Abstract
pH-triggered membrane permeabilizing peptides could be exploited in a variety of applications, such as to enable cargo release from endosomes for cellular delivery, or as cancer therapeutics that selectively permeabilize the plasma membranes of malignant cells. Such peptides would be especially useful if they could enable the movement of macromolecules across membranes, a rare property in membrane permeabilizing peptides. Here we approach this goal by using an orthogonal high-throughput screen of an iterative peptide library to identify peptide sequences that have the following two properties: (i) little synthetic lipid membrane permeabilization at physiological pH 7 at high peptide concentration and (ii) efficient formation of macromolecule-sized defects in synthetic lipid membranes at acidic pH 5 and low peptide concentration. The peptides we selected are remarkably potent macromolecular sized pore-formers at pH 5, while having little or no activity at pH 7, as intended. The action of these peptides likely relies on tight coupling between membrane partitioning, α-helix formation, and electrostatic repulsions between acidic sidechains, which collectively drive a sharp pH-triggered transition between inactive and active configurations with apparent pKa values of 5.5–5.8. This work opens new doors to developing applications of peptides with membrane permeabilizing activities that are triggered by physiologically relevant decreases in pH.
INTRODUCTION
Membrane-permeabilizing peptides could have utility in a variety of biotechnological and clinical applications due to their ability to breach the barrier imposed by lipid bilayers1–10. But to enable their practical application, they will first need to be designed to function only in specific environments, or only in response to specific triggers. One potentially useful trigger is pH, which varies in spatially and temporally specific ways in cellular organelles, and also varies locally in tissues under some pathological conditions, including cancer11–13. As an example application, pH-sensitive membrane permeabilizing peptides could be triggered upon endosomal acidification to promote the release of uptaken polar molecules from endosomal compartments into the cell cytosol1,14–16. Such an application would eliminate a long-standing barrier to the delivery of generic polar compounds, especially proteins and other macromolecules, to cells14,17,18. Indeed, while efficient methods exist to deliver oligonucleotides to cells16,19, most other types of macromolecules are more difficult to deliver. These macromolecules can be directed to existing cellular uptake mechanisms20,21, but in the absence of endosome permeabilization or disruption, they often get trapped within the classical pathways that lead to their lysosomal degradation or recycling without significant entry into the cytosol22,23. As a second example application, pH-sensitive, membrane permeabilizing peptides could potentially be used in cancer therapies to selectively permeabilize the plasma membranes of cancer cells. This is possible because the environmental milieu in the vicinity of solid tumors is often acidic due to their high rate of mostly glycolytic metabolism13. In support of this application, the locally acidic pH of solid tumors has already been shown in mice to trigger the pH-sensitive insertion of peptides into membranes, although not for peptides that cause permeabilization11,24.
Some progress has been made in the discovery or design of pH-triggered pore-forming peptides25,26, including some that we designed rationally27, and other pH triggered membrane active peptides16,28. The state-of-the art in the field has been trial and error-based addition of protonatable residues such as aspartate (D), glutamate (E) and histidine (H). Two well-studied examples are pHLIP, which inserts across membranes at pH<5.5 without permeabilization11, and GALA, which permeabilizes synthetic membranes at pH < 5.529. However, none of the known pH-sensitive peptides have all of the properties needed for the applications described above. For example, pHLIP does not form pores, although it can deliver small polar molecules that are covalently attached to it by insertion across the membrane30. GALA, and others, do form pores in membranes, but only small pores28,29,31–33 with limited utility for cellular delivery, especially for macromolecules.
A uniquely useful combination of properties would be pH-triggered membrane permeabilization that enables the movement of macromolecules across membranes. Until recently, even non-triggered macromolecular poration activity that occurs at low peptide concentration has been rare in pore-forming peptides34,35. However, we recently used high throughput screening of a peptide library to discover a unique peptide, called MelP5, which allows macromolecules through synthetic membranes even at very low concentrations of peptide36,37. Thus, with MelP5, there is at least one sequence known that is a non pH-sensitive macromolecular pore forming peptide. And as discussed above, there are also sequences, such as GALA and pHLIP and others that have pH sensitive membrane insertion and permeabilization. However, we are aware of no sequences, other than those discussed below, that have both properties.
There is a lack of detailed molecular understanding of the sequence-structure-function relationships for membrane active peptides, which hinders their rational design. Here we take a novel high-throughput approach to the discovery of pH-sensitive, macromolecular pore-forming peptides. First, we used the sequence of the macromolecular pore former, MelP5 and sequence features found in GALA and pHLIP to design a rational combinatorial peptide library. We then developed an orthogonal high-throughput screen to identify sequences from the library that have the following two properties in synthetic bilayers; (i) little membrane permeabilization at physiological pH 7 at high peptide concentration and (ii) efficient formation of macromolecule-sized defects in membranes at acidic pH 5 and low peptide concentration. Such peptides will have no effect on membranes at normal cellular pH, but will be triggered by physiologically reasonable acidic pH to form macromolecule-sized pores. We show below that this approach successfully led to the discovery of a conserved motif for pH-triggered, macromolecule sized poration. We expect these unique sequences to ultimately form the basis for multiple applications that utilize changes in pH as a functional trigger.
MATERIALS AND METHODS
Library Synthesis
The peptide library was a one-bead-one-peptide library, synthesized using a split and recombine approach described in detail previously36,38,39. The library members were synthesized on Tentagel Megabead MB NH2 resin beads (Rapp Polymere MB300002), coupled to it by a UV-cleavable photo linker, 4-(4-[1-(9-Fluorenylmethyloxycarbonylamino)ethyl]-2-methoxy-5-nitrophenoxy) butanoic acid. After synthesis, sidechains were deprotected with a mixture of trifluoroacetic acid and scavengers40. Beads were then washed extensively and stored dry at −20°C prior to use.
To cleave the photolinker and release the library members, beads were first suspended in methanol and dispersed onto a glass plate. The beads were dried thoroughly and then exposed to UV light at 365 nm for 6 hours with illumination from plate top and bottom. One day prior to screening, beads were placed into the wells of a 96-well plate, one bead per well. Water and hexafluoroisopropanol (25 uL each) were added to each well, and the plates were exposed to 365 nm UV light for an additional 3.5 hours, releasing and extracting the peptide while also evaporating the solvent. Finally, 25 uL of water was added to each well and plates were incubated overnight for peptide solubilization. About 0.5 nmol of peptide was extracted from each bead, as quantified by tryptophan fluorescence.
Vesicle Preparation
Large unilamellar vesicles for leakage assays were prepared as previously described41 in 100 mM potassium chloride, with 10 mM sodium phosphate or 10 mM sodium acetate buffer. For small molecule leakage, the lipid 1-palmitoyl-2-oleoyl-sn-3-glycerophosphocholine (POPC) dried from chloroform, was resuspended in buffer containing: 12.5 mM ANTS, 45 mM DPX, 5 mM HEPES and 20 mM sodium chloride. For macromolecular leakage assays, lipids were resuspended in buffer with 1 mg/ml TAMRA-biotin-dextran (TBD). Lipid suspensions were frozen and thawed ten times and then extruded 10 times using a 0.1 um pore size Nuclepore polycarbonate filter42. After extrusion, external ANTS and DPX were removed by gel filtration with Sephadex G100. External TBD was removed using streptavidin agarose resin.
