Abstract
Background:
The association between the tumor necrosis factor-alpha gene (TNF-a) -238G/A polymorphism and the breast cancer has been analyzed in several studies, but the results have been inconclusive. We then performed a meta-analysis to get a precise estimation of the association.
Methods:
Eight case–control studies with a total of 37,257 cases and 39,564 controls were identified by searching the ISI Web of Knowledge database and the PubMed database up to August 2014.
Results:
Overall, no association was found between TNF-alpha-238G/A polymorphism and breast cancer in any of genetic model (additive model OR = 1.06, 95%CI: 0.94–1.21, Pheterogeneity = .02; homozygous model OR = 1.04, 95%CI: 0.83–1.30, Pheterogeneity = .98; dominant model OR = 1.06, 95%CI: 0.92–1.21, Pheterogeneity = .01; recessive model OR = 1.04, 95%CI: 0.83–1.30, Pheterogeneity = .98). Furthermore, no significant association was identified when stratified by ethnicity (Caucasian, Asian).
Conclusion:
This meta-analysis indicated that the TNF-alpha-238G/A polymorphism is not associated with breast cancer risk in the overall population.
Keywords: breast cancer, meta-analysis, polymorphism, tumor necrosis factor alpha
1. Introduction
Breast cancer (BC) is the most common cancers worldwide, and new invasive breast cancer cases and deaths are reported in developed countries.[1] BC onset and progression are multifactorial processes, which are coming from the interactions among genetic, endocrine, and external environmental factors.[2] Recently, inflammation and the host immune response in the etiology of cancers have been investigated. The tumor necrosis factor-a (TNF-alpha) is a key pro-inflammatory cytokine which is secreted by macrophages and plays important roles in the cause and development of malignant diseases. A relevant study has reported[3] that lymph node involvement in breast cancer is correlated with the chronic expression of TNF-alpha; it also suggested the role of TNF-alpha in enhancing tumor cell metastasis. Furthermore, elevated levels of TNF-alpha in the circulation in breast cancer patients have a poor prognosis; TNF-alpha thus is a useful biomarker in cancers.[4,5] Several polymorphisms in the promoter region of TNF-alpha gene have been identified and implicated in the regulation of TNF-alpha transcription, such as TNF-alpha-308G/A polymorphism; it has been found as a risk factor for gastric and hepatocellular cancers.[6,7] The association between BC risk and TNF-alpha-238 polymorphism is still controversial. However, the currently published articles only related with the unified ethnicity and a modest sample size, and each of them might not reach a reliable conclusion, which indicated that a meta-analysis is needed to collect the data. Thus, we conducted a meta-analysis to combine all available studies and to validate whether the TNF-alpha-238G/A polymorphisms associate with BC susceptibility in this study.
2. Materials and methods
2.1. Retrieval and methods
We searched the ISI Web of knowledge and the PubMed database to identify all eligible articles that examined the association between TNF-a-238G/A polymorphism and breast cancer risk up to the August 2014. We searched the following keywords “tumor necrosis factor-a” or “TNF-a” or “TNF-alpha” or “TNF-a-238G/A” or “rs361525” and “polymorphism” or “SNP” or “gene mutation” or “genetic variants” and “breast cancer” or “breast tumor” or “breast carcinoma.” References of cited articles were reviewed to identify additional studied not indexed by Medline.
2.2. Inclusion criteria
The studies included in the meta-analysis were required to meet the following criteria: (1) a case–control study, (2) study examined the association between TNF-a-238G/A and breast cancer risk, (3) available genotype of both cases and controls, (4) genotype distribution of the control population must be in Hardy–Weinberg equilibrium (HWE).
2.3. Ethics statement
As this study is meta-analysis, so the ethical approval is not required, and the ethics committee of our hospital request that meta-analysis does not require ethical approval.
