Abstract
RNA interference (RNAi) refers to the set of molecular processes found in eukaryotic organisms in which small RNA molecules mediate the silencing or down-regulation of target genes. In insects, RNAi serves a number of functions, including regulation of endogenous genes, anti-viral defense, and defense against transposable elements. Despite being well studied in model organisms, such as Drosophila, the distribution of core RNAi pathway genes and their evolution in insects is not well understood. Here we present the most comprehensive overview of the distribution and diversity of core RNAi pathway genes across 100 insect species, encompassing all currently recognized insect orders. We inferred the phylogenetic origin of insect-specific RNAi pathway genes and also identified several hitherto unrecorded gene expansions using whole-body transcriptome data from the international 1KITE (1000 Insect Transcriptome Evolution) project as well as other resources such as i5K (5000 Insect Genome Project). Specifically, we traced the origin of the double stranded RNA binding protein R2D2 to the last common ancestor of winged insects (Pterygota), the loss of Sid-1/Tag-130 orthologs in Antliophora (fleas, flies and relatives, and scorpionflies in a broad sense), and confirm previous evidence for the splitting of the Argonaute proteins Aubergine and Piwi in Brachyceran flies (Diptera, Brachycera). Our study offers new reference points for future experimental research on RNAi-related pathway genes in insects.
Keywords: evolution, RNA interference, r2d2, argonaute, dicer
Introduction
RNA interference (RNAi), also known as RNA silencing, refers to a set of molecular processes in which small RNA (sRNA) molecules (i.e., siRNA, miRNAs, and piRNAs) target and silence or down-regulate the expression of specific nucleic acids (Ha and Kim 2014). The core components of RNAi pathways are Argonaute proteins, which associate with the sRNAs and silence specific target nucleic acids (Meister 2013). The Argonaute and sRNA complex is termed the RNA induced silencing complex (RISC). The RISC uses complementary base pairing of the sRNA to identify the target RNA molecules. Argonaute proteins can silence their targets, certain Argonautes cleave the target mRNA while others affect their targets using alternative mechanisms (Ketting 2011). RNAi pathways differ in number of ways including the exact proteins involved, sRNAs involved, and target RNAs. For instance the siRNA pathway targets dsRNA of viral origin while the piRNA pathway primarily targets transposons (Meister 2013; Czech and Hannon 2016).
RNAi interference pathways are found throughout eukaryotic organisms and are thought to be present in the last common ancestor of extant eukaryotes. RNAi may have originated as a means of anti-viral defense (Shabalina and Koonin 2008). Other RNAi functions, such as gene regulation, are thought to have evolved later (Shabalina and Koonin 2008). While the basic structure of RNAi pathways and involved proteins are similar throughout eukaryotes, substantial gene duplication and gene loss has occurred in multiple lineages (for examples see: Campbell et al. 2008; Tomoyasu et al. 2008; Jaubert-Possamai et al. 2010; Lewis et al. 2016). In insects, three main RNAi pathways are involved in gene regulation and defense against viruses and transposable elements (Obbard et al. 2009). The origin and evolution of the genes involved in these three pathways is not well documented. Therefore, we screened transcriptome assemblies of 100 insect species for ten core RNAi pathway genes and present the most comprehensive overview of the evolution and distribution of these core RNAi pathways in insects and related arthropods. In addition to the ten core RNAi genes, we also searched for transcripts of Sid-1, a gene associated with the systemic spread of RNAi between the cells of Caenorhabditis elegans (Winston et al. 2002).
Studies on model organisms show that duplication and loss of core RNAi pathway genes have occurred multiple times. For instance, the number of paralogous genes coding for Argonaute proteins varies throughout eukaryotes: humans have eight genes coding for Argonaute proteins, Drosophila melanogaster has five, Arabidopsis thaliana has ten, while the nematode C. elegans has 26 Argonaute proteins (Hutvagner and Simmard 2008; Siomi and Siomi 2009). This observed duplication of core RNAi pathway genes might be correlated with a diversification (Hutvagner and Simmard 2008) and functional specialization of the RNAi pathways (Mukherjee et al. 2013). In insects, the duplication of core RNAi genes led to three largely separate RNAi pathways, each using different proteins and sRNA molecules (Obbard et al. 2009). Each of the three RNAi pathways has a particular class of sRNAs that associates with a specific Argonaute protein to form a RISC, which targets and silences specific gene expression. The three insect RNAi pathways are briefly outlined below.
(1) The micro-RNA (miRNA) pathway is involved in the regulation of gene expression. miRNA molecules originate in the nuclear genome. Immature miRNAs are processed by the proteins Drosha and Pasha in the nucleus and then exported to the cytoplasm (Ghildiyal and Zamore 2009). In the cytoplasm, the miRNAs are further processed by Dicer1 and its co-factor Loquacious (Ghildiyal and Zamore 2009). The fully mature miRNAs are loaded into Argonaute1 to form the RISC of the miRNA pathway.
(2) The small-interfering-RNA (siRNA) pathway, sometimes referred to as just RNAi, has two functions. The first is a means of anti-viral defense. Here dsRNA of viral origin (produced either inside or outside of the cell) is processed by the protein Dicer2 and the dsRNA binding protein R2D2 into small interfering RNAs (siRNAs) (Meister 2013). Subsequently, the siRNAs are loaded into Argonaute2 to form a RISC, which silences viral gene expression. The second function of the siRNA pathway is as a defense against transposable elements (e.g., transposons) in the genome. The transcribed transposon RNA is processed by Dicer2 and Loquacious (rather than R2D2) to form mature siRNAs (Czech et al. 2008). The siRNAs form RISC with Argonaute2, which silences the expression of transposons to prevent their further transposition in the genome (Czech et al. 2008).