Small molecule Leakage Assay
Lipid vesicles with entrapped ANTS and DPX were diluted into wells of a 96 well plate containing peptide. After 60 minutes, vesicle permeabilization was measured by an increase in ANTS fluorescence, with excitation at 350 nm and emission at 519 nm. Fractional leakage was quantified using Equation 1:
Here I is the intensity at 60 min, Ibackground is the intensity of a control with vesicles only, and Itriton is the intensity in the presence of vesicles and 0.4% v/v of the detergent Triton X-100 added to solubilize the vesicles and release the ANTS.
Macromolecular Leakage Assay
As described previously37, 20 nM Alexafluor488-labelled streptavidin (SA) was added to a solution of vesicles with entrapped TAMRA-biotin-dextran, TBD. This solution was added to wells of a 96-well plate containing peptide, and the plate was incubated for 60 min. Release of the 40 kD dextran enables TBD-SA complex formation, which leads to quenching of the AF488 fluorescence (excitation 495 nm, emission at 519 nm) by TAMRA. The fraction of macromolecule leakage was determined by Equation 2:
where I is the intensity at 60 min, Ibackground is the intensity of a vesicle control with no peptide, and Itriton is the intensity in the presence of vesicles and 0.1% v/v of the detergent Triton X-100, added to solubilize the vesicles and release the ANTS.
High throughput screening
In the orthogonal high throughput screen, we combined the two assays described above and used them in 96-well format as follows. Step 1: Individual beads were separated into the wells of a plate and peptides were extracted into a small volume of water as described above. Each bead releases about 0.5 nmol of one peptide sequence from the library. Step 2: 1 mM lipid vesicles in 100 μl of sodium phosphate buffer at pH 7 were added to the peptides in water in the plate in Step 1. These vesicles contained the 350 Da fluorophore ANTS and its quencher DPX, entrapped at mM concentrations. After this step, the nominal peptide concentration was 5 μM and the peptide to lipid ratio was roughly 1:200. Leakage, if it occurs, causes an increase in fluorescence of ANTS. A few wells contained controls: vesicles only, 1 nmol of the peptide MelP5 (P:L = 1:100), 4 nmol of MelP5 (P:L = 1:25), or 0.4% v/v of the detergent Triton X-100. The latter two should completely permeabilize the vesicles. Step 3: After 60 minutes of incubation, ANTS fluorescence was measured for each well to quantitate small molecule release using equation 1. Step 4: Next, to the same wells, a different preparation of vesicles was added. These vesicles have entrapped 40kD TAMRA-biotin-dextran (TBD) and external Alexafluor488 streptavidin. The TBD vesicles were in sodium acetate buffer at pH 4 so that the final pH in each assay well was 5.0. This second addition of vesicles increased the total volume to 200 μl which decreased peptide to 2.5 μM and decreased ANTS vesicle concentration 0.5 mM. The new TBD vesicles were present at 1.5 mM so that the total peptide to total lipid ratio was roughly about 1:800. Step 5: After 60 minutes, the intensity of the AF488-streptavidin was measured to quantitate macromolecule release, which is calculated with equation 2. This orthogonal and sequential screen gives two measurements that are used to identify peptides with the desired properties: i) Small molecule release at pH 7 and P:L = 1:200; ii) Macromolecule release at pH 5 and P:L=1:800.
Circular Dichroism Spectroscopy
Circular dichroism was measured in a JASCO 810 spectropolarimeter. Samples in buffers of various pH were prepared by mixing peptide and vesicle solutions, each prepared at the needed pH. Spectra were collected on samples of 1 mM vesicles and 5 μM peptide in 1 mm path rectangular quartz cuvette after 1 hour of equilibration.
Tryptophan fluorescence
Membrane partitioning was assessed at various pH values using tryptophan fluorescence in a Fluorolog-3 fluorometer (Jobin Yvon). Peptide and lipid solutions were prepared at individual pH values and tryptophan fluorescence spectra were measured after 60 minutes of incubation. Excitation was fixed at 280 nm (slit 5 nm) and emission was collected from 300 to 450 nm (slit 5 nm).
RESULTS
Peptide Library Design
The peptide MelP5 is a highly potent, gain-of-function variant of the cytolytic bee venom peptide melittin that was discovered in a high-throughput screen of a library that used melittin as a template36. While the melittin library was screened only for dramatically increased potency of small molecule release, we later showed that MelP5, the most potent gain-of-function peptide discovered, also releases macromolecules from vesicles at low peptide concentration 37. Among the many known membrane permeabilizing peptides, Melp5 is unique in its ability to induce the passage of dextrans up to a molecular weight of 40,000 through bilayers. MelP5 is unstructured in solution, but folds into an amphipathic α-helix in the presence of membranes, into which it inserts, leading to macromolecule-sized membrane disruption at low peptide-to-lipid ratios, P:L ≤ 1:500.
Previously, we attempted to rationally design pH sensitive, macromolecular pore-forming peptides27 by encoding pH-sensing motifs, based on the sequences of the pH-sensitive membrane active peptides GALA29 and pHLIP11, into the pH-insensitive, macromolecular pore-forming motif in MelP537. The designed peptides, named MelP5_Δ4 and MelP5_Δ6 (see Table 1), had four or six of the residues along the polar face of the putative amphipathic helix changed to glutamate or aspartate to impart pH sensitivity. Placement and spacing of the acidic residues were based on their helical spacings in GALA and pHLIP. We found that these rationally designed peptides gained pH sensitivity, as they permeabilized membranes only at pH < 5.0. But at the same time, they lost the ability to form macromolecular-sized pores27, demonstrating that the properties of MelP5, GALA, and pHLIP are only partially additive.
Table 1.
