Abstract
Varieties of the economically important ornamental species chrysanthemum have been bred to fit a number of market niches, but the genetic basis and evolutionary relationships among various cultivated types are poorly understood. Here, a DNA marker-based analysis of 199 chrysanthemum entries representing each of the five cultivated types is presented. A set of >90,000 single nucleotide polymorphisms (SNPs) associated with a minor allele frequency of at least 5% was defined, and used to perform a phylogenetic analysis which corresponded well with the phenotypic classification. The analysis revealed that the small-flowered types, spray cut chrysanthemum (SCC) and potted and ground chrysanthemum (PGC), are more closely related to the wild progenitor species (WC) than are the large-flowered ones, disbud cut chrysanthemum (DCC) and traditional chrysanthemum (TC); and the PGC type was closest. Some 550 genetic regions appeared to have experienced selection in the separation of potted and ground-cover types from disbud cut types, and that between potted and ground-cover types from traditional types. A genome-wide association analysis revealed that seven SNPs lying within six genes were predictive of three important traits (ray floret type, cultivated type and flower shape), but no association with flower color was detected. The study has provided a number of novel insights into evolutionary relationships, the population structure and the genetic basis of some key ornamental traits.
Keywords: chrysanthemum, evolutionary relationship, genetic differentiation, single nucleotide polymorphisms, association analysis
Introduction
Chrysanthemum (Chrysanthemum morifolium Ramat.) is one of the most popular ornamental species. Its commercial production is concentrated in East Asia and to a lesser extent in western Europe (Zhang et al. 2011). During the past 1,600 years of breeding activity, chrysanthemums have developed some major cultivated types, i.e. traditional (TC) types, spray cut (SCC) types, disbud cut (DCC) types, potted and ground-cover (PGC) types, wild chrysanthemums (WC), diverse in plant architecture and inflorescence traits (Li and Shao 1990; Zhang, Dai, et al. 2014; Li et al. 2016). Specifically, the DCC type bears a single, large flower per stem, typically produced by removing side bud prior to flowering; and the SCC type bear several small flowers per stem, which were often more densely packed than those borne by the DCC type. The TC type is somewhat like DCC in strait stem and large flowers, but more diversified in flower shapes that feature Chinese traditional culture. The flower of the PGC type is as small as that of the SCC type, whereas the former type often grows into ball-like plant architecture. In addition, the WC type is loose in plant architecture, simply with small yellow or white single flowers. The wild ancestors of the modern chrysanthemum are believed to be one or more of C. indicum, C. vestitum, C. nankingense and C. lavandulifolium (Chen et al. 1996; Dai et al. 1998; Liu et al. 2012). As a result of its out-breeding habit and self-incompatibility (Drewlow et al. 1973), the cultivated chrysanthemum is highly heterozygous, with a complex genetic background. European chrysanthemum cultivars (cut, pot and garden types) were hardly grouped either by their common origin, or by similarities in phenotypes (Klie et al. 2013). In a panel of 480 Chinese traditional chrysanthemum cultivars, however, the UPGMA (unweighted pair-group method with arithmetic means) clustering, based on genomic SSR-derived Nei’s genetic distances mostly was consistent with the horticultural classification (Zhang, Dai, et al. 2014). A latest research in cut chrysanthemum failed to reflect known variation in either provenance or inflorescence type (Li et al. 2016). Despite an enormous amount of research effect into genetic characterization of modern chrysanthemum, the phylogenetic relationships harbored in various cultivated types and the wild progenitors remain obscure as yet. Part of the reason behind this lack of clarity is that the marker data upon which prior analyses have been based have been limited in scope. With the development of high-throughput DNA sequencing technology, this limitation no longer exists.
As a species, chrysanthemum displays a wealth of morphological variation, but the genetic basis of this variability has been at best only sporadically characterized. Past attempts to reveal the genetics of economically important traits have relied on biparental linkage mapping, which has successfully uncovered a number of quantitative trait loci governing plant architecture, inflorescence type and other floral characteristics (Zhang et al. 2011, 2012, 2013). Constructing the necessary mapping populations is, however, a time-consuming process, and the variation which is released is limited to the set of allelic differences between the population’s two parents. In contrast, the genotype/phenotype association concept, as exemplified by the genome-wide association study (GWAS) approach, provides an effective means of linking genotype to phenotype from a pre-established set of germplasm. A feature of the GWAS approach is that many more than two alleles will be segregating in the germplasm, although the true effect of those which are present at a low frequency will be difficult to detect. This approach has been used to dissecting genetic basis of quantitatively inherited traits in a number of plant species (Huang et al. 2010; Kump et al. 2011; Morris et al. 2013; Zhang, Song, et al. 2014). The use of GWAS to date in chrysanthemum has been restricted to the genetic analysis of a few horticultural traits (Klie et al. 2016; Li et al. 2016).
The present study describes a comprehensive analysis of the genome-wide sequence variation present in a set of 199 diverse chrysanthemums, including representatives of all five common cultivated types. The genotypic component, relevant to phylogenetic relationship and population differentiation, was provided by the acquisition of a large number of single nucleotide polymorphism (SNP) loci. The GWAS analysis focused on four important traits (ray floret type, flower color, flower shape, and cultivated type), with a view to revealing their genetic basis, knowledge of which should enhance breeding efficiency.