2.4. Data extraction
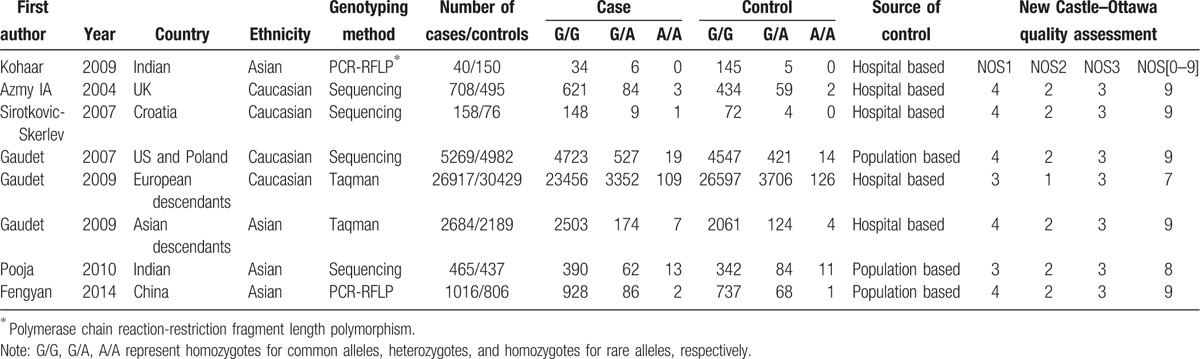
Two authors (QZ and GSZ) extracted carefully independently the information from all eligible studies based on the inclusion criteria. They discussed and resolved disagreement. If the 2 authors could not reach a consensus, a third author was consulted to resolve the disagreements. The following information of included studies was collected: first author's name, year of publication, country, ethnicity of individuals, genotyping method, number of cases/controls, numbers of genotyped cases and controls, and source of control.
The quality of the studies was evaluated according to Newcastle-Ottawa quality assessment scale.[8] In brief, 3 parameters were evaluated with a “star system”: the selection of the study groups (0 to 4 “stars”), the comparability of the groups (0 to 2 “stars”) and the ascertainment of either the exposure or outcome of interest for case–control or cohort studies respectively (0 to 3 “stars”). A questionnaire with multiple choices was associated with each parameter to define the number of stars. The maximum total score was 9 “stars” and represented the highest quality.
2.5. Statistical analysis
The meta-analysis evaluated the overall association between TNF-alpha-238G/A and the risk of breast cancer. Combined ORs and corresponding 95% confidence interval (CI) were calculated for an additive model (A vs G), a homozygous model (AA vs GG), a dominant model (AA + GA vs GG), and a recessive model (AA vs GA + GG). The combined OR was estimated using fixed effects (FE) models (Mantel-Haenszel) and random-effects (RE) models (DerSimonian and Laird). Heterogeneity was quantified by I2 metric, which is independent of the number of studies in the meta-analysis (I2 < 25% no heterogeneity; I2 = 25–50% moderate heterogeneity; I2 > 50% extreme heterogeneity). If there was a statistical difference in terms of heterogeneity (P < .1), a random-effect model was selected to pool the data. A fixed-effect model, otherwise, was employed. Because the number of included studies was < 10, we did not assess the publication bias. The Hardy–Weinberg equilibrium (HWE) was tested by the chi-square test. Meta-analysis and sensitivity analysis were carried out using the Review Manager 5.0 software, and P < .05 was considered a statistically significant result.
3. Results
3.1. Included studies
Eight relevant studies with a total number of 37,257 patients and 39,564 controls were included in this analysis.[9–15] Characteristics of the included studies are shown in Table 1. All studies used healthy volunteers or blood donors as control subjects. All the studies were in HWE (P > .05). Populations were categorized into Caucasian, Asian.
Table 1.
Characteristics of the included studies in the meta-analysis.
3.2. Effect of allele and subgroup analysis
Summary results of this meta-analysis for the association between the TNF-alpha-238G/A polymorphism and BC are shown in Table 2. Overall, no significant association was found between TNF-alpha-238G/A polymorphism and the risk of breast cancer (Figs. 1 and 2).
Table 2.
Summary of OR for TNF-a-238G/A and breast cancer.