Sequences of the peptides studied here. Top panel: Sequences of the natural bee venom pore-former melittin and its synthetically evolved gain-of-function variant, MelP537 which enables macromolecules to cross bilayers at low P:L. Using patterns from pH-sensitive membrane active peptides, we previously designed two variants MelP5_Δ4 and MelP5_Δ6, which exhibit pH-triggered membrane activity, but do not induce macromolecular-sized poration27. Bottom Panel: All of the positive peptides identified in the screen (see Figure 3). Peptides are named after the plate and well in which the positive bead was identified. For each peptide we list the number of acidic residues, and the number of helical spacings of i to i+3, i to i+4, and i to i+7 between acidic residues.
| Peptide | Sequence | # acidic residues | # helical spacings |
|---|---|---|---|
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | 0 | 0 |
| MelP5 | GIGAVLKVLATGLPALISWIKAAQQL | 0 | 0 |
| MelP5_Δ4 |
|
4 | 3 |
| MelP5_Δ6 |
|
6 | 5 |
| pHD Peptides | |||
| pHD15-30 |
|
6 | 9 |
| pHD24-52 |
|
5 | 6 |
| pHD34-20 |
|
5 | 5 |
| pHD54-73 |
|
5 | 5 |
| pHD63-38 |
|
5 | 6 |
| pHD101-77 |
|
6 | 9 |
| pHD108-47 |
|
5 | 6 |
| pHD118-85 |
|
5 | 7 |
| pHD145-40 |
|
5 | 5 |
| pHD187-4 |
|
5 | 7 |
Here, we approached the same goal with combinatorial chemistry and high throughput screening, instead of rational design. Specifically we designed an iterative 18,432-member library using MelP5 as a template (Figure 1), creating a second generation library that we screened orthogonally for peptides that release macromolecules from lipid vesicles at pH 5, but have little or no membrane permeabilizing activity at pH 7.
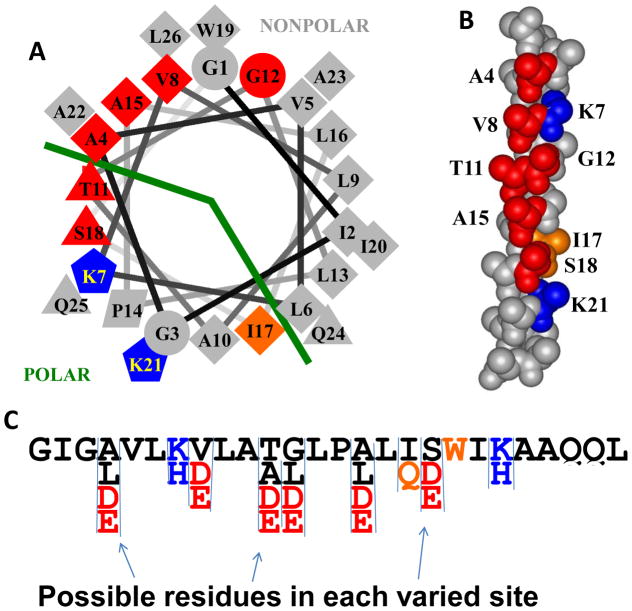
Figure 1.
Design of an iterative peptide library. The library is shown in helical wheel and space-filling representations. The library was based on the non pH-sensitive macromolecular pore-former MelP5 which is the sequence shown. Residue shape indicates type in the template sequence of MelP5, with diamonds representing hydrophobic residues and triangles representing polar residues, for example. Colors show residues that were varied in the library. Possible variations present in the library are shown at the bottom. Red positions could be the native residue, or aspartate or glutamate. In some cases, a fourth hydrophobic residue was also possible, as indicated. Blue residues are lysine in MelP5 and could be lysine or histidine in the library members. Position 17 could have the native hydrophobic isoleucine or a polar glutamine. The most important aspect of the library is in the incorporation of 6 possible protonatable acidic residues, shown in red, that align along one face of the helix, as shown.
In the library, we varied the amino acids at nine positions (Figure 1). Unlike MelP5_Δ4 and MelP5_Δ6, which had a fixed number and pattern of helically spaced acidic residues, the library contained peptides with 0 to 6 acidic residues distributed in all possible patterns along one face of the MelP5 amphipathic alpha helix. Our criteria for placement of acidic residues were as follows: i) maximize the number of i to i+3, i to i+4 and i to i+7 α-helical spacings between acidic groups to maximize the electrostatic repulsion between them at neutral pH, and prevent helix formation. For some library members, we expected that such electrostatic repulsion among the acidic sidechains will prevent binding and membrane insertion at pH 7 but allow it at pH 5. This pattern also enables library members to form amphipathic helices in which the protonation state of the acidic residues can make a significant contribution to helical propensity and membrane insertion; ii) retain basic residues at positions 7 and 21 because we hypothesized that changing them to acidic residues in MelP5_Δ4 and MelP5_Δ627 may have altered the peptide function; iii) retain overlap between the polar face of the MelP5 helix and the positions of the new acidic residues, iv) reduce hydrophobicity as little as possible to maintain the propensity of the helix to partition into membranes; v) avoid replacing any residues known to be critical to the activity of MelP5, specifically A10, P14, L16, and A2336; vi) avoid replacing tryptophan 19 because it is a useful for concentration determination and as an optical probe of structure. With these criteria in mind, we elected to allow both glutamate and aspartate to appear at six positions: A4, V8, T11, G12, A15 and S18, giving the distribution shown in Figure 1. We also allowed the native residue in each of these positions. We allowed hydrophobic leucines to occur at positions A4, G11 and A18 and a somewhat hydrophobic alanine at T11 to modulate hydrophobicity. To potentially compliment the pH sensitivity of the acidic residues, we allowed the native lysines at positions 7 and 21 to also be histidine. Lysine will be cationic at all pH values below 8.5 whereas histidine will be cationic only below its pKa of ~6.5.
Previously, we showed that the presence or absence of polar residues at the boundary of the polar-nonpolar faces is a critical feature of MelP536. Specifically, the native T10 in the first generation library was replaced with alanine, which narrows the polar face substantially (Figure 1). In the current library we preserved A10 on the N-terminal half of the peptide, and allowed position 17, which defines the cutoff between the polar and non-polar faces on the C-terminal half of the helix, to vary between hydrophobic isoleucine and polar glutamine.
There are 18,432 unique, 26-residue, MelP5 variants in the library. All library members share at least 17 residues of 26 in common with MelP5 such that the minimum identity is 73%. If we assume that D and E are equivalent, there are 64 different patterns of acidic residues in the library. If D and E are unique, there are 729 different patterns. From the library design in Figure 1, we calculate the following abundance values: 2.8% of library members have 6 acidic residues (1 pattern of 64), 13.8% have 5 acidic residues (6 patterns), 28.5% have 4 acidic residues (14 patterns), 30.6% have 3 acidic residues (20 patterns), 18.1% have 2 acidic residues (14 patterns), 5.6% have 1 acidic residue (6 patterns) and 0.7% have 0 acidic residues (1 pattern).
The library was synthesized as a one-bead, one-peptide library using a well-established split and recombine approach36,38,39. Quality control for the synthesis was done using HPLC, MALDI mass spec, and Edman degradation on peptide extracted from multiple individual beads. These methods showed that every bead examined contained predominantly a single pure sequence and that each sequence observed was, in fact, an expected member of the library. Each 0.3 mm polystyrene solid phase peptide synthesis bead releases about 0.5 nmol of one single sequence as described above.