(3) The piwi-interacting RNA pathway is involved in defense against the transposition of transposons in the germline (Siomi et al. 2011). In Drosophila, this pathway involves multiple Argonaute proteins of the Piwi sub-clade (i.e., Argonaute3, Aubergine, and Piwi) (Aravin et al 2007). Primary piRNAs are generated through cleavage transposon transcripts by the nuclease zucchini, thereby generating Piwi-interacting RNAs (piRNAs). These primary piRNAs are loaded into the Piwi proteins, resulting in transposon transcripts being further targeted and silenced. This creates a feedback loop, in which the cleavage of a transcript generates secondary piRNAs that target the same transcript (Meister 2013). This is called “the ping–pong amplification loop” (Aravin et al. 2007; Siomi et al. 2011).
RNAi effects were first observed in the 1990s (Napoli et al. 1990) with an explanatory mechanism proposed in 1998 (Fire et al. 1998) (for a historical overview see Sen and Blau 2006). An RNAi system in an organism can be exploited by the experimental introduction of double-stranded RNA. This allows researchers to silence specific genes and elucidate their function (Bellés 2010). Furthermore, RNAi-based technologies have great potential applications as tools for the management, control, and even protection of important insect species (Scott et al. 2013). Further applications of RNAi include novel therapies against disease (Bumcrot et al. 2006) and development of crops that are resistant to pest insects (Baum et al. 2007; Mao et al 2007; Price and Gatehouse 2008; Huvenne and Smagghe 2010). Although experimentally induced RNAi has been shown to silence the target genes in many insect species, the efficacy of RNAi is known to vary significantly between species (Terenius et al. 2011).
Differences of RNAi efficacy among insects could be partially explained by diversity in the RNAi pathway genes present in different lineages. Studies on insects whose genomes have been sequenced show that the number of core RNAi pathway genes varies between different major insect groups, along with gene duplications apparently occurring in several lineages. For example, in mosquitoes multiple Argonaute paralogous gene copies have been identified. Both Aedes aegypti (two copies of Argonaute1) and Culex pipiens (two copies of Argonaute2) have multiple copies of Argonaute genes (Campbell et al. 2008). The red flour beetle, Tribolium castaneum, also has two paralogs of both, Argonaute2 and R2D2 (Tomoyasu et al. 2008). In the pea aphid, Acyrthosiphon pisum, multiple copies of miRNA (gene-regulatory) pathway genes have been described (two paralogs each of Argonaute1, Loquacious, and Dicer1, and four paralogs of Pasha) (Jaubert-Possamai et al. 2010). While the genes, proteins, and overall mechanism of the RNAi system are well studied in model insect species, the distribution and evolution of core RNAi genes across the broad scale diversity of insects will be explored in this study.
Material and Methods
Data Used
To infer the distribution, duplication, and loss of core RNAi pathway genes in insects, we screened assemblies of transcriptomes of 100 insect species (supplementary table S3, Supplementary Material online), a subset of the transcriptomes published by Misof et al. (2014) for ten RNAi pathway genes involved in the three main insect RNAi pathways. We selected genes coding for three major protein families involved in insect RNAi: Argonaute proteins, Rnase III proteins, and dsRNA binding proteins. Additionally, we also searched for Sid-1, a gene associated with the systemic spread of RNAi between cells. We follow Misof et al. (2014) and use “insect/s” as a synonym for all hexapods, including the orders Protura (coneheads), Diplura (two-pronged bristletails), and Collembola (springtails). We additionally searched the official gene sets (proteins) of seven arthropod species—five insects, one chelicerate, and one crustacean: Apis mellifera and Nasonia vitripennis (Hymenoptera), Acyrthosiphon pisum (Hemiptera), Bombyx mori (Lepidoptera), Tribolium castaneum (Coleoptera), Ixodes scapularis (Chelicerata), and Daphnia pulex (crustaceans, Branchiopoda) (supplementary table S1, Supplementary Material online).
We substantiated the hypothesis of R2D2 being a derived feature of pterygote insects by screening the draft genomes of Hrabe’s Jmping Bristletail (Machilis hrabei; Archaeognatha; https://www.hgsc.bcm.edu/arthropods/hrabes-jumping-bristletail-genome-project; last accessed November 30, 2016) and Silvestri’s Northern Forcepstail (Catajapyx aquilonaris; Diplura; https://www.hgsc.bcm.edu/arthropods/silvestris-northern-forcepstail-genome-project; last accessed November 30, 2016).
Gene Identification
To identify putative orthologs of the ten RNAi-related pathway genes and Sid-1, we first translated the assembled transcripts of each transcript library into all six possible reading frames using the exonerate tool fastatranslate (Slater and Birney 2005; version 2.2). We subsequently used resulting amino acid sequences to create BLAST-searchable databases in Geneious 7.1.5 (Biomatters, Auckland, New Zealand; Kearse et al. 2012). We additionally obtained the official gene sets (protein sets) of seven arthropod species, for which full genomes are available, and generated seven separate BLAST-searchable databases in Geneious (for details, see supplementary table S1, Supplementary Material online). To determine the timing of duplication of Dicer genes we also searched the genomes of two spider species (Sanggaard et al. 2014), the African social velvet spider (Stegodyphus mimosarum) and the Brazilian white-knee tarantula (Acanthoscurria geniculata), and one centipede (Strigamia maritima) (Chipman et al. 2014) for Dicer orthologs. To determine if Sid-1/Tag-130 homologs were present in Diptera we BLAST searched (tBLASTn) the genome assemblies of three dipteran species. Species selected were: Drosophila pseudoobscura (GenBank assembly accession: GCA_001014495.1), Aedes aegypti (GCA_001014885.1), and Anopheles gambiae (GCA_001542645.1).
