Summary
Single cell sequencing approaches are needed to characterize the genomic diversity of complex tumors, shedding light on their evolutionary paths and potentially suggesting more effective therapies. In this issue of Cancer Discovery, Francis et al., develop a novel integrative approach to identify distinct tumor subpopulations based on joint detection of clonal and sub-clonal events from bulk tumor and single nucleus whole genome sequencing, allowing them to infer a subclonal architecture. Surprisingly, the authors identify convergent evolution of multiple, mutually exclusive, independent EGFR gain of function variants in a single tumor. This study demonstrates the value of integrative single cell genomics and highlights the biological primacy of EGFR as an actionable target in GBM.
Text
Tumors are greater than the sum of their parts. Cancers are not merely a collection of cells, but rather a complex ecosystem composed of cells that mutate and adapt; interact and compete. The overall behavior of the tumor is shaped by the balance of cooperation and competition between individual cells that can vary in genetic, phenotypic and functional properties (1,2). Tumors evolve in response to treatments, including targeted therapies, through classical Darwinian selection and by cellular adaptations at the DNA, RNA or protein level. Determining the landscape of mutations shared between tumor cells and identifying mutations that arise to provide selective advantage to subclones could provide critical insight into the mechanisms of tumorigenesis, progression and evolution. Further, dissecting the mutational hierarchy of a tumor may identify the pre-existing seeds of drug resistance, thus becoming a key component of more effective, personalized cancer treatment.
Glioblastoma (GBM), the most common malignant primary brain cancer of adults, is one of the most molecularly characterized forms of cancer (3,4). Mutations in coding regions that occur at greater than 5% above background are likely to have already been detected (4). A compelling picture from these studies has emerged pointing directly at EGFR as a target. Fifty-seven percent of GBMs show evidence of mutation, rearrangement, alternative splicing or focal amplification of EGFR (3). A number of independent, structural rearrangements, including EGFRvIII and extracellular domain point mutations that confer gain of function and enhancement of tumorigenicity have been identified (3). Despite the strong evidence, skepticism regarding the importance of EGFR in GBM still remains, largely due to the failure of EGFR tyrosine kinase inhibitors (TKIs) in the clinic. Resistance to EGFR TKIs may be attributed to a number of factors, including: 1) sub-therapeutic dosing of EGFR TKIs (5,6); 2) bypass signaling pathways including through other receptor tyrosine kinases (7) and 3) reversible loss of extrachromosomal EGFRvIII DNA (8), all of which suggest the need for developing better EGFR-targeted treatments. Another, less well-understood aspect, is the role of intratumoral heterogeneity of EGFR mutants in GBM, and its impact on resistance.
GBM is one of the most heterogeneous of all cancers. Individual tumor cells vary in shape, size, and expression levels of signaling proteins including EGFRvIII (7). Fluorescent in situ hybridization studies also show that other oncogenic receptor tyrosine kinases (RTKs), including PDGFRα and MET can be co-amplified with EGFR, either in the same or different tumor cells (9,10). Genomic assessment of bulk tumor tissue cannot resolve the landscape of driver mutations at the single cell level. At present, it is unclear whether multiple EGFR mutations occur in individual cells, and whether they co-exist with amplification of wild type EGFR and/or other RTKs. Single cell sequencing techniques are crucial for answering these questions.
In this issue of Cancer Discovery, Matthew Meyerson’s and Keith Ligon’s laboratories present a novel single cell sequencing approach to identify unique, non-overlapping sub-clonal alterations from archived clinical samples, shedding new light on the clonal diversity of EGFR alterations within GBM. Francis et al. (11) examined bulk RNA sequencing analysis of 76 TCGA GBM samples with validated focal EGFR amplification and detected focal EGFR amplification coexisting with at least one EGFR variant, including structural alterations and/or missense extracellular domain mutations, in 71% of the samples. Deeper joint DNA and RNA sequencing analysis of an additional 25 cases revealed remarkable diversity of EGFR alterations. Transcripts encoding the same EGFR mutant were shown to possibly arise from distinct DNA templates within the tumor and some tumors contained multiple EGFR structural variants and missense mutations. In one particularly illuminating example, the possibility of up to 32 different possible clonal combinations based on five distinct EGFR genomic lesions was raised. It is possible that other mutational targets also have considerable diversity, suggesting that the overall clonal diversity could be much larger.
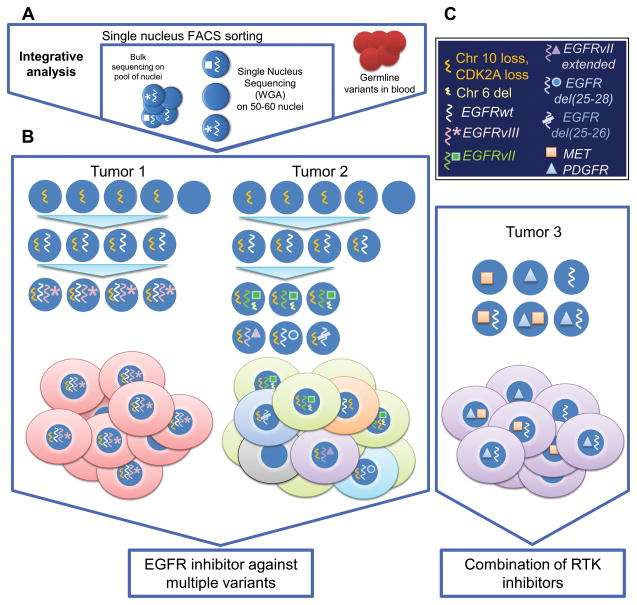
To begin to dissect the clonal and subclonal diversity of EGFR, Francis and colleagues developed an integrative method to study two GBM samples with focal EGFR amplification in depth (11). This novel integrative approach, summarized in Fig. 1A, coupled single nucleus sequencing with bulk tumor analysis, enabling the authors to estimate the clonality of somatic mutations and to infer a subclonal architecture of the tumors.
