.
Fig. 2.—
Chromosomes show general conservation among the three Apis species, indicated by the color coding of markers with homologous sequences in the A. mellifera genome. Most single markers in the A. florea and A. dorsata linkage groups that corresponded to sequences in non-homologous chromosomes were likely assigned to the different chromosomes in error due to the presence of multiple homologous sequences in A. mellifera. Markers without significant homology to A. mellifera or with homologs in “ungroups” were left white.
Compared to the marker order within the chromosomes of the A. mellifera reference genome, the most likely marker order in the A. florea linkage groups suggested at least 28 inversions and 19 single markers that violated a syntenic order. In A. dorsata the most likely marker order indicated at least 46 inversions and 14 nonsyntenic single markers. Reordering the markers according to the corresponding order of the homologous sequences in the A. mellifera genome resulted in a 33.9% map expansion in A. florea and 30.1% in A. dorsata. All of the 14 rearranged chromosomes in A. florea and 14 of 16 rearranged chromosomes in A. dorsata were substantially (>10%) expanded in linkage map size and the originally determined marker order was significantly more likely in all of these cases (supplementary table S2, Supplementary Material online).
Discussion
Our genome-wide recombination rate estimates for dwarf and giant honey bees exceed 20 cM/Mb, providing unequivocal evidence that high recombination rates are the rule in the honey bee genus Apis. The presented results confirm a preliminary study of A. florea (Meznar et al. 2010) and complement the findings of high recombination in the cavity-nesting honey bee species A. mellifera (Beye et al. 2006) and A. cerana (Shi et al. 2013) with data from both remaining sub-genera, Megapis and Micrapis. Thus, a common explanation for the evolution and/or evolutionary maintenance of high recombination rates in all honey bees is most parsimonious. Even though the species of honey bees differ in many details of their biology and have presumably diverged over 20 million years ago (Ramírez et al. 2010; Cardinal and Danforth 2013), they share a basic social organization with one polyandrous reproductive female and thousands of functionally sterile workers cooperating in perennial colonies that reproduce by fission (Oldroyd and Wongsiri 2006). The reason for the evolutionary origin and maintenance of the exceptional recombination rates of honey bees is presumably related to these fundamentals and it is most likely that the increase in recombination rate occurred only once before the divergence of the three honey bee lineages. However, we can neither specify the timing and particular biological characteristic, nor exclude a mechanistic and potentially non-adaptive explanation.
The genome-wide estimate of recombination rate in A. dorsata is substantially higher than any comparable value among Metazoans (Wilfert et al. 2007). Two studies that report a higher recombination rate for A. mellifera, estimating a genome average of 37 cM/Mb (Liu et al. 2015) and 26.0 cM/Mb (Wallberg et al. 2015), differ substantially in methodology. Specifically, both were based on a much higher (>100×) marker density. A higher marker density typically results in higher recombination estimates because it allows for the detection of crossover events that may be dismissed as genotyping errors or missed entirely with lower marker densities (Liu et al. 2015). All studies with comparable marker densities and analyses have estimated a lower value for the recombination rate of A. mellifera (Ross, DeFelice, et al. 2015) than our value for A. dorsata. This finding provides evidence against a trade-off between polyandry and recombination rate, as predicted by the genetic diversity hypothesis, because A. dorsata also exceeds all other Apis species in queen mating frequency (Tarpy et al. 2004). Likewise, our results contradict the prediction of the genetic diversity hypotheses for A. florea to exhibit a higher recombination than other Apis species. Despite its lowest queen mating frequency in the genus (Tarpy et al. 2004), A. florea does not show a higher recombination rate than the other Apis species. Instead, our results are consistent with the hypothesis that the high recombination rates of all honey bees determined so far reflect shared ongoing selection processes or the legacy of an evolutionary history of small effective population sizes and strong divergent selection pressures (Kent and Zayed 2013). Our study does not present a conclusive test because the number of independent data points does not allow for statistical testing and the positive correlation between polyandry and high recombination could be due to the influence of other confounding variables. However, it is interesting to note that the other highly recombining social insect genomes in ants and wasps (Sirviö et al. 2006; Sirviö, Johnston, et al. 2011; Sirviö, Pamilo, et al. 2011) are also coinciding with polyandry. Based on our data, we can also not determine whether the high recombination rates in Apis are due to current selection or represent the evolutionary legacy of early honey bee social evolution. The closest relatives to honey bees with a recombination rate estimate are bumblebees with a considerably lower recombination rate (Stolle et al. 2011), suggesting that the very high recombination rates reported for Apis is genus-specific.