We used ten amino acid sequences involved in RNAi pathways known from Drosophila melanogaster and one amino acid sequence (Sid-1), which is absent in Drosophila, but is known from B. mori as query sequences (supplementary table S2, Supplementary Material online). All sequences were downloaded from the NCBI protein database. We used each of the eleven amino acid sequences as a query for blastp (BLAST program suite, Altschul et al. 1990) and searched within Geneious against local BLAST databases created from the 100 transcriptomes and the seven official gene sets. We removed false positives (nonorthologous homologs) by searching each hit with blastp against the NCBI nonredundant protein database (http://blast.ncbi.nlm.nih.gov/Blast.cgi; last accessed November 30, 2016). We only considered a transcript to be an ortholog and derived from a given RNAi pathway gene when it was found as best reciprocal hit.
Generation of Gene Trees
All identified amino acid sequences of a given RNAi pathway protein were aligned using the Geneious alignment tool (using the Geneious alignment algorithm; Kearse et al. 2012). We used the deer tick (I. scapularis, Chelicerata) and the crustacean branchiopod (D. pulex) as outgroups. Short sequences (< 50% of the proteins consensus length) were removed from the alignments. We visually inspected the alignments and manually corrected them for obvious misalignments. For six alignments, we inferred a gene tree applying the maximum likelihood optimality criterion as implemented in PhyML (Guindon et al. 2010; version 3.0) with the following parameters: substitution model: WAG + G, proportion of invariant sites: 0 (fixed), substitution rate categories: 4, alpha-shape parameter: estimated, optimization parameters: topology/length/rate. Statistical tree robustness was assessed in PhyML via bootstrapping (1,000 bootstrap replicates).(supplementary figures S1–S6, Supplementary Material online).
Sid-1/Tag-130 Identification
It has been suggested that putative insect orthologs of C. elegans Sid-1 are in fact orthologous with the C. elegans protein tag-130 (Tomoyasu et al. 2008). To test this, we recreated the multiple sequence alignment using C. elegans Sid-1 and tag-130 amino acid sequences. Using the multiple sequence alignment, we recreated the Sid-1 gene tree to determine if our putative insect orthologs clustered more closely to C. elegans Sid-1 or tag-130. The method used was the same as for the other gene trees.
Ancestral State Reconstruction
To infer gains and losses of orthologs of core RNAi pathway genes throughout insect evolutionary history, we used the ancestral state reconstruction package of Mesquite (Maddison and Maddison 2008; version 3.02). We used the number of genes found in each species as character states and a phylogenetic tree adapted from Misof et al. (2014). The ancestral states were reconstructed using maximum parsimony. Note that Mesquite does not allow ancestral state reconstruction under the Dollo parsimony (Maddison and Maddison 2008) optimality criterion, which penalizes the loss and subsequent regain of a character. Thus, certain figures (supplementary figs. S7–S17, Supplementary Material online) appear to show the loss of gene in one lineage and its subsequent re-evolution in a descendant lineage.
To independently infer contraction and expansion of Argonaute genes we used the CAFE 3.0 (Han et al. 2013). As input we used selected Argonaute1, Argonaute2, Piwi/Aubergine, and Argonaute3 as gene families and provided the number of homologs belonging to each gene family and a ultrametric phylogenetic tree of all species (adapted from Misof et al, 2014). We specified that CAFE 3.0 search for an optimal λ value. We did not specify that λ varies.
Testing for Evidence of Positive Selection in Specific Genes
To determine whether or not positive selection was acting on certain core RNAi pathway genes, we used the package codeML in the program PAML (version 4.8; Yang 2007). codeML calculates the ratio of nonsynonymous substitutions to synonymous substitutions (ω).
We selected two genes to test. The first was R2D2 in beetles (Coleoptera). Duplicate copies of R2D2 previously identified in Tribolium castaneum were identified in three beetle species. We tested for evidence of positive selection in all branches of the beetle clade comprising Gyrinus marinus, Aleochara curtula, and Meloe violaceus (note that we only detected one copy of R2D2 in Lepicerus sp.).
R2D2 was not found in several Lepidoptera transcriptomes suggesting that it was lost in members of this group. We hypothesized that the double-stranded RNA binding protein Loquacious may fulfill the role of R2D2 in species which have lost R2D2. We tested Loquacious for evidence of positive selection in the branches within the Lepidoptera clade comprising Nemophora degeerella, Yponomeuta evonymellus, Zygaena fausta, and Parides eurimedes. Evidence of positive selection in Loquacious in specific branches of Lepidoptera would suggest that it underwent rapid evolution and may taking the role ordinarily taken by R2D2.
For both genes (i.e., R2D2 and Loquacious), we generated multiple sequence alignments on the nucleotide level with the amino acid alignments as guidance using PAL2NAL (version 14) (Suyama et al. 2006). We applied a branch site model, in which ω is allowed to vary among both sites and branches, to test for positive selection in specified branches. For both genes we used the gene trees created above as input trees for the codeML analyses. We used two models: one in which ω varies on our branch of interest (alternative model) and one in which ω is fixed for each branch (null models). Models settings for null model were: model = 2, NSsites = 2, fix_kappa = 0, kappa = 2, fix_omega = 1, omega = 1. Model settings for alternative model were: model = 2, NSsites = 2, fix_kappa = 0, kappa = 2, fix_omega = 0, omega = 1. We tested for statistically significant difference between the two models using a Likelihood Ratio Test (LRT) with one degree of freedom.