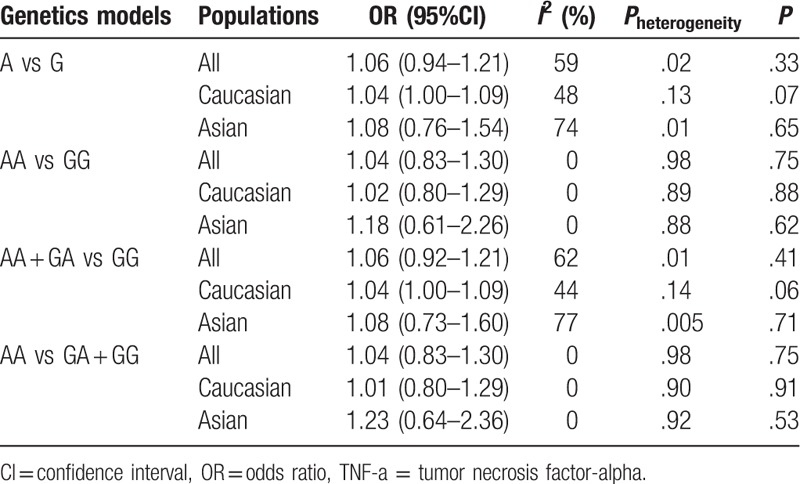
Figure 1.
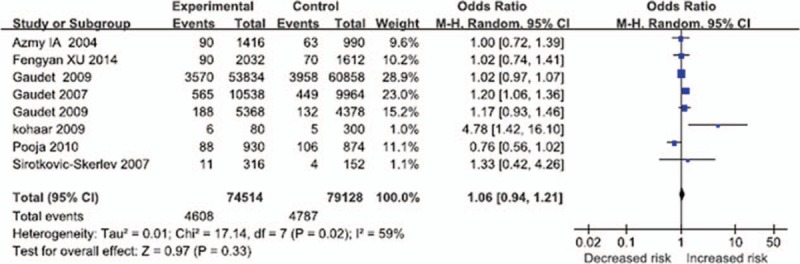
The association between tumor necrosis factor alpha-238G/A and the breast cancer in total populations (A vs G: OR = 1.06, 95% CI = 0.94–1.21, P = .33; P = .02, I2 = 59% for heterogeneity; random-effects model). CI = confidence interval.
Figure 2.
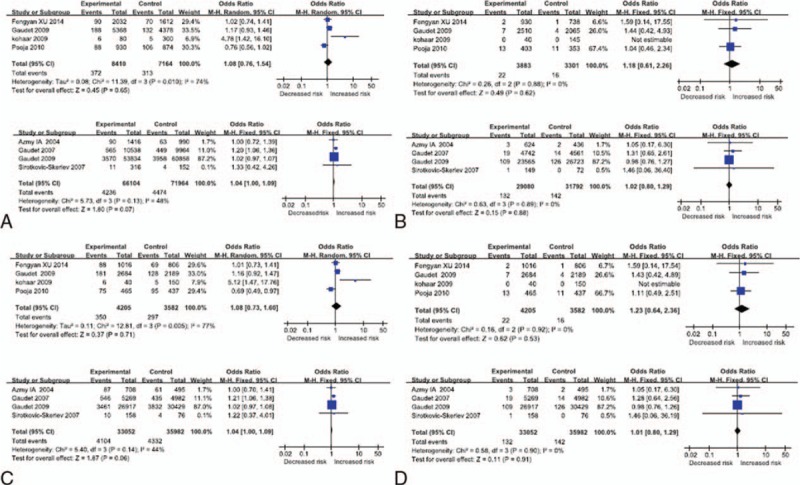
Forest plots for the TNF-a-238G/A polymorphism and breast cancer risk. (A) Additive model (A vs G) Asian, Caucasian; (B) homozygous model (AA vs GG) Asian, Caucasian; (C) dominant model (AA + GA vs GG) Asian, Caucasian; (D) recessive model (AA vs GG + GA) Asian, Caucasian. CI = confidence interval, TNF-a = tumor necrosis factor-alpha.
3.3. Test of heterogeneity
Overall comparisons were used to assess the heterogeneity. No heterogeneity existed between the included studies examined with the recessive model and the homozygote contrast; the heterogeneity P values were both 0.98. A fixed-effects model was used to combine the results obtained from the above models. Heterogeneity existed in the dominant model and the additive model (Pheterogeneity = .01 and .02, respectively); the random-effects model was used to combine the results (dominant model, P = .75, OR = 1.04, 95% CI = 0.83–1.30; additive model P = .33, OR = 1.06, 95% CI = 0.94–1.21). However, in the further stratified analyses by study populations, the summary ORs were 1.04 (95% CI = 1.00–1.09; P = .07, I2 = 48% for heterogeneity; fixed-effects model) and 1.04 (95% CI = 1.00–1.09; P = .06, I2 = 44% for heterogeneity; fixed-effects model) in Caucasian populations under the same comparisons, respectively. Heterogeneity existed in Asian populations in the additive model and the dominant model.