A consistently high genomic recombination rate in the genus Apis throughout its evolutionary history is most likely. Therefore, the generated data sets also allowed us to study the long-term consequences of high recombination rates for genome evolution. We found no substantial evidence of inter-chromosomal translocations between the three analyzed Apis genomes. The small number of markers that were identified as non-homologous in the linkage groups of A. florea and A. dorsata were presumably due to misidentification of homology among several similar sequences in the A. mellifera genome. In no case were multiple linked markers found on a nonhomologous linkage group. Thus, the high recombination rate in honey bees appears not to increase the likelihood of crossovers between nonhomologous chromosomes, although both processes depend on double strand breaks and are potentially linked (Sargent et al. 1997; Puchta 1999; Stankiewicz and Lupski 2002; Berg et al. 2010). Conversely, the absence of inter-chromosomal translocations supports the notion that homologous chromosome tetrads are stabilized by crossovers during meiosis (Baker et al. 1976). In contrast, many intra-chromosomal inversions during Apis evolution were suggested by our data and the statistical support for inversions was high in most chromosomes. Due to the limited marker density of our study, it is likely that many of the nonsyntenic single markers also indicate inversions and that the majority of inversions remained undetected (Feuk et al. 2005). The causation of intra-chromosomal rearrangements by several simultaneous crossover events is mechanistically plausible (Sharakhov et al. 2006). Such inversions facilitate differentiation within species and speciation by locally suppressing recombination (Hoffmann and Rieseberg 2008), which might be particularly important in social insect species (Linksvayer et al. 2013; Wang et al. 2013). However, it remains to be tested which of the chromosomal inversions is associated with functionally important gene clusters and signatures of accelerated evolution in honey bees.
Material and Methods
A total of 71 drones and 110 workers were collected from a colony of Apis dorsata, located on the grounds of the Agricultural Research Station Tenom (Sabah, Malaysia) in a tree about 20 m above ground. To nondestructively sample bees from one colony, individuals were captured with a net mounted on a pulley system directly next to the top of the comb (Koeniger et al. 2010). To preferentially sample drones, the net was only dropped during the mating flight time in the evening (Koeniger et al. 1994) of February 19–28, 2007. Individuals were killed by chilling and head and thorax stored in RNAlater® (Life Technologies). Genomic DNA was extracted using a modified CTAB protocol (Hunt and Page 1995) and diluted to 100 ng/µl after quantification with a Nanodrop 1000 spectrophotometer (Thermo Scientific). To verify that they were derived from the same mother, samples were genotyped at three microsatellite loci on a DNAnalyzer (LiCor), using previously described methods (Meznar et al. 2010).
Apis florea was sampled from Thailand, King Mongkut's University of Technology Thonburi, Ratchaburi Campus on May 24, 2012. A section of drone comb was cut from a single colony and all emerged, adult bees were removed. A cohort of capped drone pupae near emergence was collected using ethanol-washed forceps. Drone heads and thoraces were cut in half along the sagittal plane with a sterile razor and stored immediately in RNAlater® (Life Technologies). Genomic DNA extraction was performed using the Wizard® Genomic DNA Purification Kit (Promega) and diluted to 100 ng/µl after quantification with a Nanodrop 1000 spectrophotometer (Thermo Scientific).