Transcriptome Completeness Assessment
To assess transcriptome assembly completeness, we used BUSCO version 1.1b (Simão et al. 2015) to search for a set of 2,675 conserved genes that are near-universal single-copy orthologs in arthropods. These genes serve as a benchmark for genome or transcriptome completeness and are found as single copies in the majority (95%) of arthropod genomes in the OrthoDB database (Kriventseva et al. 2015). BUSCO uses a combination of BLAST (Camacho et al. 2009), profile Hidden Markov Models generated with HMMER 3 (Eddy 2011), and a gene model refinement procedure (Stanke et al. 2004) to identify and discriminate genes which are present, duplicated, fragmented, or missing in the searched transcriptome. As transcriptomes only contain a subset of the total genes present in the genome we expect that not all 2,675 BUSCO genes will be found.
Results
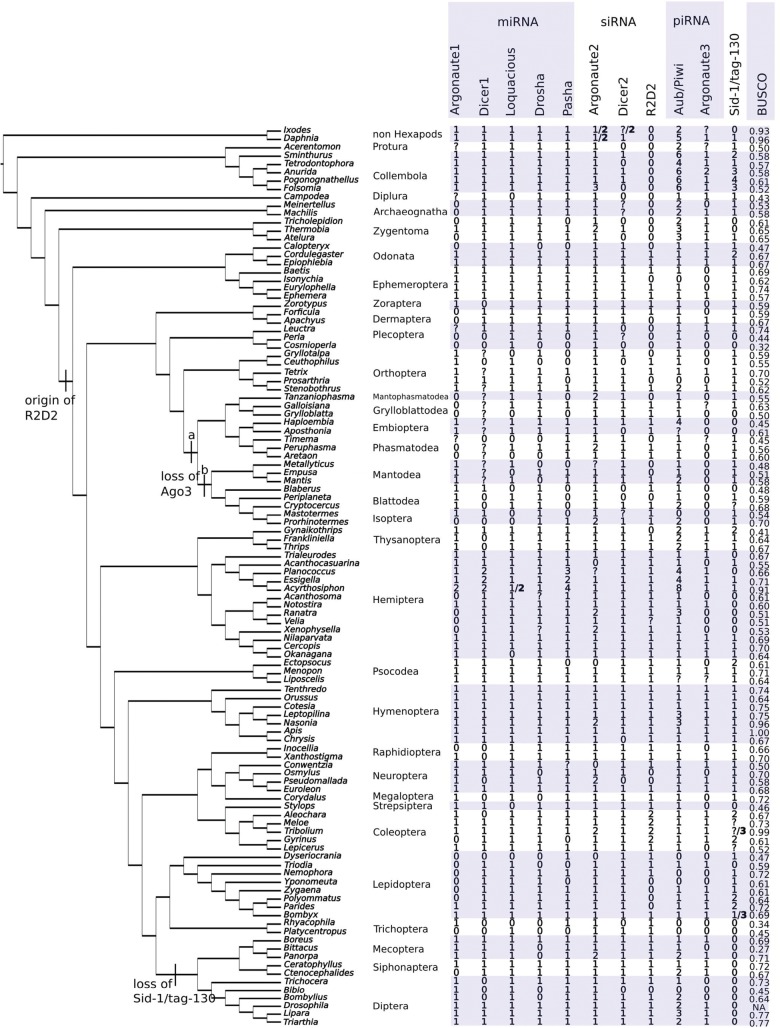
Our systematic search for core genes directly involved in RNA interference pathways (five in the miRNA pathway, three in the siRNA pathway, and two in the piRNA pathway) in whole-body transcript libraries of 100 insect species revealed putative orthologs of at least one gene from each of the three RNA silencing pathways in all 32 studied insect orders. We found a complete set of ten genes in 13 of all studied orders. We furthermore found putative orthologs of Sid-1, a gene associated with systemic RNAi, in 25 out of the 32 insect orders. Finally, analysis of the 100 transcriptomes indicated gene duplication and gene loss events in multiple lineages and species (fig. 1). While transcriptomes can be used to identify the RNAi genes, they do not allow us to conclusively state that a gene is missing from the genome.
Fig. 1.—
Distribution of ten core RNA interference genes and the systemic RNA interference gene Sid-1 in insects. The number of copies of each gene found using our methodology is noted in the table. Genes whose presence could not be conclusively verified or denied are marked with a question mark (?). Genes which were not found are marked with a zero (0). In some genome species known genes were not recovered. Where this is the case we include the known number of genes in bold after the number we have identified. Tree topology is based on that of Misof et al. (2014). Note that Blattodea is considered paraphyletic.
miRNA Pathway Genes
We identified orthologs of five miRNA pathway genes known from Drosophila (Obbard et al. 2009) in our studied insect species: we found Argonaute1, Dicer1, Loquacious, Drosha, and Pasha in the transcriptomes of 67, 66, 87, 79, and 80 insect species, respectively, representing all major lineages (table 1). Consistent with this observation, ancestral reconstruction using Mesquite (v. 3.02) suggests that all five miRNA pathway genes were present in the last common ancestor of insects. Possible duplicates of Dicer1 and Pasha were found in the transcriptomes of Planococcus citri (Hemiptera; two Dicer 1 and three Pasha) and Essigella californica (Hemiptera; two Dicer1 and two Pasha) (fig. 1).