Materials and Methods
The Germplasm Panel
The set of 199 chrysanthemum entries was obtained from the Nanjing Agricultural University Chrysanthemum Germplasm Resource Preserving Centre. It included representatives of each of the five major cultivated types: 81 entries were spray cut (SCC) types, 28 were disbud cut (DCC) types, 50 were potted and ground-cover (PGC) types, 30 were traditional (TC) types and 10 were wild relatives (WC) (supplementary table S1, Supplementary Material online and fig. 1). The provenance of the collection included China, Japan, South Korea and western Europe. The full collection was planted in late May in two consecutive years (2011 and 2012), with each entry (of 40 plants, planted in four rows) replicated 3 times. The inter-row separation was 15 cm, and neighboring plants within a row were planted 10 cm apart from one another. The whole experiment was set out in a greenhouse as a randomized complete block. The conditions in the greenhouse were controlled to give a 12-h photoperiod provided by light of flux density ∼160 µmol m −2 s −1. The temperature during the light and dark periods was, respectively, 25 °C and 18 °C. Each entry was scored for flower color, flower shape, ray floret type and cultivated type.
Fig. 1.—
The morphology produced by each of the five chrysanthemum cultivated types. (A) DCC, “Shunfa”; (B) SCC, “Nannong Feizi”; (C) TC, “D1045”; (D) PGC, “Jinling Hongxia” and (E) WC, Chrysanthemum dichrum.
DNA Extraction and Specific Locus Amplified Fragment (SLAF) Sequencing
Genomic DNA was extracted from young leaves of each entry using a slightly modified CTAB-based procedure (Murray and Thompson 1980). DNA quality and concentration were assessed using an OD-1000 spectrophotometer (OneDrop Technologies, Inc) and by electrophoretic separation through a 1% agarose gel. About 500-ng genomic DNA from each entry was processed using the SLAF-seq technique (Sun et al. 2013), with minor modifications. Briefly, a pilot SLAF experiment was performed to establish the conditions and appropriate restriction enzymes that optimize SLAF yield and maximize SLAF-seq efficiency. Then, the SLAF library was constructed based on the result of the pilot experiment as following. Genomic DNA from each sample was incubated at 37 °C with EcoRV (New England Biolabs (NEB), Ipswich, MA), T4 DNA ligase (NEB), ATP (NEB), and EcoRV adapter. The reaction was inactivated at 65 °C, and then digested with another restriction enzyme ScaI at 37 °C. PCR was performed using the diluted restriction-ligation samples, dNTP, Taq DNA polymerase and EcoRV primer containing barcode 1. PCR products were purified using an E.Z.N.A. Cycle Pure Kit (Omega Bio-Tek Inc, Norcross, GA) and pooled. After treating the pooled sample with EcoRV, T4 DNA ligase, ATP and Solexa adapter (Illumina, Inc., San Diego, CA) at 37 °C, the sample was purified using a Quick Spin column (Qiagen, Hilden, Germany), then electrophoresed through a 2% agarose gel. Fragments in the size range 264–294 bp were excised using a Gel Extraction kit (Qiagen). The purified fragments were once more subjected to a PCR based on Phusion Master Mix (NEB) and the Solexa amplification primer mix (Illumina). The PCR settings were those recommended by the supplier. Finally the amplicon was gel-purified and the 264–294 bp range was extracted, purified and submitted for pair-end sequencing with a HiSeq 2500 device (Illumina, Inc., San Diego, CA) at Biomarker Technologies Corporation in Beijing.
SNP Calling and Data Analysis
SLAF-seq raw reads were separated by barcodes, and reads with quality scores <30 were discarded. The remaining SLAF pair-end reads with clear index information were clustered on the basis of their sequence similarity, detected via a one-to-one alignment by BLAT (Kent 2002). Sequences sharing >98% identity were grouped to form one SLAF locus. Alleles were determined in each SLAF by the MAF evaluation. SLAF loci harboring from two to four different tags were classified as polymorphic. Due to the lack of a chrysanthemum reference sequence, the highest depth tag in each SLAF was taken as reference sequence tag. SNPs were called using the BWA software package (Li and Durbin 2009). The GATK (www.broadinstitute.org/gatk/; last accessed November 15, 2016) Realigner Target Creator and InDel-Realigner (McKenna et al. 2010) were used to realign indels and Unified Genotyper was used to call genotypes across the 199 entries using the default parameters. All data filter processing was performed following the “best practices” workflow described by McKenna et al. (2010). Variable sites were detected using both GATK and Samtools (www.htslib.org; last accessed November 15, 2016) routines. SNPs associated with a minor allele frequency (MAF) across the germplasm set of <5% were discarded. If the depth of the minor allele was larger than one-third of total sample depth, the locus was considered to be in the heterozygous state.
Phylogenetic Analysis and the Evaluation of Population Structure
SNP genotype was used to calculate the genetic distances between the 199 entries, using the Jin and Nei (1990) p-distance method, and this was followed by a phylogenetic analysis based on the neighbor-joining method, as implemented in the MEGA5 package (Tamura et al. 2011). A bootstrap consensus tree was formed from a sample of 1,000 replicates. Population structure was revealed by applying the ADMIXTURE program (Alexander et al. 2009), in which the K value was constrained to within the range 2–10. Then the value of cross-validation (CV) error of population for each K value was calculated, and the K value with a minimum value of CV error was most suitable. Entries assigned to membership of a group at a probability <0.80 were considered to belong to an admixed group (AD). Relative pairwise kinships based on the SNP genotype data were calculated using SPAGeDi software (Hardy and Vekemans 2002), and a principal component analysis (PCA) was performed using Cluster software (de Hoon et al. 2004).