Orthogonal High-Throughput Screen Results
The library was screened for peptides that simultaneously i) cause little or no membrane permeabilization at pH 7, even for small molecule reporters and high peptide concentrations, and ii) cause macromolecule passage across bilayers at pH 5, at low peptide concentrations. Two different assays, described above, were used in tandem to achieve these aims. One was an assay for release of ANTS, a small molecule (Figure 2A), performed at pH 7. The second was an assay for release of a 40,000 Da TAMRA-biotin-dextran, conducted at pH 5 (Figure 2B). These two assays can be performed in parallel or in series in the same samples because there is no relevant spectral overlap between ANTS fluorescence (ex/em 350/519), and AF488 fluorescence (ex/em 488/505 nm). In the high-throughput screen described in Materials and Methods, the ANTS/DPX assay was conducted at a nominal peptide to lipid ratio of 1:200 at pH 7, and the dextran leakage assay was performed at nominal P:L = 1:800, at pH 5. Because the peptide release from individual beads varies, P:L could vary between individual wells by a factor of 2 or more.
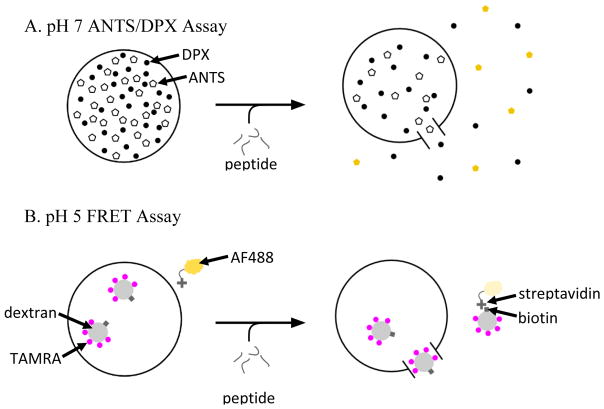
Figure 2.
Two different leakage assays are used to measure the pore-forming activity of the peptides and screen for the desired activities. A: To evaluate leakage of small molecules we co-encapsulate ANTS, a small molecule dye and DPX, its obligate quencher, inside lipid vesicles52. Membrane destabilization results in release of ANTS and DPX and recovery of ANTS fluorescence. B: To evaluate leakage of macromolecules, we used a recently published assay37 based on FRET detection. In this case, a 40 kDa dextran co-labelled with biotin and the acceptor fluorophore, TAMRA, is encapsulated within vesicles. Streptavidin labeled with the donor fluorophore, AlexaFluor488, is on the outside of the vesicles. Upon macromolecular permeation, the TAMRA-biotin-dextran (TDB) can escape and form a complex with streptavidin, allowing FRET to occur. In our high-throughput screen, we measure leakage of ANTS/DPX at pH 7 and nominal P:L = 1:200 and also leakage of TBD at pH 5 and nominal P:L = 1:800.
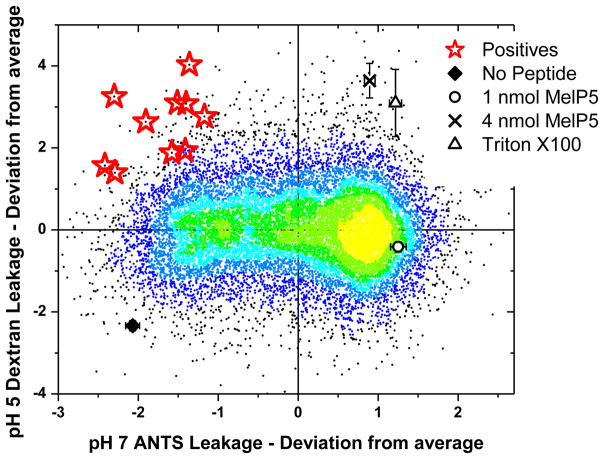
We screened 15,000 library members using the orthogonal high throughput screen, covering about 80% of the library’s sequence space. In Figure 3 we show the ability of each screened peptide to cause small molecule leakage at pH 7 and macromolecular leakage at pH 5. We present the results in the form of a scatterplot colored according to point density from yellow (highest density) to black (lowest density). Because there were vesicle batch-to-batch variations in the raw intensity values for each assay along the duration of the screen, we have plotted all values in Figure 3 as plate-by-plate Z-values; the points plotted are the number of standard deviations from the plate mean, on each axis. The density of points on the Y-axis is centered on zero because the distribution of dextran leakage has a symmetrical Gaussian shape. On the other hand, the distribution of points on the X-axis is offset from zero because ANTS leakage at pH 7 is asymmetric; many library members cause > 80% leakage of ANTS at pH 7.
Figure 3.
Results of the screen. The serial, two part screen described in Figure 2 was used to assay 15,000 randomly selected library members. Vesicles were made from 100% POPC. The two activities for each of the 15,000 library members are shown as points on a temperature scale, where the point color is determined by point density, from yellow (most dense) to black (least dense). The results of each assay are shown as Z-values, or the difference of each point from the plate mean expressed as standard deviations. This approach normalizes for batch-to-batch variations in lipid vesicle intensities in the two assays. About 0.5 nmol of each library member was assayed. For comparison, we also show the values for 1 and 4 nmol MelP5, for Triton-X-100 detergent, which solubilizes vesicles, and for buffer only. The center of the yellow area corresponds to ~85% ANTS leakage and ~30% dextran leakage. Most library members are similar to the template MelP5, but there are outliers in all four quadrants. The peptides we seek, with low ANTS leakage at pH 7 and P:L=1:200 and high TBD release at pH 5 and P:L=1:800, are found in the upper left corner. Ten positive sequences, highlighted with red stars, were selected and sequenced using Edman degradation. Their sequences are shown in Table 1.
Four points provide useful landmarks. The activity of 1 nmol and 4 nmol of the template MelP5 under the conditions of the screen are indicated in Figure 3, to compare with 0.5 nmol of each library peptide. We also show values for blank wells with no peptide and for 0.1% v/v Triton X100, which solubilizes all vesicles. One nmol of MelP5 causes high small molecule permeabilization and partial macromolecule release at all pH values. Four nmol of MelP5 (P:L = 1:100) and 0.1% v/v of Triton X-100 each cause essentially complete release of both types of probes. The activities of most library members are centered on the yellow area in Figure 3; on average they have MelP5-like high leakage of ANTS at pH 7 and moderate leakage of dextran at pH 5. The center of the yellow area corresponds to ~85% ANTS leakage and ~30% dextran leakage.
The activity we seek in this work is found in the peptides closest to the upper left corner of Figure 3. These peptides have low small molecule permeabilization at pH 7 (P:L=1:200) and high macromolecule permeabilization at pH 5 (P:L=1:800). Ten library members, shown by red stars in Figure 3, were selected from within this region, and these peptides were sequenced using Edman degradation. Their sequences are shown in Table 1. Below we demonstrate that these selected peptides have exactly the properties we sought in the screen. Thus, our strategy was successful.