3.4. Sensitivity analysis
Sensitivity analysis was performed by deleting the single study that was involved in the meta-analysis each time, as well as the cumulative statistics from all comparisons in all patients, and the subgroups to explore the effect of the individual data on the combined ORs. For the TNF-alpha-238G/A, when the study of Gaudet et al[13] was removed in Caucasian populations, the P-value became significant in the dominant and additive models (P < .05); the Pheterogeneity value was insignificant (I2 = 0% and P > .10). The other results pattern was not impacted by any single study in all groups and subgroups in different genetic models (data not shown).
3.5. Publication bias
Because the number of included studies was <10, we did not assess the publication bias.
4. Discussion
The appearance of the breast cancer is determined by a combination of exogenous factors and genetics.[16–18] TNF-alpha, the most crucial inflammatory cytokine, has been involved in both the development and progression in experimental and human cancers. For the TNF-a study, A allele at -238G/A position in the promoter region was reported to influence TNF gene expression,[19] and these studies and related variant studies result in the increasing in BC study in recent decades. However, the results from these studies were ambiguous, for their unified ethnicity and small sample size; thus a meta-analysis is needed to achieve a more comprehensive and reliable conclusion on both variants in order to provide further insights into this debated subject.
We performed a meta-analysis of eight studies with a total of 37,257 breast cancer cases and 39,564 controls. Our results suggested that TNF-alpha-238G/A polymorphism is not associated with breast cancer risk in the overall population or when the analysis was stratified according to ethnicity. This is consistent with the findings of previous meta-analyses. But previous meta-analysis contained the limited studies, for instance.
Shen et al[20] reported only 3 studies which could not provide sufficient information. We identified further studies that were suitable for inclusion. Our meta-analysis leads us to draw a conclusion that our results tend to be more precise estimate of the relationship between the TNF-alpha-238G/A polymorphism and breast cancer risk than those of all previous ones. Heterogeneity existed in the dominant and additive models in all populations; we choose the random-effects models in dominant and additive models, which make the results relatively conservative. So we should cautiously explain the combined results of this analysis. Considering that the polymorphism frequencies might differ among ethnic groups, we performed a stratification analysis by ethnicity. In the ethnicity-specific meta-analysis, a separate analysis was performed in Caucasian and Asian populations. The results demonstrated that the TNF-alpha-238G/A polymorphism may not play an important role in breast cancer among different ethnicities.[21] No heterogeneity existed in Caucasian populations in the different genetic models. Heterogeneity existed in the additive model and the dominant model in Asian populations. Due to the Kohaar[9] small sample size, or other potential confounding factors, this research could be sources of heterogeneity.
Furthermore, this retrospective analysis has some limitations. First, there were only 8 studies that met our inclusion criteria, and the sensitivity analysis showed the negative results were unstable. Second, the genotyping method, such as the sequencing and PCR-RFLP, and experimental design (population-based controls and hospital-based controls) were different. Third, the allele frequency of mutation and the incidence rate were low, with particular regard to the absence of the TNF-alpha allele in some populations.[21]
In conclusion, our meta-analysis suggests that the TNF-alpha-238G/A polymorphism is not associated with the genetic susceptibility to breast cancer in the overall population. Further large, well-designed, and epidemiological studies are necessary to better clarify the role of the TNF-alpha -238G/A polymorphism and breast cancer susceptibility.
Footnotes
Abbreviations: BC = breast cancer, CI = confidence interval, FE = fixed effects, HWE = Hardy–Weinberg equilibrium, RE = random-effects, TNF-a = tumor necrosis factor-alpha.
QZ and GSZ equally contributed to this study and are co-first authors.
Funding: This study was supported by a grant from the National Natural Science Foundation of China (21173217) and The Health Authority of Dalian (2014142).