The Estimation of Genetic Parameters
The PopGen package (https://cran.r-project.org/src/contrib/Archive/popgen/; last accessed November 15, 2016) was employed to calculate the nucleotide diversity (π) and the Waterson nucleotide polymorphism (θ) genetic diversity parameters (Tajima 1983), along with FST (Wright 1949). The latter parameter was derived using a 100-kb sliding window with 10–kb steps. Where the mean FST fell within the upper 5% of the empirical distribution of FST, the sliding window in question was defined as an FST outlier.
Signatures of selection were revealed by comparing the PGC with the DCC types and also the PGC with the TC types, using a comparison of FST values, as suggested by Lam et al. (2010): genomic regions in which FST reached a 5% significance level were taken as being associated with the determination of cultivated type. A similar criterion was applied for π values. The annotation of genes present in these differentiated genomic regions was obtained from the variety “Jinba” transcriptome. The candidate genes were finally functionally assigned via the gene ontology method (http://geneontology.org/page/go-enrichment-analysis; last accessed November 15, 2016).
GWAS
The validated set of SNPs was also used to perform GWAS. The parameters applied were a MAF threshold of 0.05 and the integrity of each SNP of 50%. The analysis assumed the mixed linear model (Yu et al. 2006) as implemented in TASSEL 4.0 software (Bradbury et al. 2007). The Q and kinship matrices were included as fixed and random effects, respectively. The P value (adjusted by the Bonferroni correction) of either 0.01 or 0.1 (corresponding to a probability of, respectively, 1.07 × 10 −7 and 1.07 × 10 −6) was applied as a threshold to determine whether a significant association existed. An attempt was made to identify potential candidate genes underlying these associations by using the BlastX algorithm to align the identified variable SLAF sequences with the chrysanthemum cv. “Jinba” transcriptome.
Results
SLAF Sequencing of the Association Panel
The SLAF sequencing generated a mean of 2,591,719 paired-end reads per entry, with a mean quality score of 83.7%. The mean number of SLAFs per entry was 107,597, equivalent to a sequencing depth of 17.9× (supplementary table S2, Supplementary Material online). In all, 15,8510 of the SLAFs displayed polymorphism across the 199 entries. From the total number of 480,592 SNPs, 92,830 met the thresholds imposed for MAF and SNP integrity (supplementary table S3, Supplementary Material online). Across the set of entries, heterozygosity accounted for 20% of the polymorphisms. The analysis of SLAFs and SNPs according to cultivated type is reported in table 1. The number of SLAFs recovered from the DCC types (111,789) was somewhat higher than from any of the other four cultivated types. SNP frequency (MAF > 0.05) was similar in the SCC, DCC, PGC and TC types, but was much lower in the WC type, probably reflecting a much smaller representation of entries present (10 as opposed to between 28 and 81).
Table 1.
SLAF and SNP Statistics in the Five Chrysanthemum Types
| Accession Types | SCC | DCC | PGC | TC | WC |
|---|---|---|---|---|---|
| Reads suma | 214,423,576 | 75,340,402 | 133,976,021 | 76,260,867 | 15,751,198 |
| SNP number (before filter) | 176,808 | 174,911 | 179,487 | 172,877 | 125,700 |
| SNP number (filter by MAF >0.05) | 70,380 | 70,182 | 70,399 | 69,886 | 49,650 |
| SNP heterozygosity (filter by MAF >0.05) | 20.60% | 20.65% | 20.79% | 19.82% | 10.08% |
| SLAF number | 109,116 | 111,789 | 110,129 | 107,978 | 69,749 |
| SLAF depth | 18.16 | 18.12 | 18.11 | 17.70 | 15.11 |
Reads sum is the sum of the samples reads in each group, other indicators are average values within each group.
SCC, spray cut chrysanthemums; DCC, disbud cut chrysanthemums; PGC, potted and ground-cover chrysanthemums; TC, traditional chrysanthemums; WC, wild chrysanthemums species.