Sequence analysis of positives
The positive sequences have many features in common with one another, demonstrating that we have identified a family of closely related sequences with a unique, shared activity. P-values were calculated against null hypotheses determined by the abundance of the particular residue, class, or motif in the library, using exact binomial statistics. Every positive peptide has 5 or 6 acidic residues out of six possible (p<10−5). Positions 4 and 8 are acidic in all positives (p=0.002 and 0.007, respectively). In the remaining four positions that could have acidic groups, 11, 12, 15 and 18, eight of the ten peptides have 3 acidic residues and one other non-acidic residue and two have six acidic residues. Of the 9 possible i to i+3, i to i+4 and i to i+7 helical spacings of acidic groups available, the positive peptides have an average of 6.5 (range 5–9). The constancy of the acidic residue abundance supports the hypothesis, discussed earlier, that pH-triggered membrane activity is determined mostly by the coupling between electrostatic repulsion, and the formation of the amphipathic helix as it relates membrane binding (see measurements of helix content below).
Among the 52 selected acidic residues, there was a strong preference for glutamate with 36 glutamates selected compared to 16 aspartates (p=0.007). The preference for E over D is even more striking in the first two and last two possible positions, where 26 of 33 acidic residues were glutamate (p=0.001). We do not currently know why this preference exists, but speculate that the longer sidechain of glutamate enables more conformational flexibility of the sidechains by which the electrostatic effects can be modulated, either between acidic groups or in the interactions of acidic residues with basic residues. When an acidic residue was possible but not selected, the selected residues included both native residues and the more hydrophobic residues possible, indicating no strong preference for the native residues at positions 11, 12, 15 and 18. Similarly, 9 lysines and 11 histidines were selected in positions 7 and 21, with no obvious preference or pattern. In fact, all possible patterns (KK, HH, HK, and KH) were observed in the 10 positives. We conclude that the identity of the basic sidechains and their charge state at pH 7 are of little consequence to the function of these peptides.
Finally, in position 17, where the native, hydrophobic isoleucine and the polar glutamine were possible, we found that the native residue was replaced with glutamine in 10 of 10 peptides (p=0.002). The current lack of explicit structure-function relationships in these membrane active peptides makes it difficult to know exactly how this glutamine contributes to activity at this time, but we speculate that its hydrogen bonding capabilities may enable lateral interactions between peptides in the bilayer. We will investigate this and other structural hypotheses in the near future.
It is interesting to compare the positive sequences selected from this library, which have the desired property of macromolecular poration at pH 5.5–5.8, to the rationally designed sequences MelP5_Δ4 and MelP5_Δ6, which have a lower pKa for permeabilization and do not allow macromolecules across membranes at any pH. The rational and selected peptides both had 4–6 acidic residues with helical spacings. The acidic residues replacing V8, T11 and S18 are present in both families. The rationally designed peptides have 3 or 5 helical spacings between acidic residues, while the selected peptides have 5–9, but there is overlap. In any case, fewer helical spacings should theoretically lead to a higher pKa whereas the designed peptides actually had a lower pKa than the selected ones. We replaced one or both basic residues in the designed peptides whereas these positions only contained basic residues in the library because we hypothesized that favorable electrostatic interactions with the basic residues could abrogate acidic repulsions in the library-selected peptides. Furthermore, the designed peptides always had isoleucine in position 17, while the selected peptides always had glutamine at position 17. Isoleucine was available at position 17 in the library, but was never selected. While we can list the differences between our screening successes and our design failures, it is currently not possible to explain or predict these behaviors in molecular terms, effectively demonstrating the power of synthetic molecular evolution. We hope that structural and computational scientists will endeavor to explore these mechanistic questions more deeply in the near future.
Verification of Positive Peptides
Since the selected peptides are very similar to each other, we chose to synthesize and purify a subset of them for detailed validation. Tested peptides included four representative sequences with five acidic residues, pHD24, pHD34, pHD108 and pHD118 as well as pHD15, one of the two with six acidic residues. Because we are interested in pH-triggered macromolecule release at low pH, we validated the positive peptides with both the ANTS assay and with the macromolecular release assay using 40 kDa dextran and 53 kD streptavidin, described above.
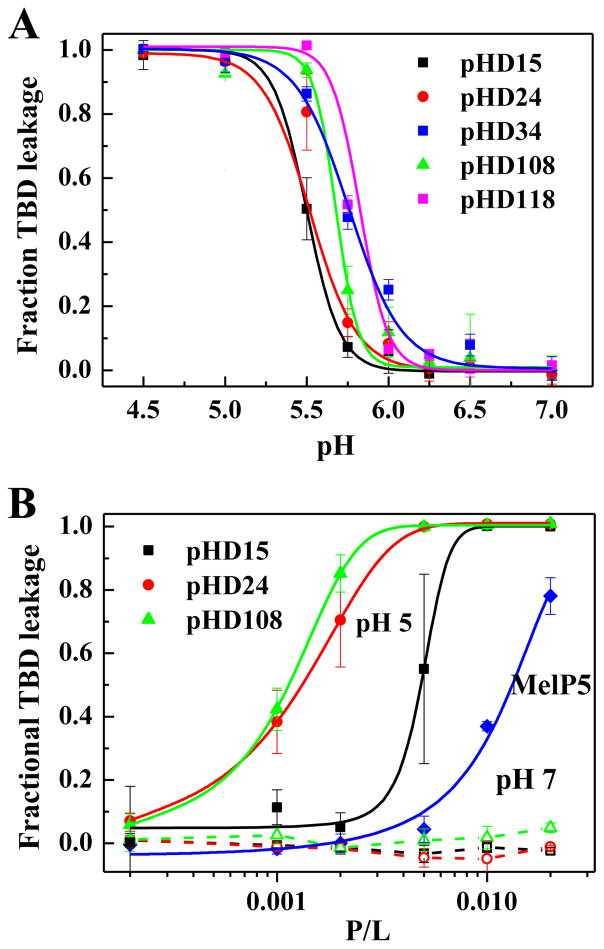
Dextran leakage was measured as functions of peptide concentration and pH using vesicles made from phosphatidylcholine lipids. The leakage of 40 kDa dextran from lipid vesicles at P:L=1:200 as a function of pH is shown for these peptides in Figure 4A. At this concentration, all of the selected peptides cause 100% dextran release at pH 5 and no leakage at pH 7, as desired. Activity occurs only as pH is decreased into the range of pH 6 to 5.5. The apparent pKa values for the five peptides are similar, ranging from 5.5 to 5.8, with pHD15 having the lowest apparent pKa value, consistent with it having one more acidic residue. Consistent with this result, and with the screen, we also observe <5% ANTS leakage at at pH 7 (P:L = 1:200) and ~100% leakage at pH 5 (not shown). We note that the macromolecule leakage activity measured in Figure 4 is unique and remarkable, as there are no other peptides known, except for MelP5 37, that have been shown to release macromolecules from lipid vesicles at such low P:L ratios under any conditions. These synthetically evolved peptides have a pH-triggered version of this activity, and release macromolecules at pH 5 even better than MelP5 does at any pH.
Figure 4.