Table 1.
Orthologs of the Members of the Three Different RNAi Pathways Identified in 100 Investigated Insect Transcriptomes (subset of data published by Misof et al. 2014)
| Gene | Pathway | Present | Duplicates |
|---|---|---|---|
| Argonaute1 | miRNA | 67 | 0 |
| Dicer1 | miRNA | 66 | 2 |
| Loquacious | miRNA | 87 | 0 |
| Drosha | miRNA | 79 | 0 |
| Pasha | miRNA | 80 | 2 |
| Argonaute2 | siRNA | 94 | 6 |
| Dicer2 | siRNA | 80 | 0 |
| R2D2 | siRNA | 68 | 3 |
| Aubergine/Piwi | piRNA | 89 | 28 |
| Argonaute3 | piRNA | 51 | 1 |
| Sid-1/Tag-130 | Systemic RNAi | 68 | 7 |
Note.—The present column shows the number of transcriptomes (out of 100) in which a putative ortholog was found. The duplicates column shows the number of transcriptomes (out of 100) in which more than one putative ortholog for a given gene was identified. For this study, we used assembly version 2 of all transcriptomes, released in October 2015.
siRNA Pathway Genes
Of all three currently known core genes involved in the siRNA pathway of Drosophila, we identified orthologs of Argonaute2, Dicer2, and R2D2 in the assembled transcripts of 94, 80, and 68 species, respectively (table 1), again representing the major insect lineages. However, we did not find R2D2 in any of the primary wingless insect (nonpterygote) species. We found possible duplicates of Argonaute2 in the transcript assemblies of the following species: Tanzaniophasma sp. (Mantophasmatodea), Peruphasma schultei (Phasmatodea), Prorhinotermes simplex (Isoptera), Xenophysella greensladeae (Hemiptera), Pseudomallada prasinus (Neuroptera), and Panorpa vulgaris (Mecoptera). We identified two copies of R2D2 in Meloe violaceus, Aleochara curtula, and Gyrinus marinus (Coleoptera). Ancestral state reconstruction using Mesquite (v. 3.02) suggests that R2D2 was present in the last common ancestor of Pterygota. Ancestral state reconstruction using CAFE 3.0 indicates that Argonaute2 was present in two copies in the last common ancestor of insects. Subsequently, in winged insects one copie was lost while in wingless insects Argonaute2 was duplicated.
piRNA Pathway Genes
The piRNA system of Drosophila melanogaster involves three Argonaute proteins of the Piwi family (Argonaute3, Piwi, and Aubergine). We identified both Piwi and Aubergine only in Diptera (three species: Bombylius major, Lipara lucens, and Triarthria setipennis) (fig. 1). Outside of Diptera, we found orthologs of either Piwi/Aubergine in the transcript assemblies of 85 species (table 1), representing all major insect lineages. Consistent with this observation, ancestral state reconstruction generated with Mesquite (v. 3.02) suggests that homologs of both Piwi/Aubergine and Argonaute3 were present in the last common ancestor of insects, with Piwi/Aubergine present in multiple copies (integers between two and five were equally likely). Ancestral state reconstruction with CAFE 3.0 indicates that two copies of Piwi/Aubergine were present in last common ancestor of insects as well as two copies of Argonaute3. Furthermore the duplications of Piwi/Aubergine in several insect clades (e.g., Diptera and Hemiptera) were suggested to be independent gene expansions. We found multiple copies of Piwi/Aubergine in the transcriptomes of 25 nondipteran species (fig. 1). We found orthologs of Argonaute3 in transcriptome data of 51 species, representing major insect lineages except many polyneopteren groups encompassing Isoptera, Blattodea, Mantodea, Grylloblattodea, Mantophasmatodea, Phasmatodea, and Embioptera. While we found a possible transcript of Argonaute3 in one species of the insect order Grylloblattodea (ice crawlers), Grylloblatta bifratrilecta, the length of the transcript was too short to unambiguously assess orthology. Finally, we found multiple copies of Argonaute3 in Anurida maritima (Collembola).
Systemic RNAi
Phylogenetic analysis of putative insect Sid-1 orthologs indicates that they form a clade distinct from C. elegans Sid-1 and Tag-130. We also identified Tag-130 protein domains in many insect putative Sid-1 orthologs. We identified putative orthologs of Sid-1/Tag-130 in the transcriptomes of 68 species, representing almost all major insect lineages except species belonging to Antliophora (i.e., Diptera, Mecoptera, and Siphonaptera). We found multiple copies of Sid-1/Tag-130 in the transcriptomes of 13 species, in particularly in Collembola, with two present in Sminthurus viridis, three in Folsomia candida, four in Pogonognathellus sp., and three in Anurida maritima. Multiple copies of Sid-1/Tag-130 were also identified in Cordulegaster boltonii (Odonata), Gynaikothrips ficorum (Thysanoptera), Ectopsocus briggsi (Psocodea), Aleochara curtula and Gyrinus marinus (both Coleoptera), and Polyommatus icarus and Parides eurimedes (both Lepidoptera). Ancestral state reconstruction suggests that Sid-1/Tag-130 was present in the last common ancestor of insects.
Dicer Genes in Other Arthropods
For both spider species we found multiple contigs homologous with insect Dicer proteins (see supplementary material). Both Dicer1 and Dicer2 returned many of the same contigs. Therefore we could not conclusively determine if the spider Dicers were orthologs of insect Dicer1, Dicer2, or orthologous with both. In both spiders, the resulting sequences had a higher identity with Dicer1. In the centipede we found two sequences homologous with insect Dicers. Both sequences were returned as BLAST hits for both Dicer1 and Dicer2 queries. Both hits shared a higher identity with Dicer1.