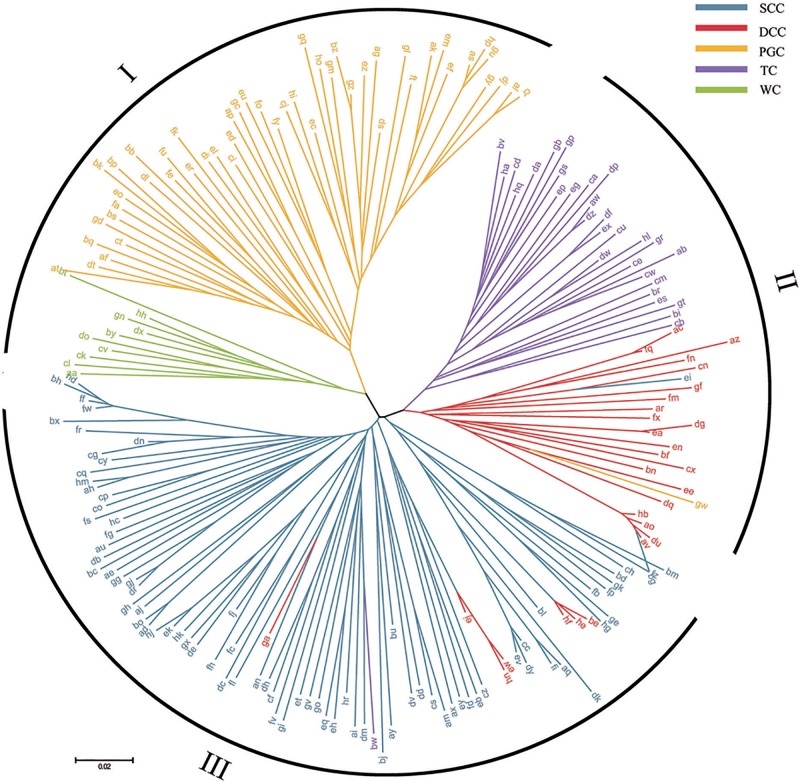
Phylogeny and Population Structure of the Germplasm Panel
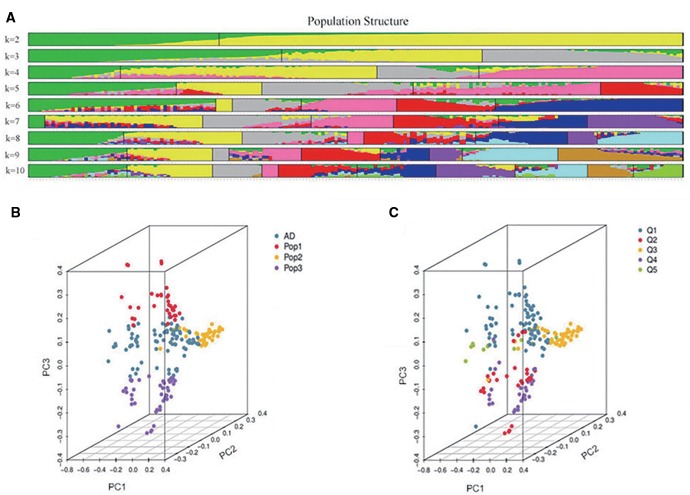
The phylogenetic relationships between the 199 entries, based on the set of 92,830 SNPs, are illustrated in figure 2. The separation between the five cultivated types is mirrored in the genotype-based clustering, although the number of major clades recognized was three rather than five; this was because the WC and PGC types clustered with one another into clade I, the DCC and TC types formed clade II (both these types form a single, large inflorescence per stem), leaving the SCC types (small-flowered plants) on their own in clade III. The output of the ADMIXTURE program, based on minimizing the value of CV error associated with K (supplementary fig. S1, Supplementary Material online), suggested that the germplasm panel formed three major sub-populations (fig. 3A). There was a distinct relationship between cultivated type and sub-population: all 34 entries in sub-population Q1 were SCC types; sub-population Q2 (41 entries) included 41 of the 50 PGC types; and sub-population Q3 included 17 of the 28 DCC, 24 of the 30 TC types, and each one PGC and SCC entry. The remaining 81 entries (41% of the total) were assigned to the AD group. The predicted population structure is relatively consistent with that predicted by the phylogenetic analysis based on genetic distances (Q1 = clade III, Q3 = clade II), but unlike in clade I, none of the WC types grouped with the PGC types in Q2. The corresponding Q matrix (K = 3) was used for the subsequent genomewide association. The SNP genotypes were used to derive the set of pairwise kinship values, which averaged 0.013. Almost 68% of the pairwise kinship values were zero and nearly a quarter lay in the range 0–0.05 (supplementary fig. S2, Supplementary Material online). The 9% of estimates of >0.05 represented the minimum relative kinship among samples and could not cause further complexity in subsequent genome-wide association analysis. The PCA suggested a four cluster structure (fig. 3B), with the AD group being in the middle of these three sub-populations, defined by the two axes PC2 and PC3 (supplementary fig. S3A, Supplementary Material online). There was a good level of discrimination between the five cultivated types (fig. 3C), although there was some intermingling of the DCC and SCC types and the other TC group (supplementary fig. S3B, Supplementary Material online).
Fig. 2.—
A phylogenetic analysis of the 199 entry germplasm panel, based on 92,830 SNPs. SCC types shown in blue, DCC in red, PGC in yellow, TC in purple and WC in green.
Fig. 3.—
The population structure of the 199 entry germplasm panel. (A) Population structure as generated by ADMIXTURE software. Each color represents one sub-population. Individual entries are represented by vertical bars; the length of each colored segment in a given vertical bar represents inferred membership to the given number of sub-populations (K). (B and C) PCA scatter plots, where each dot represents an entry. (B) Sub-populations as defined by ADMIXTURE analysis. Pop1: sub-population 1, Pop2: sub-population 2, Pop3: sub-population 3, AD: admixed sub-population, and (C) entries grouped according to cultivated type. Q1: SCC, Q2: DCC, Q3: PGC, Q4: TC, Q5: WC.