A: Macromolecule leakage versus pH. A representative set of positive peptides from Table 1 were synthesized and assessed for their ability to promote leakage of a 40,000 Da TAMRA-biotin-dextran (Figure 2B) at P:L=1:200 and 1 mM POPC vesicles. Changes in pH lead to sharp transition in macromolecular poration. The apparent pKa values from the curve midpoints range from 5.5 to 5.8. B: Macromolecule leakage versus concentration. The peptides assessed for their ability to promote leakage of a 40,000 Da TAMRA-biotin-dextran (Figure 2B) at pH 5 and pH 7. The peptides exhibit no activity at pH 7, as desired, even at peptide to lipid ratios as high as 1:50 (dashed lines). At pH 5 all peptides cause potently cause TBD release with curve midpoints that range from P:L ~1:900 to 1:600.
Dextran release at pH 5 and pH 7 as functions of peptide concentration (expressed as peptide to lipid ratio, P:L) is shown in Figure 4B, for pHD 15, pHD 24, and pHD 108. The dashed lines show that there is no activity at pH 7, as desired, even at peptide to lipid ratios as high as 1:50. However, at pH 5 the selected peptides induce substantial macromolecule leakage with 50% leakage activity at peptide to lipid ratios of 1:900 for pHD108 and 1:750 for pHD24. pHD15, which has six acidic is the least active of the positives tested, with 50% release at P:L = 1:600. While MelP5 has been shown to release a 10,000 Da dextran at similar concentrations37, it releases the 40,000 Da dextran used in this work at ~P:L = 1:100. Taken together, these results show that we have successfully identified peptides that are significantly more active than the unique MelP5, and are triggered to act only in acidic pH environments.
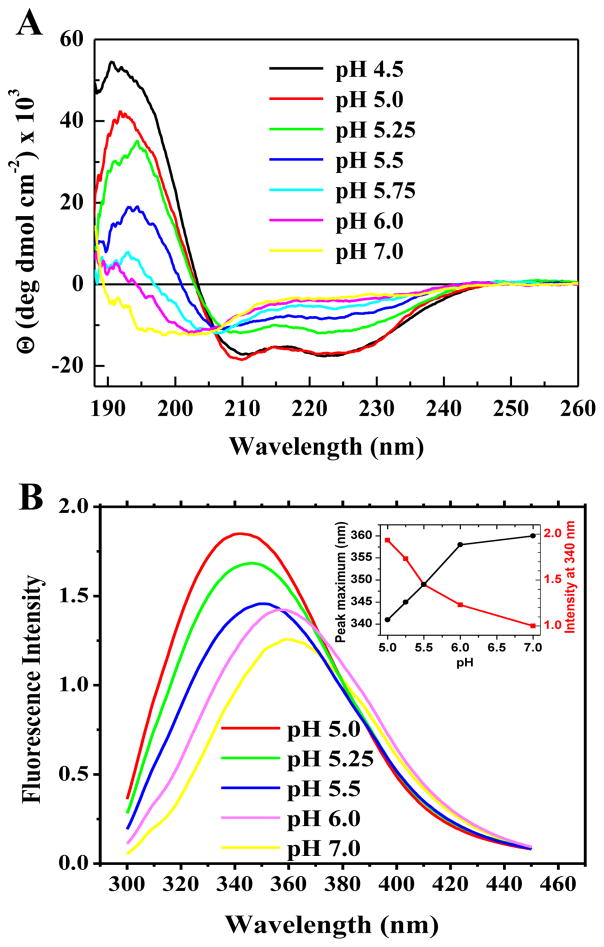
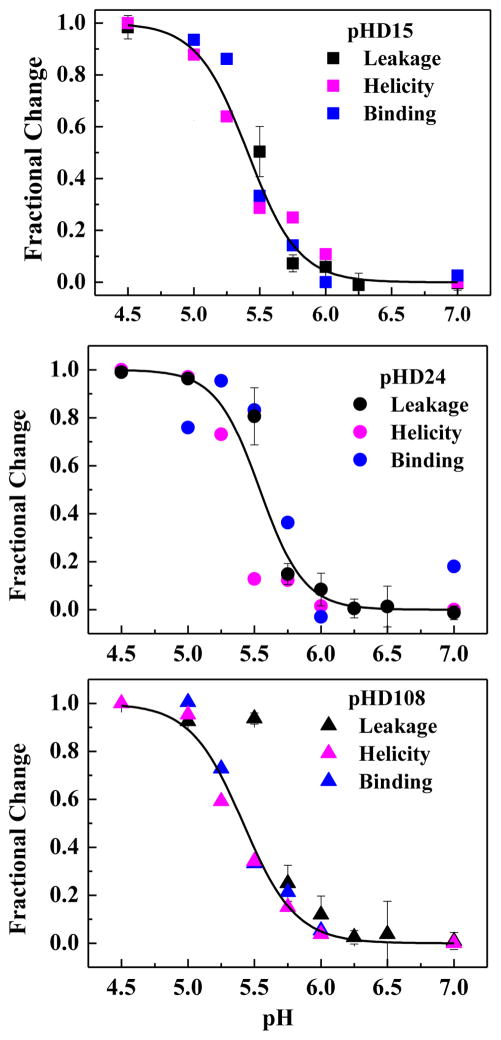
We also studied the peptide secondary structure in the presence of vesicles as a function of pH by circular dichroism spectroscopy. Example CD spectra for pHD108 at P:L = 1:200 (Figure 5A) show a pH-triggered transition from random coil to α-helical structure with an effective pKa around 5.5. Finally, we measured tryptophan fluorescence as a function of pH at 1:200 which provides a measure of membrane partitioning. Like the circular dichroism spectra, example fluorescence spectra for pHD108 (Figure 5B) also show a sharp transition from lower intensity emission at 360 nm to higher intensity and 340 nm emission maximum, consistent with a transition from weak to strong membrane partitioning over the expected pH range of 5–6. In Figure 6 we directly compare the pH-dependence of macromolecule leakage, helicity and tryptophan fluorescence for three peptides at P:L = 1:200, and show that they are very similar, with pKa of 5.5–5.8, consistent with our hypothesis that leakage, helicity, and binding are coupled for macromolecule release.
Figure 5.
Example binding and folding data. A: Circular dichroism spectra of pHD108 versus pH at P:L=1:200 in 1 mM POPC vesicles. Separate samples are made at each pH and equilibrated for 30 minutes prior to the measurement. The spectra show a structural transition from random coil at pH 7 to classical α-helix at pH < 5.75. B: Tryptophan fluorescence spectra of pHD108 at P:L=1:200 in 1 mM POPC vesicles. Separate samples are made at each pH and equilibrated for 30 minutes prior to the measurement. The spectra show a transition from a more polar, water-exposed environment to a less polar, buried environment consistent with peptide that has partitioned into bilayers.
Figure 6.