The authors have no conflicts of interest to disclose.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Welch DR, Steeg PS, Rinker-Schaeffer CW. Molecular biology of breast cancer metastasis. Genetic regulation of human breast cancer carcinoma metastasis. Breast Cancer Res 2000;2:408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Leek RD, Landers R, Fox SB, et al. Association of tumour necrosis factor alpha and its receptors with thymidine phosphorylase expression in invasive breast carcinoma. Brit J Cancer 1998;77:2246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ferrajoli A, Keating MJ, Manshouri T, et al. The clinical significance of tumor necrosis factor-alpha plasma level in patients having chronic lymphocytic leukemia. Blood 2002;100:1215–9. [PubMed] [Google Scholar]
- [5].Garcia-Tunon I, Ricote M, Ruiz A, et al. Role of tumor necrosis factor-alpha and its receptors in human benign breast lesions and tumors (in situ and infiltrative). Cancer Sci 2006;97:1044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang Y, Luo C, Feng R, et al. The TNF-alpha, IL-1B and IL-10 polymorphism and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol 2011;137:947–52. [DOI] [PubMed] [Google Scholar]
- [7].Lu PH, Tang Y, Li C, et al. Meta analysis of association of tumor necrosis factor alpha-308 gene promoter polymorphism with gastric cancer. Zhonghua Yu Fang Yi Xue Za Zhi 2010;44:209–14. [PubMed] [Google Scholar]
- [8].Stang A. Critical evaluation of the Newcastle–Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [9].Kohaar I, Tiwari P, Kumar R, et al. Association of single nucleotide polymorphisms in TNF-LTA locus with breast cancer risk in Indian population. Breast Cancer Res Treat 2009;114:347–55. [DOI] [PubMed] [Google Scholar]
- [10].Azmy IA, Balasubramanian SP, Wilson AG, et al. Role of tumour necrosis factor gene polymorphisms ((308 and (238) in breast cancer susceptibility and sevetity. Breast Cancer Res 2004;6:R395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sirotkovic-Skerlev M, Cacev T, Krizanac S, et al. TNF alpha promoter polymorphisms analysis in benign and malignant breast lesions. Exp Mol Pathol 2007;83:54–8. [DOI] [PubMed] [Google Scholar]
- [12].Gaudet MM, Egan KM, Lissowska J, et al. Genetic variation in tumor necrosis factor and lymphotoxin-alpha (TNF-LTA) and breast cancer risk. Hum Genet 2007;121:483–90. [DOI] [PubMed] [Google Scholar]
- [13].Gaudet MM, Milne RL, Cox A, et al. Five polymorphisms and breast cancer risk: results from the Breast Cancer Association Consortium. Cancer Epidemiol Biomarkers Prev 2009;18:1610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pooja S, Francis A, Bid HK, et al. Role of ethnic variations in TNF-α and TNF-β polymorphisms and risk of breast cancer in India. Breast Cancer Res Treat 2011;126:739–47. [DOI] [PubMed] [Google Scholar]
- [15].Xu F, Zhou G, Han S, et al. Association of TNF-α, TNFRSF1A and TNFRSF1B gene polymorphisms with the risk of sporadic breast cancer in northeast Chinese Han women. PLoS One 2014;9:e101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Simonds NI, Ghazarian AA, Pimentel CB, et al. Review of the gene–environment interaction literature in cancer: what do we know? Genet Epidemiol 2016;40:356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Benna C, Helfrich-Förster C, Rajendran S, et al. Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget 2017;8:23978–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Maroni P, Matteucci E, Bendinelli P, et al. Functions and epigenetic regulation of Wwox in bone metastasis from breast carcinoma: comparison with primary tumors. Int J Mol Sci 2017;18:pii: E75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shih CM, Lee YL, Chiou HL, et al. Association of TNF-alpha polymorphism with susceptibility to and severity of non-small cell lung cancer. Lung Cancer 2006;52:15–20. [DOI] [PubMed] [Google Scholar]
- [20].Shen C, Sun H, Sun D, et al. Polymorphisms of tumor necrosis factor-alpha and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 2011;126:763–70. [DOI] [PubMed] [Google Scholar]
- [21].Xu L, Yuan W, Sun H, et al. The polymorphisms of the TNF-α gene in multiple sclerosis?—a meta-analysis. Mol Biol Rep 2011;38:4137–44. [DOI] [PubMed] [Google Scholar]