Genome-Wide Divergence among Five Cultivated Types
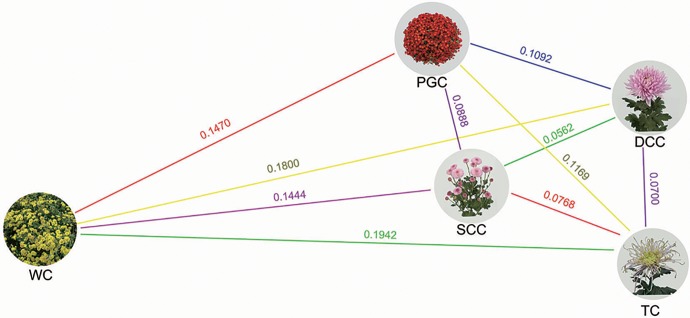
Several population genetics parameters were computed to quantify the genetic diversity within and the extent of differentiation between the five cultivated types. The global π value was 0.0055 and the global θ value was 0.0045. Within cultivated type, diversity varied only marginally, being lowest among the WC types and highest among the SCC types (supplementary table S4, Supplementary Material online); this variation may be in part influenced by group size (10 WC entries, 81 SCC entries). The size of the FST among five cultivated types varied from 0.0562 to 0.1942. The FST values computed from the comparison between the WC type with the PGC and SCC types were estimated to be 0.147 and 0.144, respectively. While markedly higher FST values were obtained from the WC type with the TC (0.194) and DCC (0.180) types (fig. 4). On this basis, the PGC and SCC types were taken as being genetically closer to the WC type than were either the DCC or the TC types. Applying the p-distance method, the prediction was that the WC type was closer to the PGC type than to any of the other four types (table 2). The overall conclusion was that of the four cultivated types, the PGC type was the closest and the TC type the most distant from the WC type.
Fig. 4.—
Pairwise FST values between the five cultivated types, based on genetic distances.
Table 2.
Genetic Distances Separating the Five Chrysanthemum Types
| Types | WC | TC | DCC | SCC |
|---|---|---|---|---|
| TC | 0.1112 | |||
| DCC | 0.1093 | 0.0918 | ||
| SCC | 0.1080 | 0.1018 | 0.0976 | |
| PGC | 0.0997 | 0.1042 | 0.1041 | 0.1030 |
Genomic Regions Associated with Artificial Selection among Different Cultivated Types
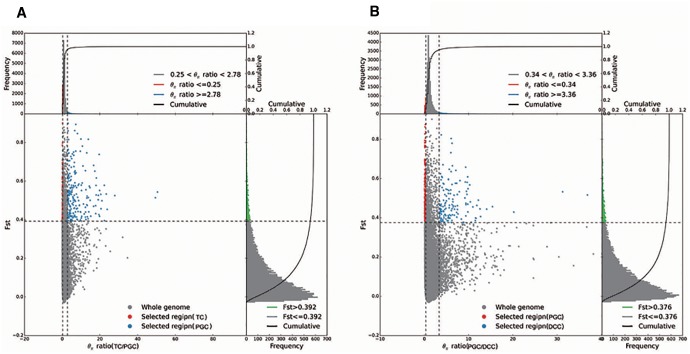
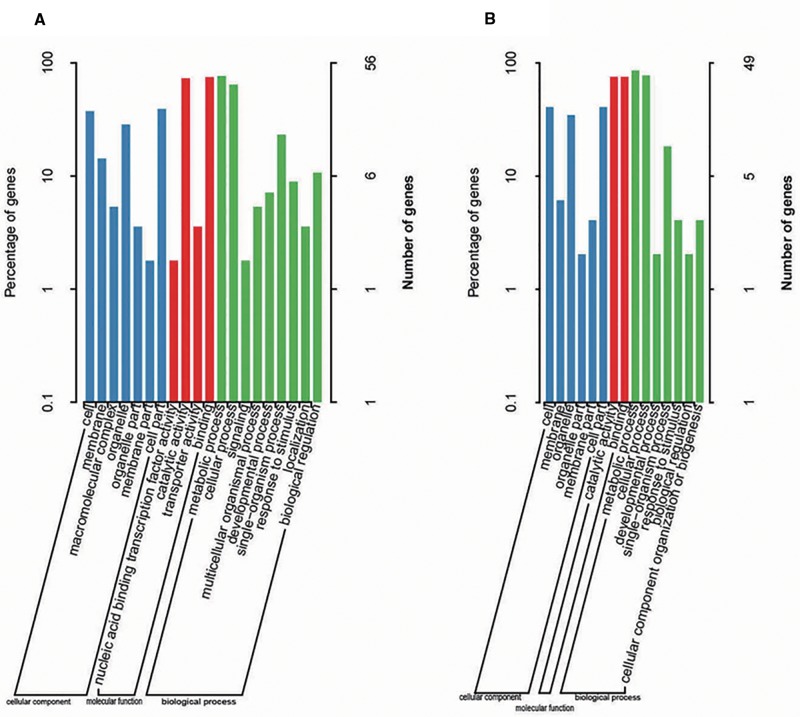
The two comparisons PGC versus TC and PGC versus DCC were used to identify differentiated genomic regions potentially associated with artificial selection. In the former, 277 candidate regions emerged at a FST threshold of 0.390 (fig. 5A), and the equivalent in the latter contrast was 294 regions (FST > 0.376) (fig. 5B). Gene ontology classifications were possible for 56 genes in the former contrast and 49 in the latter. For this gene set, within the category “biological process”, the two most common functions were “cellular process” and “metabolic process”, and the genes were active predominantly in the “cell”, “organelle” or “cell part”. In terms of molecular function, most of the genes were involved in either “catalytic activity” or “binding” proteins (fig. 6A and B).
