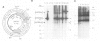Abstract
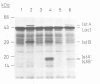
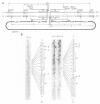
The bacterial 2.1 kb insertion sequence IS21 occurs as a tandem repeat [=(IS21)2] on the broad host range plasmid R68.45. In (IS21)2, the two IS21 elements are separated by 3 bp termed junction sequence. Plasmids carrying (IS21)2 form cointegrates with other replicons at high frequencies. The two IS21 genes, istA and istB, were found to be necessary for cointegrate formation in vivo. Since the outer ends of (IS21)2 are dispensable for cointegrate formation, we favor a transposition model according to which a plasmid carrying (IS21)2 is cleaved at the junction sequence; the opened plasmid is then inserted into a target replicon. Here we show that Escherichia coli cell extracts, which contained over-produced IstA protein, nicked a supercoiled (IS21)2 plasmid precisely at the inner 3' termini of IS21; the resulting staggered cut generated 5' protrusions. The istA gene, but not the istB gene, was required for in vitro cleavage of an IS21-IS21 junction. Because of this cleavage and our previous findings (generation of 4 bp target duplications and loss of the junction sequence after cointegrate formation in vivo) we propose that plasmids with (IS21)2 produce cointegrates by a mechanism which involves joining of the inner 3' ends of IS21 to the 5' ends of the target.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benjamin H. W., Kleckner N. Intramolecular transposition by Tn10. Cell. 1989 Oct 20;59(2):373–383. doi: 10.1016/0092-8674(89)90298-5. [DOI] [PubMed] [Google Scholar]
- Bolton B. J., Schmitz G. G., Jarsch M., Comer M. J., Kessler C. Ksp632I, a novel class-IIS restriction endonuclease from Kluyvera sp. strain 632 with the asymmetric hexanucleotide recognition sequence: 5'-CTCTTC(N)1-3' 3'-GAGAAG(N)4-5'. Gene. 1988 Jun 15;66(1):31–43. doi: 10.1016/0378-1119(88)90222-3. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Correct integration of retroviral DNA in vitro. Cell. 1987 May 8;49(3):347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987 Nov 6;51(3):493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- Currier T. C., Morgan M. K. Direct DNA repeat in plasmid R68.45 is associated with deletion formation and concomitant loss of chromosome mobilization ability. J Bacteriol. 1982 Apr;150(1):251–259. doi: 10.1128/jb.150.1.251-259.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Grindley N. D. Replicative and conservative transposition in bacteria. Cell. 1986 Nov 7;47(3):325–327. doi: 10.1016/0092-8674(86)90586-6. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci U S A. 1989 May;86(9):3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988 Aug 12;54(4):497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Mumm S. R. Unraveling retrovirus integration. Cell. 1990 Jan 12;60(1):3–4. doi: 10.1016/0092-8674(90)90707-l. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Haas D., Holloway B. W. R factor variants with enhanced sex factor activity in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Mar 30;144(3):243–251. doi: 10.1007/BF00341722. [DOI] [PubMed] [Google Scholar]
- Haas D., Riess G. Spontaneous deletions of the chromosome-mobilizing plasmid R68.45 in Pseudomonas aeruginosa PAO. Plasmid. 1983 Jan;9(1):42–52. doi: 10.1016/0147-619x(83)90030-6. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Watson J. M., Haas D., Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984 May;11(3):206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Craigie R. Mechanism of bacteriophage mu transposition. Annu Rev Genet. 1986;20:385–429. doi: 10.1146/annurev.ge.20.120186.002125. [DOI] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H., Kutter E., Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179(2):421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Grindley N. D. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981 Sep;25(3):721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- Reimmann C., Haas D. IS21 insertion in the trfA replication control gene of chromosomally integrated plasmid RP1: a property of stable Pseudomonas aeruginosa Hfr strains. Mol Gen Genet. 1986 Jun;203(3):511–519. doi: 10.1007/BF00422078. [DOI] [PubMed] [Google Scholar]
- Reimmann C., Haas D. Mode of replicon fusion mediated by the duplicated insertion sequence IS21 in Escherichia coli. Genetics. 1987 Apr;115(4):619–625. doi: 10.1093/genetics/115.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C., Moore R., Little S., Savioz A., Willetts N. S., Haas D. Genetic structure, function and regulation of the transposable element IS21. Mol Gen Genet. 1989 Feb;215(3):416–424. doi: 10.1007/BF00427038. [DOI] [PubMed] [Google Scholar]
- Reimmann C., Rella M., Haas D. Integration of replication-defective R68.45-like plasmids into the Pseudomonas aeruginosa chromosome. J Gen Microbiol. 1988 Jun;134(6):1515–1523. doi: 10.1099/00221287-134-6-1515. [DOI] [PubMed] [Google Scholar]
- Riess G., Holloway B. W., Pühler A. R68.45, a plasmid with chromosome mobilizing ability (Cma) carries a tandem duplication. Genet Res. 1980 Aug;36(1):99–109. doi: 10.1017/s0016672300019704. [DOI] [PubMed] [Google Scholar]
- Riess G., Masepohl B., Puehler A. Analysis of IS21-mediated mobilization of plasmid pACYC184 by R68.45 in Escherichia coli. Plasmid. 1983 Sep;10(2):111–118. doi: 10.1016/0147-619x(83)90063-x. [DOI] [PubMed] [Google Scholar]
- Sancar A., Wharton R. P., Seltzer S., Kacinski B. M., Clarke N. D., Rupp W. D. Identification of the uvrA gene product. J Mol Biol. 1981 May 5;148(1):45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- Sherman P. A., Fyfe J. A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A., Holloway B. W. A mutant sex factor of Pseudomonas aeruginosa. Genet Res. 1972 Feb;19(1):91–108. doi: 10.1017/s0016672300014294. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Bandyopadhyay P. K. Model for how type I restriction enzymes select cleavage sites in DNA. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Crowther C., Holloway B. W. The insertion sequence IS21 of R68.45 and the molecular basis for mobilization of the bacterial chromosome. Plasmid. 1981 Jul;6(1):30–52. doi: 10.1016/0147-619x(81)90052-4. [DOI] [PubMed] [Google Scholar]