All samples were treated with RNaseA (1µl of a 10ng/µl solution), precipitated with 3N sodium-acetate and ethanol, and resuspended in molecular grade water (G-Biosciences). Subsequently, all samples were re-evaluated by Nanodrop spectrophotometry and a small aliquot was analyzed for DNA quality by electrophoresis. Genomic DNA samples that were of sufficient quality and quantity (A. florea: 82 drones, A. dorsata: 71 drones and 24 workers) were sent to SNPSaurus (Eugene, OR) for genotyping by sequencing libraries using nextRAD markers (http://snpsaurus.com/nextrad-genotyping/; last accessed November 14, 2016), a modified, proprietary RAD-tag sequencing protocol (Baird et al. 2008).
The resulting 5,374 A. florea SNP markers were quality-filtered, excluding all markers with a minor allele frequency < 0.2 and > 50% missing genotypes. All heterozygous worker genotypes in A. dorsata were coded as missing data because these genotypes could not be unambiguously assigned a maternal allele. Of the original 6,193 A. dorsata SNP markers only markers with a minor allele count of >19 in drones were retained. Linkage maps were generated de novo because neither species’ genome is currently assembled at the chromosome level. Linkage analyses were performed “phase unknown” (Sirviö et al. 2006) because the grandparents were not available for phase determination. Data were processed and analyzed with “r/QTL” (Broman et al. 2003) in the “RStudio v. 0.98.501” environment. Data sets were rechecked for quality of markers and individuals and markers with identical genotype information (duplicate markers) were eliminated. Pairwise recombination frequencies were calculated with est.rf and linkage groups were formed based on pairwise linkage results with formLinkageGroups with a maximum recombination frequency (rf) of 0.15 and a minimum LOD score of 8 for A. florea and rf ≤ 0.20 and LOD ≥ 6 for A. dorsata. For A. dorsata this step was repeated without the 24 female offspring, because preliminary analyses indicated that the inclusion of these individuals inflated the genetic map. Resulting linkage groups were symmetrical and one half of these were dismissed to account for the duplication of markers before mapping “phase unknown” (Rueppell et al. 2004). The marker order in the remaining linkage groups was determined using the orderMarkers command, except for the largest linkage groups where this was not possible due to memory limitations. These linkage groups were partitioned into smaller subgroups by using more stringent linkage criteria (maximum recombination frequency reduced by 5 cM and minimum LOD score threshold increased by two). Preliminary linkage maps for these subgroups and the smaller linkage groups were computed. Based on these maps all markers were eliminated that were within 0.01 cM of each other because we regarded their information as largely redundant and potentially inflating the map size. Subsequently, all linkage groups were searched for linkage gaps of >20 cM and the flanking markers of these gaps and markers at the end of the linkage groups were investigated by manually inspecting their LOD scores with all other such “flanking” markers to identify the correct linkage patterns.
Resulting linkage groups were tested with two ripple commands to adjust local marker order where necessary. First, ripple was run with a window size of seven based on total crossover counts. The second ripple run for each chromosome was based on likelihood, restricting the window size to four. Suggested marker orders from both ripple runs were compared and the order selected that resulted in the shortest overall map length. In addition, markers near remaining linkage gaps (>20 cM) were tested manually for linkage with all markers in the data set to search for better linkages or single markers located in the gaps. Chromosomes were tested with the allchrsplits command. Map expansion by individual markers was assessed with the droponemarker command and internal markers that expanded the map by >6 cM were eliminated. Marker order was rechecked with ripple analyses before linkage map calculation, using Kosambi’s mapping function (Kosambi 1943). In a second step, double crossover events around a single marker were eliminated by recoding the specific genotype as missing data because such double crossovers are considered improbable in honey bees due to local crossover interference (Solignac et al. 2007). The relatively low density of markers does not allow us to distinguish a true double crossover from a genotyping error or a local gene conversion event. Thus, the elimination of the questionable data was the most conservative approach.