Fig. 5.—
The empirical distribution of π and FST and selected regions of chrysanthemum genome. (A and B) The x and y axes represent, respectively, the empirical distributions of (πTC/πPGC and πPGC/πDCC and the FST value. Red and blue points lying above the horizontal dashed line (the 5% right and left tails of the empirical π ratio distribution) and green points lying above the dashed line (the 5% right tail of the empirical FST distribution) indicate regions experiencing selection during the diversification of cultivated type (PGC vs. TC, PGC vs. DCC).
Fig. 6.—
Candidate gene analysis. Annotated genes mapping within the regions associated with the difference between (A) the PGC and TC, and (B) the PGC and DCC types, sorted according to gene ontology. From left to right, “cellular components” (shown in blue), “molecular function” (red), “biological process” (green).
Genome-Wide Association Analyses and Candidate Gene Identification
When GWAS was used to identify SNPs associated with the four traits, i.e. flower shape, cultivated type, ray floret type and flower color, 97 associated SNPs were identified based on a P threshold of 1.07 × 10 −7 (supplementary table S5, Supplementary Material online). The number rose to 188 when the threshold was lowered to 1.07 × 10 −6. Of the former set, 48 SNPs were associated with flower shape, 44 with cultivated type, five with ray floret type and none with flower color.
When the SLAF sequences harboring one or more of the above 188 SNPs were aligned with cv. “Jinba” transcriptome, six (harboring seven of the SNPs) were found to reside within an annotated gene (table 3). The SNP associated with both flower shape and cultivated type lay within a gene encoding an F-box protein, predicted to be involved in protein binding and ubiquitin-protein transferase activity. Proteins of this type are known to contribute to photomorphogenesis (Yang et al. 2005), the defense response (Gou et al. 2009), the hormone response (Xie et al. 2002) and the regulation of cell division (Menges et al. 2002). The other three SNPs associated with flower shape lay within genes encoding TBP-associated factor 12, a bHLH48 transcription factor and a Sin3-like 2 protein. TBP-associated factor 12 is a component of RNA polymerase II machinery. Although TBP-associated factors have been extensively characterized in yeast, fruit fly and human, little is known of their function in plants. Lago et al. (2004) have suggested that they are ubiquitously expressed, which would be consistent with their having a role in basal cellular mechanisms. The bHLH48 transcription factor belongs to a large family of plant transcription factors which contribute to many processes in cell and tissue development as well as in metabolism (Heim et al. 2003). The Sin3-like 2 protein belong to the SNL protein family, which is known to be important for both development and growth (Wang et al. 2013). The two SNPs present in the sequence associated with ray floret type lay within a homolog of the Arabidopsis thaliana gene AtEML3, the product of which is involved in determining the timing the swtich from vegetative to reproductive growth (Tsuchiya and Eulgem 2011). The final sequence marked by a SNP associated with cultivated type lay within a homolog of the A. thaliana gene AtSZF2, the product of which is involved adjusting metabolism and development to a changing environment.
Table 3.
SNPs within SLAF Sequences Associated with Phenotype, and the Candidate Gene Affected
| Trait | SLAF tag | SNP position (bp) | P value | Candidate gene | Functional annotation |
|---|---|---|---|---|---|
| Ray floret type | Marker 7769 | 163 and 186 | 3.53E-11 | Unigene7275_All | Protein EMSY-LIKE 3; AtEML3 |
| Flower shape | Marker 32989 | 71 | 1.16E-10 | Unigene51632_All | F-box protein CPR30; Protein CONSTITUTIVE EXPRESSER OF PR GENES30 |
| Flower shape | Marker 51400 | 130 | 3.99E-08 | CL12590.Contig1_All | Transcription initiation factor TFIID subunit 12; TBP-associated factor 12 |
| Flower shape | Marker 27312 | 118 | 9.68E-12 | CL13983.Contig4_All | Paired amphipathic helix protein Sin3-like 2 |
| Flower shape | Marker 12776 | 73 | 1.59E-08 | Unigene19865_All | Transcription factor bHLH48; AtbHLH48 |
| Cultivated type | Marker 13753 | 34 | 1.33E-10 | CL5208.Contig1_All | Zinc finger CCCH domain-containing protein 29; AtSZF2 |
| Cultivated type | Marker 32989 | 71 | 8.76E-07 | Unigene51632_All | F-box protein CPR30; Protein CONSTITUTIVE EXPRESSER OF PR GENES30 |
Discussion
The advent of high-throughput DNA sequencing technologies has invigorated both gene discovery and genome exploration (Renaut et al. 2013; Moura et al. 2014). Sequence data has been used to reveal the mechanics of crop domestication (He et al. 2011; Han et al. 2016) and is increasingly being exploited as a resource for developing functional markers (Pujolar et al. 2014; Hess et al. 2015). Here, the SLAF-seq technique has been applied to study phenotypic diversification in chrysanthemum and, in conjunction with GWAS, to identify candidate genes underlying key horticultural traits. An advantageous feature of the SLAF-seq technique is that it is applicable to species which lack a reference genome sequence (Sun et al. 2013).