Evidence of Positive Selection
We found no evidence for positive selection in the two candidate genes R2D2 and Loquacious along any of the investigated branches (i.e., branches within Coleoptera and branches within Lepidoptera). However, it is important to note that evidence for positive selection may have been missed due to the small number of nucleotide sequences analyzed.
Discussion
RNAi is an important biological process in insects (and other eukaryotes) and serves a range of biological functions. Manipulation of RNAi systems is a potentially lucrative field of research with numerous applications. Our results show that the genes of the three major insect RNAi pathways identified in Drosophila melanogaster are present in all insect orders. Our analysis indicates that in different insect lineages RNAi-related pathway genes have been duplicated and, in some cases, have potentially been lost more frequently than previously known. Duplications may lead to subfunctionalization or neofunctionalization in RNAi pathways and could explain observed differences in the efficacy of RNAi across different insect groups. Loss of core RNAi-related genes may also explain observed decreases in RNAi efficacy in certain lineages.
Using whole-body transcriptomes of mostly adult insects (supplementary table S3, Supplementary Material online) to detect presence or absence of genes has limitations. As the transcriptome only contains genes expressed at the time of the insect’s death (e.g., frozen with liquid Nitrogen), the respective transcriptome may lack genes only expressed at specific developmental stages. Moreover, gene expression restricted to specific tissues could have caused low transcript abundance in whole-body transcriptomes. We therefore cannot distinguish between a gene which may have been lost and one that was not (or very lowly) expressed. Therefore, we also searched for the eleven genes in several published official gene sets (supplementary table S1, Supplementary Material online).
Our results indicate/imply that the evolution of RNAi pathways in insects is a gradual and complex process. Insects inherited a complete RNAi system from their common ancestor and, over time, diversified and expanded this original system. One striking example of this is the evolution of the dsRBP R2D2 in the winged insects. This provided winged insects with two complementary and parallel RNAi pathways—miRNA and siRNA. We infer numerous expansions of argonaute proteins involved in the piRNA pathway in insects. Duplicate copies of Piwi/Aubergine were found in 28 of 100 transcriptomes. In comparison, we did not identify any duplicates of Argonaute1 (argonaute protein of the miRNA pathway) in a single transcriptome. In flies a similar pattern has been observed in which multiple copies of Piwi/Aubergine are frequently observed while Argonaute1 duplications are not (Lewis et al. 2016). As we used transcriptomes we cannot conclusively state that a gene is lost from a species (the gene in question may not have been expressed at the time the transcriptome was generated). However, we do observe several intriguing patterns which suggest that certain components have indeed been lost in specific lineages. One example is Sid-1/Tag-130 which appears to have been lost in flies and their close relatives (i.e., Antliophora). Another putative loss event is observed in a large clade of hemimetabolous insects, the Dictyoptera (Mantodea, Blattodea, and Isoptera) which appear to have lost Argonaute3. Like Piwi/Aubergine, Argonaute3 is involved in the piRNA pathway and its apparent loss poses a curious counter example to the multiple expansions of this pathway observed in other lineages. Our results underscore the diversity of RNAi systems observed in insects and hint at the complex evolutionary histories which must have brought them into being.
Origin of R2D2
The three core proteins of the anti-viral RNAi pathway are Argonaute2, Dicer2, and R2D2. The siRNAs involved in this pathway originate from exogenous dsRNA (e.g., from viruses). The pathway is, therefore, sometimes termed the exo-siRNA pathway. It is the pathway exploited when RNAi is experimentally induced. R2D2 is a double-stranded RNA binding protein (dsRBP) necessary for loading siRNAs into RISC (Liu et al. 2003, 2006). Orthologs of R2D2 have been identified in several insects including Drosophila (Liu et al. 2003), Tribolium (Tomoyasu et al. 2008), and the crop pest Bemisia tabaci (whitefly) (Uphadhyay et al. 2013). To date, R2D2 has not been identified outside of insects. We identified orthologs of R2D2 in all orders of winged insects (Pterygota). However, we neither found R2D2 in apterygote insect orders (12 transcriptomes in total) nor in outgroup taxa (Ixodes scapularis and Daphnia pulex). Ancestral state reconstruction correspondingly suggests that R2D2 is a derived feature (autapomorphy) of Pterygota. It also suggests that core RNAi proteins duplicated gradually and involved a series of independent gene duplication events rather than a single whole-scale duplication of the RNAi pathway.
While R2D2 is seemingly absent in primary wingless insects, the other core siRNA pathway genes (i.e., Argonaute2 and Dicer2) are present. Additionally, we could not find R2D2 in the draft genomes of Hrabe’s Jumping Bristletail (Machilis hrabei) and Silvestri’s Northern Forcepstail (Catajapyx aquilonaris). These two genomes have been sequenced and are currently analyzed by researchers of the i5K initiative (i5K Consortium 2013). The absence of R2D2 does not necessarily mean that these species lack a functional exo RNAi pathway. It is possible that the corresponding gene from the miRNA pathway (Loquacious) could compensate for R2D2 in these species.