Next, we tested maps for synteny with the Apis mellifera genome by blast searches of the marker sequences against the Amel_4.5 reference assembly. We used NCBI’s Megablast with default parameters, including repeats and low complexity regions. The chromosome identity and position of the best Blast match was recorded and compared to the marker order of the A. florea and A. dorsata linkage maps. Excluding markers that matched other chromosomes or unplaced genome fragments (Ungroups), inversions were counted when the linkage map order of ≥3 markers was inverted relative to the A. mellifera order on the homologous chromosome. Misplaced single markers that were directly adjacent to the inversion points were disregarded but all other single markers that differed from A. mellifera in their relative position to other markers were counted as “non-syntenic single markers”. All A. florea and A. dorsata markers with significant matches in the homologous A. mellifera chromosomes were reordered according to their position in A. mellifera. The likelihood of this new, syntenous order was compared to the originally determined order (Meznar et al. 2010), and the size between alternative orders was compared, limiting any linkage gap between adjacent markers to 50 cM.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Gudrun Koeniger, Orawan Duangphakdee, Evelyn Hunggims, and all members of the social insect group at UNCG for their generous practical support. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15 GM102753. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was further supported financially by the National Institute of Food and Agriculture, US Department of Agriculture, under Agreement No. 2010-65104-20533 and the National Research University Project of Thailand's Office of the Higher Education Commission.
Literature Cited
- Baird NA, et al. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Carpenter ATC, Esposito MS, Esposito RE, Sandler L. 1976. The genetic control of meiosis. Annu Rev Genet. 10:53–134. [DOI] [PubMed] [Google Scholar]
- Berg IL, et al. 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 42:859–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beye M, et al. 2006. Exceptionally high levels of recombination across the honey bee genome. Genome Res. 16:1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890. [DOI] [PubMed] [Google Scholar]
- Cardinal S, Danforth BN. 2013. Bees diversified in the age of eudicots. Proc R Soc. B Biol. Sci. 280:20122686.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S. 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8:e1002905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. 2007. An evolutionary view of human recombination. Nat Rev Genet. 8:23–34. [DOI] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2008. Evolution of the genomic rate of recombination in mammals. Evolution 62:276–294. [DOI] [PubMed] [Google Scholar]
- Fahrenhorst H. 1977. Uniform chromosome numbers (N = 16) in 4 species of Apis. Apidologie 8:89–100. [Google Scholar]
- Feuk L, et al. 2005. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 1:e56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GJ, Page RE. 1995. Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics 139:1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CM, Huang W, Mackay TF, Singh ND. 2016. The genetic architecture of natural variation in recombination rate in Drosophila melanogaster. PLoS Genet. 12:e1005951.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent CF, Zayed A. 2013. Evolution of recombination and genome structure in eusocial insects. Commun Integr Biol. 6:e22919.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeniger N, Koeniger G, Tingek S. 2010. Honey bees of Borneo: exploring the centre of Apis diversity. Kota Kinabalu: Natural History Publications (Borneo; ). [Google Scholar]
- Koeniger N, Koeniger G, Tingek S, Kalitu A, Mardan M. 1994. Drones of Apis dorsata (Fabricius 1793) congregate under the canopy of tall emergent trees in Borneo. Apidologie 25:249–264. [Google Scholar]
- Kosambi DD. 1943. The estimation of map distances from recombination values. Ann Eugen. 12:172–175. [Google Scholar]
- Linksvayer TA, Busch JW, Smith CR. 2013. Social supergenes of superorganisms: do supergenes play important roles in social evolution? Bioessays 35:683–689. [DOI] [PubMed] [Google Scholar]