The sequence data acquired from the germplasm panel yielded over 92,000 high-quality SNPs, ∼20% of which reflected a locus in the heterozygous state, underlining the extensive heterozygosity present in chrysanthemum. The overall level of sequence diversity in the panel, as measured by π, was 0.0055, a level somewhat lower than that estimated for A. thaliana (0.0071) (Schmid et al. 2005), but also substantially higher than those in tomato (0.0016), soybean (0.0019) or rice (0.0024) (Huang et al. 2012). π varied only to a minor degree among the five cultivated types of chrysanthemum (0.0052–0.0056). It is more plausible to suggest that π is low in tomato, soybean and rice because all three species experienced a major genetic bottleneck during their domestication, which was probably not the case for chrysanthemum. The identification of a large number of SNPs allowed for a robust phylogenetic analysis to be conducted, and most of the branches forming the phylogenetic tree were associated with a bootstrap support level of >50%. The analysis revealed that the five phenotypically based cultivated types reflect major differences in DNA sequence (fig. 2). A previous study, based on a different type (and rather limited number) of marker along with a smaller sample of germplasm, came to the conclusion that inflorescence size, rather than petal type or color, was the major driver of the clustering (Li et al. 2013). There were a small number of discrepancies between horticultural classification and SNP-based phylogenetic cluster: notably, seven of the DCC types shared the same clade as the large group of SCC types. These exceptions probably reflect examples of gene flow induced by deliberate crossing between different cultivated types. The clustering of the WC with the PGC types implies that the latter is the cultivated type most closely related to the wild progenitor.
The population structure of chrysanthemum germplasm has not been addressed in any depth till now. An analysis by Klie et al. (2013) of a set of 81 European varieties implied a lack of any detectable structure, which was thought to reflect a history of repeated inter- and backcrossing, along with active germplasm exchange between breeders. By including the full range of cultivated types in the germplasm panel, however, a strong element of structure was clearly present. Three distinct sub-populations were revealed, but just under half (41%) of the entries belonged to the admixed group. The best interpretation of this structure is that strong selection for cultivated type has homogenized genetic variation in key regions of the genome, but that the breeding history also likely involved some deliberate (or possibly unintentional) introgression events. The three sub-populations mapped reasonably well on to the three phylogenetically based clades, and the PCA provided further confirmation of the structure.
Based on RAPD fingerprinting, an early and now rather discredited marker type, Dai et al. (1998) proposed that the cultivated chrysanthemum’s wild progenitor was one of C. indicum, C. vestitum or C. nankingense. No attempt was made in the Dai et al. (1998) study to separately determine the origin of each of the four cultivated types SCC, DCC, PGC and TC. Here, in contrast, the combined use of the FST statistic, measures of genetic distance and phylogenetic clustering led to the conclusion that the small-flowered varieties (PGC and SCC types) are more closely related to the wild progenitor than are the large-flowered ones (TC and DCC types), that PGC was closer to the WC than was SCC, and that TC was more distant than DCC.
Differential adaptation to environments and artificial selection have led to the high differentiation between these chrysanthemums (the PGC vs. DCC type and also the PGC vs. TC type) at the phenotypic trait of plant architecture, inflorescence type and the like. Here, the distinction between the PGC type and the TC type and the DCC types reflected sequence variation in a large number (>550) of genomic regions, some (perhaps all) of which must have experienced artificial selection during the species’ breeding history. The SNPs in these differentiated regions will be valuable to develop molecular markers for the marker-assisted selection of important traits during chrysanthemum breeding.
In chrysanthemum, Klie et al. (2016) have used GWAS to identify a number of AFLP markers and candidate gene-related alleles associated with shoot branching traits and Li et al. (2016) have uncovered considerable dominant markers linked to certain plant and inflorescence traits. The present attempt was based on a large collection of SNP assays, so was more informative than the Li et al. (2016) study, which worked with a very limited number of gel-based markers. The outcome of the GWAS was a set of 97 SNPs significantly associated with one or more of the key traits. However, none of the SNPs marked flower color, as was also the outcome of similar studies performed in two other crops (Spencer et al. 2009; Stranger et al. 2011). Finally, six potential candidates were revealed (table 3). The variant in one of these genes was associated with both flower shape and cultivated type, suggesting that the F-box protein CPR30 may be a regulator of plant morphogenesis and development. A future research goal will be to determine the actual function of these genes. In the meantime, it should be feasible to exploit sequence variants in these genes in the form of a marker-assisted breeding strategy aiming to improve the ornamental value of chrysanthemum.
Additionally, to validate the associated SNPs in cultivar classification, we constructed three phylogenetic trees using the neighbor-joining method, based on the associated 97 SNPs dataset, the cultivated type-related 44 SNPs and the flower shape-related 48 SNPs, respectively (supplementary fig. S4, Supplementary Material online). The result showed that none of the trees could sharply distinguish the cultivars according to flower shapes or cultivated types. This confirms the difficulty in classifying chrysanthemums of complex backgrounds with limited molecular markers.
Supplementary Data
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was partially supported by the grant from the National Natural Science Foundation of China (grant nos. 31425022, 31370699, 31372092, 31471900, and 31572159), the Natural Science Foundation of Jiangsu Province (grant no. BK2015657), and Research Funds for the Central Universities (grant no. KYZ201507).