Coupling of binding, structure and activity. For three pHD peptides we plot changes in TBD leakage, changes in α-helicity from CD, and changes in tryptophan fluorescence as pH is varied. All measurements are at P:L = 1:200. Curves represent the global fit for each peptide of a cooperative transition using all three data sets. There is little or no detectable difference between the pKa values for leakage, structure and binding, consistent with our hypothesis that they are coupled.
DISCUSSION
Several thousand membrane destabilizing peptides, including antimicrobial peptides and other classes of pore-forming peptides, have been described and investigated over past decades35,43–47. Many of these peptides are cationic and destabilize anionic bilayers in a manner driven by strong electrostatic interactions. Efficient permeabilization of zwitterionic phosphatidylcholine (PC) bilayers at low P:L (≤1:100) is uncommon and efficient release of macromolecules from PC vesicles at low P:L ratios was essentially unknown until we reported the discovery of a novel peptide, MelP536. This peptide is unique in its ability to allow the efficient passage of macromolecules through membranes, even at very low peptide-to-lipid ratios (P:L ≤ 1:500 when using 10,000 Da dextran as a probe) where detergent-like vesicle solubilization is unlikely33,37. Here, we used MelP5 as a template for an iterative peptide library that we designed and screened to select peptides that cause macromolecular permeabilization of a 40,000 Da dextran at low P:L, like MelP5, but in a pH-dependent manner. The peptides that were identified are remarkably potent, macromolecular sized pore-formers at pH 5, while having little or no membrane activity at all at pH 7. Thus, we succeeded in “evolving” MelP5 for gain of function. All of the selected peptides possess exquisite pH sensitivities, with activities transitioning from ~0 to ~100% over one pH unit, centered on apparent pKa values of 5.5 to 5.8. This success in achieving a goal that we previously failed to achieve using a completely rational design approach27 highlights the power of synthetic molecular evolution.
Mechanism of action
All 10 of the positive peptides identified in the library have 5 or 6 acidic residues out of the 6 possible, despite the fact that only 17% of library members have 5 or 6 acidic residues. The peptide with six charges that we tested is somewhat less potent and has a lower pKa than the peptides with five charges (Figure 4B), suggesting that five negative charges arranged with helical spacing on an amphipathic helix are optimal for the observed pH-triggered activity. The fact that we observed various patterns of 5 acidic residues in the selected positives indicates that the pH sensitivity is due to the physical chemistry of folding and membrane binding. Yet, the specific preference of glutamate compared to aspartate, the overabundance of acidic residues is several positions, and the 100% conservation of glutamine at position 17 suggests that sequence-specific interaction may also play a role in this activity.
According to the Henderson-Hasselbalch equation, an equilibrium that simply reflects protonation of glutamate or aspartate should transition from 10% to 90% complete over two pH units and 1% to 99% over four pH units. However, we observe transitions in dextran leakage, secondary structure and binding by the pHD peptides from near 0% to near 100% over one pH unit (Figure 6), indicative of highly cooperative behavior. Furthermore, the apparent pKa values that we measure for the selected peptides range from 5.5 to 5.8, much higher than the pKa of ~ 3.5 to 4.0 expected for the free side chains for Glu or Asp. We hypothesize that the tight coupling between membrane partitioning, α-helix formation, and electrostatic repulsion between acidic sidechains drives both the upward shift in pKa, relative to that of the free sidechains, and the unexpectedly sharp transition. Potentially favorable electrostatic interactions between the basic lysine and histidine residues and some of the acidic sidechains may also contribute to the pH sensitivity.
To estimate the protonation state of the peptide, we used the Membrane Protein Explorer (MPEx)48 to predict free energies of membrane partitioning of the pHD peptides compared to MelP5. Assuming that bound peptides are 75% α-helix (36, Figure 5A), protonation of at least 4 or 5 of the acidic sidechains in the pHD peptides would be required for strong membrane partitioning that we observed at pH 5. Thus, we hypothesize that most of the acidic sidechains in the pHD peptides are cooperatively protonated with an apparent pKa of 5.5–5.8.
Implications for peptide design and applications
In previous work, we attempted to engineer pH sensitivity into the unique macromolecular poration activity of MelP5, but failed. Here we achieved the goal of pH-triggered macromolecular poration using a fundamentally different approach: synthetic molecular evolution, which is accomplished with orthogonal screening of a rationally designed, iterative, combinatorial peptide library. Despite being able to list similarities and differences, between the rationally designed and molecularly evolved peptides, we are not able to identify obvious logical flaws in the rational design approach based on any molecular principle. Specific design principles are not yet apparent from sequence comparison alone. Given the lack of complete understanding of structure-function relations for membrane active peptides in general, our work shows that synthetic molecular evolution is a powerful, and necessary, method to drive the discovery of novel peptides with specific membrane activities.
The peptides we have selected cause pH-triggered macromolecular poration, a property that could potentially be exploited in multiple ways. For example, there are applications in medicine where acidic environmental pH could be used to trigger the activity of the new peptides. One example is the acidic environment around solid tumors13 where tumor selective cytolytic activity could be triggered. A second example are acidified organelles, such as endosomes and lysosomes17,49–51, where selective cargo delivery into the cytosol could be enabled by pH-induced macromolecular poration. The work described here opens new doors to exploring such applications using peptides with activities that are triggered by a physiologically relevant decrease in pH.
Acknowledgments
This work was supported by NIH grants GM068619 (KH) and GM111824 (WCW). We thank Jing He, Taylor Fuselier, W. Berkeley Kauffman, Charles G. Starr and Hussain Badani for early brainstorming on the library design.