- Liu H, et al. 2015. Causes and consequences of crossing-over evidenced via a high-resolution recombinational landscape of the honey bee. Genome Biol. 16:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2006. The origins of eukaryotic gene structure. Mol Biol Evol. 23:450–468. [DOI] [PubMed] [Google Scholar]
- Meznar ER, Gadau J, Koeniger N, Rueppell O. 2010. Comparative linkage mapping suggests a high recombination rate in all honey bees. J Hered. 101:S118–S126. [DOI] [PubMed] [Google Scholar]
- Oldroyd BP, Wongsiri S. 2006. Asian honey bees: biology, conservation and human interactions. Cambridge: Harvard University Press. [Google Scholar]
- Puchta H. 1999. Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez SR, et al. 2010. A molecular phylogeny of the stingless bee genus Melipona (Hymenoptera: Apidae). Mol Phylogenet Evol. 56:519–525. [DOI] [PubMed] [Google Scholar]
- Ross C, DeFelice D, Hunt G, Ihle K, Rueppell O. 2015. A comparison of multiple genome-wide recombination maps in Apis mellifera In: Rychtar J, Chhetri M, Gupta SN, Shivaji R, editors. Collaborative mathematics and statistics research. New York: Springer International Publishing; p. 91–98. [Google Scholar]
- Ross L, Blackmon H, Lorite P, Gokhman V, Hardy N. 2015. Recombination, chromosome number and eusociality in the Hymenoptera. J Evol Biol. 28:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O, Meier S, Deutsch R. 2012. Multiple mating but not recombination causes quantitative increase in offspring genetic diversity for varying genetic architectures. PLoS One 7:e47220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O, et al. 2004. The genetic architecture of the behavioral ontogeny of foraging in honey bee workers. Genetics 167:1767–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent RG, Brenneman MA, Wilson JH. 1997. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 17:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhov IV, et al. 2006. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci U S A. 103:6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PW. 1979. Insect chromosome numbers and eusociality. Am Nat. 113:925–935. [Google Scholar]
- Shi YY, et al. 2013. A SNP based high-density linkage map of Apis cerana reveals a high recombination rate similar to Apis mellifera. PLoS One 8:e76459.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirviö A, et al. 2006. High recombination frequency creates genotypic diversity in colonies of the leaf-cutting ant Acromyrmex echinatior. J Evol Biol. 19:1475–1485. [DOI] [PubMed] [Google Scholar]
- Sirviö A, Johnston JS, Wenseleers T, Pamilo P. 2011. A high recombination rate in eusocial Hymenoptera: evidence from the common wasp Vespula vulgaris. BMC Genet. 12:95.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirviö A, Pamilo P, Johnson RA, Page RE, Gadau J. 2011. Origin and evolution of the dependent lineages in the genetic caste determination system of Pogonomyrmex ants. Evolution 65:869–884. [DOI] [PubMed] [Google Scholar]
- Solignac M, Mougel F, Vautrin D, Monnerot M, Cornuet JM. 2007. A third-generation microsatellite-based linkage map of the honey bee, Apis mellifera, and its comparison with the sequence-based physical map. Genome Biol. 8:R66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18:74–82. [DOI] [PubMed] [Google Scholar]
- Stolle E, et al. 2011. A second generation genetic map of the bumblebee Bombus terrestris (Linnaeus, 1758) reveals slow genome and chromosome evolution in the Apidae. BMC Genomics 12:48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpy DR, Nielsen R, Nielsen DI. 2004. A scientific note on the revised estimates of effective paternity frequency in Apis. Insectes Soc. 51:203–204. [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2:196–206. [DOI] [PubMed] [Google Scholar]
- Wallberg A, Glémin S, Webster MT. 2015. Extreme recombination frequencies shape genome variation and evolution in the Honeybee, Apis mellifera. PLoS Genet. 11:e1005189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. 2013. A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493:664–668. [DOI] [PubMed] [Google Scholar]
- Webster MT, Hurst LD. 2012. Direct and indirect consequences of meiotic recombination: implications for genome evolution. Trends Genet. 28:101–109. [DOI] [PubMed] [Google Scholar]
- Wilfert L, Gadau J, Schmid-Hempel P. 2007. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity 98:189–197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.