Literature Cited
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, et al. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. [DOI] [PubMed] [Google Scholar]
- Chen F, Chen P, Li H. 1996. Genome analysis and their phylogenetic relationships of several wild species of Dendranthema in China. Acta Hortic Sin. 23:67–72. [Google Scholar]
- Dai SL, Chen JY, Li WB. 1998. Application of RAPD analysis in the study on the origin of Chinese cultivated chrysanthemum. Acta Bot Sin. 40:1053–1059. [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. [DOI] [PubMed] [Google Scholar]
- Drewlow L, Ascher P, Widmer R. 1973. Genetic studies of self incompatibility in the garden chrysanthemum, Chrysanthemum morifolium Ramat. Theor Appl Genet. 43:1–5. [DOI] [PubMed] [Google Scholar]
- Gou M, et al. 2009. An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60:757–770. [DOI] [PubMed] [Google Scholar]
- Han Y, et al. 2016. Domestication footprints anchor genomic regions of agronomic importance in soybeans. New Phytol. 209:871–884. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2:618–620. [Google Scholar]
- He Z, et al. 2011. Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 7:e1002100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, et al. 2003. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 20:735–747. [DOI] [PubMed] [Google Scholar]
- Hess JE, et al. 2015. Use of genotyping by sequencing data to develop a high-throughput and multifunctional SNP panel for conservation applications in Pacific lamprey. Mol Ecol Resour. 15:187–202. [DOI] [PubMed] [Google Scholar]
- Huang X, et al. 2010. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 42:961–967. [DOI] [PubMed] [Google Scholar]
- Huang X, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Nei M. 1990. Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol Biol Evol. 7:82–102. [DOI] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klie M, Menz I, Linde M, Debener T. 2013. Lack of structure in the gene pool of the highly polyploid ornamental chrysanthemum. Mol Breeding. 32:339–348. [Google Scholar]
- Klie M, Menz I, Linde M, Debener T. 2016. Strigolactone pathway genes and plant architecture: association analysis and QTL detection for horticultural traits in chrysanthemum. Mol Genet Genomics 291:957–969. [DOI] [PubMed] [Google Scholar]
- Kump KL, et al. 2011. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet. 43:163–168. [DOI] [PubMed] [Google Scholar]
- Lago C, Clerici E, Mizzi L, Colombo L, Kater MM. 2004. TBP-associated factors in Arabidopsis. Gene 342:231–241. [DOI] [PubMed] [Google Scholar]
- Lam HM, et al. 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 42:1053–1059. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shao J. 1990. Investigation collection and classification of chrysanthemum cultivars in China. J Nanjing Agri Univ. 13:30–36. [Google Scholar]
- Li P, et al. 2016. Genetic diversity, population structure and association analysis in cut chrysanthemum (Chrysanthemum morifolium Ramat.). Mol Genet Genomics 291:1117–1125. [DOI] [PubMed] [Google Scholar]
- Li T, Li Y, Ning H, Sun X, Zheng C. 2013. Genetic diversity assessment of chrysanthemum germplasm using conserved DNA-derived polymorphism markers. Sci Hortic Amsterdam 162:271–277. [Google Scholar]
- Liu PL, Wan Q, Guo YP, Yang J, Rao GY. 2012. Phylogeny of the genus Chrysanthemum L.: evidence from single-copy nuclear gene and chloroplast DNA sequences. PLoS One 7:4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA. 2002. Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem. 277:41987–42002. [DOI] [PubMed] [Google Scholar]
- Morris GP, et al. 2013. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci U S A. 110:453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura AE, et al. 2014. Killer whale nuclear genome and mtDNA reveal widespread population bottleneck during the last glacial maximum. Mol Biol Evol. 31:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujolar JM, et al. 2014. Assessing patterns of hybridization between North Atlantic eels using diagnostic single-nucleotide polymorphisms. Heredity 112:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut S, et al. 2013. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat Commun. 4:1827.. [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T. 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169:1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CC, Su Z, Donnelly P, Marchini J. 2009. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 5:e1000477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Stahl EA, Raj T. 2011. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 187:367–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, et al. 2013. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8:e58700.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Eulgem T. 2011. EMSY-like genes are required for full RPP7-mediated race-specific immunity and basal defense in Arabidopsis. Mol Plant Microbe In. 24:1573–1581. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. 2013. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid–ethylene antagonism mediated by histone deacetylation. Plant Cell 25:149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1949. The genetical structure of populations. Ann Eug. 15:323–354. [DOI] [PubMed] [Google Scholar]
- Xie Q, et al. 2002. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419:167–170. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2005. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17:804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 38:203–208. [DOI] [PubMed] [Google Scholar]
- Zhang D, Song H, et al. 2014. The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet. 10:e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dai S, Hong Y, Song X. 2014. Application of genomic SSR locus polymorphisms on the identification and classification of chrysanthemum cultivars in China. PLoS One 9:e104856.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, et al. 2011. SRAP-based mapping and QTL detection for inflorescence-related traits in chrysanthemum (Dendranthema morifolium). Mol Breeding 27:11–23. [Google Scholar]
- Zhang F, et al. 2013. Genetic mapping of quantitative trait loci underlying flowering time in chrysanthemum (Chrysanthemum morifolium). PLoS One 8:e83023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Jiang J, Chen S, Chen F, Fang W. 2012. Mapping single-locus and epistatic quantitative trait loci for plant architectural traits in chrysanthemum. Mol Breeding 30:1027–1036. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.