An alternative siRNA pathway (known as the endo-siRNA pathway) involving the proteins Argonaute2, Dicer2, and the dsRBP Loquacious is known from Drosophila (Czech et al. 2008; Okamura et al. 2008). This pathway is likely involved in the down-regulation of transposons in somatic cells (Chung et al. 2008). We identified orthologs of Loquacious in all primary wingless insects but Campodea augens (Diplura). This suggests that primary wingless insects have a complete siRNA pathway. It remains to be investigated whether or not the siRNA pathway in primary wingless insects involves siRNA of exogenous (e.g., viruses) or endogenous (e.g., transposons) origin or both.
Duplication of R2D2 has been previously found in Tribolium (Tomoyasu et al. 2008). We found evidence of multiple R2D2 homologs in other beetle transcriptomes; however, we were unable to determine if duplication of R2D2 occurred once in beetles or multiple times independently. We tested R2D2 orthologs in five beetle species to infer evidence of positive selection acting on these genes. While R2D2 is one of the most rapidly evolving genes in Drosophila (Obbard et al. 2006), we did not find any evidence for positive selection in beetles.
R2D2 in Lepidoptera
An R2D2 ortholog has been identified in the silk moth (Bombyx mori). However, it is expressed at very low rates (Swevers et al. 2011). We could not identify R2D2 in the transcriptomes of four investigated species of Lepidoptera, suggesting that in these species R2D2 is either expressed at a very low level or is entirely absent. All four investigated species of Lepidoptera belong to the large group of Ditrysia, which includes the vast majority of Lepidoptera, including B. mori. The four species belong to four families within Ditrysia (Yponomeutidae, Zygaenidae, Lycaenidae, and Papilionidae). While the number of families investigated is small, they represent the broader diversity of Ditrysia. The consistent pattern observed and the congruency with published results (Swevers et al. 2011) suggests that R2D2 may be expressed at a low level or is entirely absent in all members of Ditrysia.
The low level of expression of the R2D2 gene observed in B. mori has been suggested as a response to the domestication of this species and subsequent decrease in frequency of viral infection (Swevers et al. 2011). Our results, however, suggest that the R2D2 protein is not (or is generally very lowly) expressed in members of Ditrysia (and, thus, the majority of the Lepidoptera). This implies that loss or low expression of R2D2 significantly predates the domestication of B. mori. The possibility that R2D2 is expressed at low concentrations in Ditrysia may partially explain the variable success observed in experimentally inducing RNAi in Lepidoptera under laboratory conditions (Terenius et al. 2011). It may also have implications for developing RNAi-based crop protections against pest species within Lepidoptera.
Piwi/Aubergine in Diptera
In insects, the piRNA pathway acts as a defense against transposons in the germ line. Unlike in other RNA silencing pathways (miRNA and endo- and exo-siRNA), Dicer proteins are not involved. Additionally, the piRNA pathway uses Argonaute proteins of the Piwi family rather than those of the Ago family (i.e., Argonaute1 and Argonaute2). In the model species D. melanogaster, three Piwi proteins (Piwi, Aubergine, and Argonaute3) take part in the piRNA pathway. Argonaute3 and Aubergine operate in a loop (termed the ping–pong amplification loop) which alternately are cleaving sense and anti-sense transcripts. Piwi binds to the resulting piRNAs generated by the loop (Aravin et al. 2007; Siomi et al. 2011). In Tribolium castaneum, only two Piwi proteins are present: an ortholog of Argonaute3 and one corresponding to Aubergine/Piwi (Tomoyasu et al. 2008). The mosquitoes Aedes aegypti and Culex pipiens have large expansions of Piwi proteins with seven and six copies of the Aubergine gene, respectively (Campbell et al. 2008). In mosquitoes expansion of Piwi genes has been suggested to be a response to increased transposon content in the genome (Campbell et al. 2008).
The split between Aubergine and Piwi occurred 182–156 million years ago in a common ancestor of brachyceran flies (Lewis et al. 2016). In Brachycera, Piwi plays a role in heterochromatin formation (Chambeyron and Seitz 2014). Our results are consistent with the evidence that the Piwi/Aubergine split occurred in the most recent common ancestor Brachycera. We also investigated the transcript assembly of a representative of Bibionomorpha, which are considered to be the closest relatives of the Brachycera, Bibio marci, but did not find any orthologs of Piwi and Aubergine in this species. The BUSCO value of B. marci was only 0.45 (all species mean = 0.6, all species median = 0.61) which suggests a relatively incomplete transcriptome. Because we could not identify orthologs of several other target genes in this species either, this possibly indicates that the transcriptome may have been of inferior quality. Thus, our data are inconclusive in respect of whether the split between Piwi and Aubergine occurred in the last common ancestor of Brachycera or whether it occurred earlier in the dipteran phylogeny.
In Diptera numerous independent duplications of Argonaute3 and Piwi/Aubergine have also been identified (Lewis et al. 2016). These duplications have been suggested as a response to genomic parasites (e.g., transposons) (Lewis et al. 2016). Our results suggest that the Piwi/Aubergine gene has also been duplicated numerous times independently in other insect groups such as Hemiptera, Thysanoptera, and Hymenoptera. Whether this is a response to a high frequency of transposons in the genomes of the analyzed species or whether the duplication has led to new functionality remains to be investigated. The genomes currently sequenced and analyzed in context of the i5K initiative (i5K Consortium 2013) will provide the basis for such investigations.