Figure 1. Integrative analysis of single nucleus whole genome sequencing and bulk tumor tissue enables detection of the subclonal architecture of tumors.
A) Schematic view of the analytic approach. B) A schematic view of illustrative GBM cases. Tumor 1 shows linear evolution of a GBM whose cells relatively uniformly contain both amplified wild type EGFR and EGFRvIII. Tumor 2 shows a far more complex evolutionary path, characterized by emergence of multiple, independent, mutually exclusive EGFR variants. Tumor 3 represents an alternative scenario, in which individual GBM cells vary in their amplification of EGFR, PDGFRα and MET. C) Genomic alterations: legend.
Each of the two tumors revealed a strikingly different path to oncogenic EGFR signaling (Fig. 1B). All of the tumor nuclei from tumor 1 contained a mixture of both wild type EGFR and EGFRvIII. EGFRvIII arises from an in-frame genomic deletion of exons 2–7 (7). Further, the common amplification boundaries and the EGFRvIII deletion breakpoints are shared among the tumor nuclei, suggesting that the amplification of EGFR preceded the emergence of the EGFRvIII mutant. The levels of EGFRvIII DNA varied greatly between cells, consistent with the possibility that extrachromosomal EGFRvIII DNA levels could be responsive to environmental cues (8). For the portion of the tumor sampled here, the path to oncogenic EGFR signaling was relatively linear. Based on shared clonal features in addition to EGFRvIII, the authors were able to use a chromothriptic event involving chromosomes 1, 4, 6 to define the point at which a minor subclone split from the major tumor population.
For tumor 2, integrated analysis revealed a very different evolutionary course. Multiple amplified EGFRs were detected, including: 1) wild type EGFR; 2) EGFRvII, which contains a deletion of exons 14 and 15; 3) EGFRvII-extended, a novel deletion that extends from exon 14 to include alternative exon 16; 4) a C-terminal EGFR truncation and 5) an EGFR harboring deletion of exons 25–26. Integrated analysis of these single cell EGFR variants with homozygous and hemizygous deletions detected in bulk tumor, revealed a far more remarkable, non-linear and surprising path to oncogenic EGFR. Unlike tumor 1, all of the EGFR variants were mutually exclusive of wild type EGFR amplification and of each other. This convincing demonstration of convergent evolution of independent EGFR gain of function variants in a single tumor provides compelling independent evidence for the importance of altered EGFR in GBM.
It is possible that intratumoral heterogeneity may be even greater than initially suspected. In some cases like tumor 1, the path to oncogenic EGFR is relatively linear, whereas in examples like tumor 2, convergent evolution of independent EGFR variants may drive oncogenic signaling. These findings have important clinical implications. Tumor composition may be an emergent property arising from the interplay of competition and cooperation between tumor cells differing in genetic and functional properties. This emergent balance can potentially be shaped by nutrient limitations, regional influences and the capability of tumor cells to signal to each other (7). Personalizing therapies for patients based on molecular composition may therefore depend on being able to detect these mutational hierarchies and use them to guide more effective therapies, including upfront combinations (Fig. 1B).
Like all good studies, this paper raises a number of important questions for future study. The extent of clonal diversity of EGFR mutants in GBM, and their functional and therapeutic properties needs to be further characterized. It is interesting to note that EGFRvII enhanced sensitivity to EGFR TKIs. The spectrum of EGFR variants, and extracellular domain missense mutations will need to be further characterized to determine if these lesions sensitize tumor cells to ATP competitive TKIs, EGFR-targeted antibodies, and or combinations. Further, EGFR variant co-expression and functional interaction with other RTK alterations needs to be further clarified to determine whether multiple RTK inhibitors will be needed (Fig. 1, tumor 3). This study examined samples that were less than 1cm. How diverse are tumor cells across different regions of a tumor?
EGFR mutations in GBM occur almost exclusively in the extracellular domain of the receptor. In contrast, in tumors from other parts of the body, the EGFR mutations occur primarily in the kinase domain of the receptor (4). Is there a similar convergent evolution of independent EGFR mutations in these other cancer types? Most importantly, future studies will be needed to assess how targeted treatments alter the molecular composition of tumors, possibly helping to anticipate and develop therapies that suppress drug resistance. The future of personalized cancer therapy will depend on treating tumors as an ecosystem, not as a collection of individual cells. The approach developed by Francis and colleagues (11) provides an important path forward.
Acknowledgments
Beatrice Gini is supported by The European Commission (PIOF-GA-2010-271819). Paul S. Mischel is supported by the Ludwig Institute for Cancer Research and grants from National Institute for Neurological Diseases and Stroke (NS73831), the National Cancer Institute (CA151819), The Ben and Catherine Ivy Foundation and Defeat GBM Research Collaborative, a subsidiary of National Brain Tumor Society.
Footnotes
Disclosure of Potential Conflicts of Interest:
No potential conflicts of interest were disclosed.
References
- 1.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annual review of pathology. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 2.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 3.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkovich KJ, Hariono S, Garske AL, Zhang J, Blair JA, Fan QW, et al. Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer discovery. 2012;2:450–7. doi: 10.1158/2159-8290.CD-11-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer discovery. 2012;2:458–71. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annual review of pathology. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 8.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer cell. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3041–6. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis JM, Zhang CZ, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]