Reference List
- 1.Kakudo T, Chaki S, Futaki S, Nakase I, Akaji K, Kawakami T, Maruyama K, Kamiya H, Harashima H. Biochemistry. 2004;43:5618–5628. doi: 10.1021/bi035802w. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira S, van Rooy I, Kranenburg O, Storm G, Schiffelers RM. Inter J Pharamaceut. 2007;331:211–214. doi: 10.1016/j.ijpharm.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 3.Kullberg M, Owens JL, Mann K. J Drug Target. 2010;18:313–320. doi: 10.3109/10611861003663549. [DOI] [PubMed] [Google Scholar]
- 4.Lam JKW, Liang W, Lan Y, Chaudhuri P, Chow MYT, Witt K, Kudsiova L, Mason AJ. J Control Release. 2012;158:293–303. doi: 10.1016/j.jconrel.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shai Y, Oren Z. Peptides. 2001;22:1629–1641. doi: 10.1016/s0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- 6.Kauffman WB, Fuselier T, He J, Wimley WC. Trends Biochem Sci. 2015;40:749–764. doi: 10.1016/j.tibs.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komin A, Russell LM, Hristova KA, Searson PC. Adv Drug Deliv Rev. 2016 doi: 10.1016/j.addr.2016.06.002. http://dx.doi.org/10.1016/j.addr.2016.06.002. [DOI] [PubMed]
- 8.Wimley WC, Hristova K. J Membr Biol. 2011;239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soman NR, Baldwin SL, Hu G, Marsh JN, Lanza GM, Heuser JE, Arbeit JM, Wickline SA, Schlesinger PH. J Clin Invest. 2009;119:2830–2842. doi: 10.1172/JCI38842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sani MA, Separovic F. Acc Chem Res. 2016;49:1130–1138. doi: 10.1021/acs.accounts.6b00074. [DOI] [PubMed] [Google Scholar]
- 11.Andreev OA, Engelman DM, Reshetnyak YK. Molecular Membrane Biology. 2010;27:341–352. doi: 10.3109/09687688.2010.509285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantaleo MA, Nannini M, Lopci E, Castellucci P, Maleddu A, Lodi F, Nanni C, Allegri V, Astorino M, Brandi G, Di Battista M, Boschi S, Fanti S, Biasco G. Int J Oncol. 2008;33:443–452. [PubMed] [Google Scholar]
- 13.Gillies RJ, Robey I, Gatenby RA. J Nucl Med. 2008;49(Suppl 2):24S–42S. doi: 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 14.Duncan R. Nat Rev Drug Disc. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 15.Heitz F, Morris MC, Divita G. British Journal of Pharmacology. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bechinger B. Biomol Concepts. 2012;3:283–293. doi: 10.1515/bmc-2011-0067. [DOI] [PubMed] [Google Scholar]
- 17.Varkouhi AK, Scholte M, Storm G, Haisma HJ. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Raagel H, Saalik P, Pooga M. Biochim Biophys Acta. 2010;1798:2240–2248. doi: 10.1016/j.bbamem.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Majdoul S, Seye AK, Kichler A, Holic N, Galy A, Bechinger B, Fenard D. J Biol Chem. 2016;291:2161–2169. doi: 10.1074/jbc.M115.675033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallbrink M, Floren A, Elmquist A, Pooga M, Bartfai T, Langel U. Biochim Biophys Acta. 2001;1515:101–109. doi: 10.1016/s0005-2736(01)00398-4. [DOI] [PubMed] [Google Scholar]
- 21.Muro S. J Control Release. 2012;164:125–137. doi: 10.1016/j.jconrel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiolo JR, Ottinger EA, Ferrer M. J Am Chem Soc. 2004;126:15376–15377. doi: 10.1021/ja044867z. [DOI] [PubMed] [Google Scholar]
- 23.Yu HJ, Zou YL, Wang YG, Huang XN, Huang G, Sumer BD, Boothman DA, Gao JM. Acs Nano. 2011;5:9246–9255. doi: 10.1021/nn203503h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao L, Daniels J, Wijesinghe D, Andreev OA, Reshetnyak YK. J Control Release. 2013;167:228–237. doi: 10.1016/j.jconrel.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subbarao NK, Parente RA, Szoka FC, Nadasdi L, Pongracz K. Biochemistry. 1987;26:2964–2972. doi: 10.1021/bi00385a002. [DOI] [PubMed] [Google Scholar]
- 26.Parente RA, Nadasdi L, Subbarao NK, Szoka FC. Biochemistry. 1990;29:8713–8719. doi: 10.1021/bi00489a030. [DOI] [PubMed] [Google Scholar]
- 27.Wiedman G, Wimley WC, Hristova K. Biochim Biophys Acta. 2015;1848:951–957. doi: 10.1016/j.bbamem.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fendos J, Barrera FN, Engelman DM. Biochemistry. 2013;52:4595–4604. doi: 10.1021/bi400252k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parente RA, Nir S, Szoka FC., Jr Biochemistry. 1990;29:8720–8728. doi: 10.1021/bi00489a031. [DOI] [PubMed] [Google Scholar]
- 30.An M, Wijesinghe D, Andreev OA, Reshetnyak YK, Engelman DM. Proc Natl Acad Sci U S A. 2010;107:20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura Y, Takeda K, Ezawa R, Ishii J, Ogino C, Kondo A. J Nanobiotechnology. 2014;12:11. doi: 10.1186/1477-3155-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endoh T, Ohtsuki T. Adv Drug Del Rev. 2009;61:704–709. doi: 10.1016/j.addr.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Wiedman G, Herman K, Searson P, Wimley WC, Hristova K. Biochim Biophys Acta. 2013;1828:1357–1364. doi: 10.1016/j.bbamem.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mularski A, Wilksch JJ, Hanssen E, Strugnell RA, Separovic F. Biochim Biophys Acta. 2016;1858:1091–1098. doi: 10.1016/j.bbamem.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Wimley WC. ACS Chem Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krauson AJ, He J, Wimley WC. J Am Chem Soc. 2012;134:12732–12741. doi: 10.1021/ja3042004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiedman G, Fuselier T, He J, Searson PC, Hristova K, Wimley WC. J Am Chem Soc. 2014;136:4724–4731. doi: 10.1021/ja500462s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rathinakumar R, Walkenhorst WF, Wimley WC. J Am Chem Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krauson AJ, He J, Wimley AW, Hoffmann AR, Wimley WC. ACS Chem Biol. 2013;8:823–831. doi: 10.1021/cb300598k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atherton E, Sheppard RC. Solid phase peptide synthesis. IRL Press; Oxford: 1989. [DOI] [PubMed] [Google Scholar]
- 41.Wimley WC, Hristova K, Ladokhin AS, Silvestro L, Axelsen PH, White SH. J Mol Biol. 1998;277:1091–1110. doi: 10.1006/jmbi.1998.1640. [DOI] [PubMed] [Google Scholar]
- 42.Mayer LD, Hope MJ, Cullis PR. Biochim Biophys Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 43.Ganz T, Selsted ME, Lehrer RI. Eur J Haematol. 1990;44:1–8. doi: 10.1111/j.1600-0609.1990.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 44.Lehrer RI, Ganz T, Selsted ME. Cell. 1991;64:229–230. doi: 10.1016/0092-8674(91)90632-9. [DOI] [PubMed] [Google Scholar]
- 45.Brown KL, Hancock REW. Curr Opin Immunol. 2006;18:24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Bechinger B. Crit Rev Plant Sci. 2004;23:271–292. [Google Scholar]
- 47.Dempsey CE. Biochim Biophys Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 48.Snider C, Jayasinghe S, Hristova K, White SH. Protein Sci. 2009;18:2624–2628. doi: 10.1002/pro.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akinc A, Thomas M, Klibanov AM, Langer R. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 50.Erazo-Oliveras A, Muthukrishnan N, Baker R, Wang TY, Pellois JP. Pharmaceuticals (Basel) 2012;5:1177–1209. doi: 10.3390/ph5111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Dyke RW. Subcellular Biochem. 1996;27:331–360. doi: 10.1007/978-1-4615-5833-0_10. [DOI] [PubMed] [Google Scholar]
- 52.White SH, Wimley WC, Ladokhin AS, Hristova K. Methods Enzymol. 1998;295:62–87. doi: 10.1016/s0076-6879(98)95035-2. [DOI] [PubMed] [Google Scholar]