Loss of Sid-1/Tag-130 in Antliophora
Sid-1 is a transmembrane protein associated with the systemic spread of the RNAi response in the nematode C. elegans (Winston et al. 2002). Drosophila species lack both orthologs of the gene Sid-1 and a systemic RNAi response. In other insects, such as Tribolium, Sid-1 like genes have been identified (Tomoyasu et al. 2008). The particular role of the Sid-1 protein in insects, however, remains uncertain. Our analysis could not distinguish if the insect Sid-1 like genes are orthologous with either C. elegans Sid-1 or Tag-130. We identified orthologs of Sid-1/Tag-130 in species of most insect orders, but were unable to detect transcripts of Sid-1/Tag-130 in the analyzed transcriptomes of dipteran species. This corroborates the idea that this gene is absent in flies and relatives. Intriguingly, we did not find orthologs of Sid-1/Tag-130 in other members of Antliophora (i.e., Mecoptera—scorpion flies in a broader sense—and Siphonaptera—fleas), either. This suggests that Sid-1/Tag-130 was already lost in the last common ancestor of this species rich endopterygote insect lineage.
Conclusion
Using transcriptomic data of 100 insect species, we have gained new insights into the evolution of RNAi pathways in this highly diverse animal group. We show that RNAi related pathway genes are found in all insect orders. Our results suggest several novel gene expansions and indicate the distribution of core RNAi pathway genes in numerous nonmodel organisms. Additionally, we have identified certain key evolutionary events including the origin of R2D2 in pterygote insects and the loss of Sid-1 in Diptera.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This manuscript has been enabled by the 1KITE consortium and the i5K initiative. The sequencing and assembly of the 1KITE transcriptomes were funded by the BGI through support of the China National GenBank. Alexander Donath, Bernhard Misof, Oliver Niehuis, Ralph S. Peters, and Lars Podsiadlowski furthermore acknowledge the Leibniz association for installing the graduate school Genomic Biodiversity Research, in which the present study arose. Karen Meusemann acknowledges the Schlinger Foundation for funding. We especially thank Stephen Richards and Richard Gibbs of the Baylor College of Medicine Human Genome Sequencing Center for granting access to i5K pilot data prior to their official publication.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. 2007. The piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764. [DOI] [PubMed] [Google Scholar]
- Bellés X. 2010. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 55:111–128. [DOI] [PubMed] [Google Scholar]
- Baum JA, et al. 2007. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 25:1322–1326. [DOI] [PubMed] [Google Scholar]
- Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. 2006. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2:711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Black WC, Hess AM, Foy BD. 2008. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics 18:425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Seitz H. 2014. Insect small non-coding RNA involved in epigenetic regulations. Curr Opin Insect Sci. 1:1–9. [DOI] [PubMed] [Google Scholar]
- Chipman AE, et al. 2014. The first myriapod genome sequenced reveals conservative gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12(11):e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Okamura K, Martin R, Lai EC. 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 18:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci. 41(4):324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet. 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):1–37. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 15:509–524. [DOI] [PubMed] [Google Scholar]
- Han M, Thomas GW, Lugo-Martinez J, Hahn MW. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30(8):1987–1997. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simmard MJ. 2008. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 9:22–32. [DOI] [PubMed] [Google Scholar]
- Huvenne H, Smagghe G. 2010. Mechanism of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol. 56:227–235. [DOI] [PubMed] [Google Scholar]
- i5K Consortium. 2013. The i5K initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered. 104:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert-Possamai S, et al. 2010. Expansion of the miRNA pathway in the hemipteran insect Acrythosiphon pisum. Mol Biol Evol. 27:979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF. 2011. The many faces of RNAi. Dev Cell 20(2):148–161. [DOI] [PubMed] [Google Scholar]
- Kriventseva EV, et al. 2015. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 43:D250–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Salmela H, Obbard DJ. 2016. Duplication and diversification of Dipteran Argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol Evol. 8(3):507–518.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, et al. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921–1925. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. 2006. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly siRISC complexes. RNA 12:1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. 2008. Mesquite: a modular system for evolutionary analysis. Version 3.02. http://mesquiteproject.org [Google Scholar]
- Mao Y, et al. 2007. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 25:1307–1313. [DOI] [PubMed] [Google Scholar]
- Meister G. 2013. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 14(7):447–459. [DOI] [PubMed] [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Campos H, Kolaczkowski B. 2013. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol. 30:627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen B. 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-supression of homologous genes in trans. Plant Cell 2:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. 2009. The evolution of RNAi as a defence against viruses and transposable elements. Phil Trans R Soc B. 364:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ. 2006. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 16:580–585. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. 2008. Two distinct mechanisms generate endogenous siRNA from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 15:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DRG, Gatehouse JA. 2008. RNAi mediated crop protection against insects. Trends Biotechnol. 26:393–400. [DOI] [PubMed] [Google Scholar]
- Sanggaard KW, et al. 2014. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 5:3765.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JG, et al. 2013. Towards the elements of successful insects RNAi. J Insect Physiol. 59:1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Blau HM. 2006. A brief history of RNAi: the silence of the genes. FASEB J. 20:1293–1299. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 23:578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi M. 2009. On the road to reading the RNAi code. Nature 457:396–404. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. Piwi-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 12:246–258. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32:W309–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into corresponding codon alignments. Nucleic Acids Res. 34:w609–w612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius O, et al. 2011. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 57:231–245. [DOI] [PubMed] [Google Scholar]
- Swevers L, Liu J, Huvenne H, Smagghe G. 2011. Search for limiting factors in the RNAi pathway in silkmoth tissues and the Bm5 cell line: the RNA-binding proteins R2D2 and Translin. PLoS One 6:e20250.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y, et al. 2008. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 9:R10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphadhyay SK, et al. 2013. siRNA machinery in whitefly (Bemisia tabaci). PLoS One 8:e83692.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456–2459. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.