Abstract
The discovery and confirmation of the female urinary microbiota in 2012 provided opportunities to improve insight into lower urinary tract disorders in women, including UTI and urgency urinary incontinence. Now, research in 2016 has shown that expanded culture techniques enable improved uropathogen detection and confirm that bacteria detected by culture-independent methods are alive.
Nearly every clinician assesses urinary health from time to time; for example, ordering and interpreting a urine analysis and/or urine culture to detect a UTI. Clinicians providing care for women with lower urinary tract (LUT) disorders have also relied on these traditional urinary assessments in order to evaluate, diagnose, and treat affected patients. However, all of these assessments are based on the assumption that the healthy female urinary bladder is sterile, an assumption that we now know to be incorrect. Since 2012, when microbes were detected in the female urinary bladder (the female urinary microbiota)1, additional and confirmatory evidence has clearly documented the presence of a living bacterial community in the bladders of women with and without lower urinary symptoms2–6. This major paradigm shift raises many questions regarding the role of the female urinary microbiota in lower urinary tract health and disease.
Much more is known about the microbiota of other organs than that of the bladder. For example, the well-studied gut microbiota are known to vary considerably based on dietary intake and BMI. Studies that link the gut microbiota with CNS function highlight the biological importance of microbial niches throughout the human body. The urinary microbiota holds similar potential, especially given the well-known connections between brain and bladder function7.
Three papers published in 2016 have addressed several clinically relevant questions in the field. Karstens and co-workers8 provided confirmatory evidence of the presence of bacterial communities in the bladders of women with and without lower urinary tract symptoms. In a carefully controlled study, these investigators collected catheterized urine samples from clinically well-characterized women (mean age approximately 58 years) with urgency urinary incontinence (UUI, n=10) and nine women without such symptoms. The investigators reported detection of relevant bacterial sequences in 95% of samples, and a median DNA content of 95fg/mL without group differences between women with and without symptoms. They also observed a higher number of operational taxonomic units (OTUs) in samples from women with UUI (mean = 49) compared with those without, although these data were not statistically significant (mean = 39 (P =0.2)), group differences in OTU abundance (In UUI, nine more abundant and five less abundant, compared with controls), and significant individual variation in the number of bacterial families (range 2–49) detected per sample, as well as the diversity and richness of each sample. Overall, they concluded that increased symptom distress and urgency incontinence episodes correlated with the characteristics of the urinary microbial community, which could be clinically important.
The work of Karstens and colleagues8 provides important and independent confirmation of work previously published by Pearce et al.4 who first described differences in the urinary microbiota of women with UUI compared with those without such symptoms. Despite the small size of Karstens and colleagues'8 cohorts, the findings are strengthened by the careful characterization and matching of the participants. In addition, the proportion of samples that were sequenced is high; this strength would have been further enhanced by the concomitant use of enhanced culture-based techniques2,3,5 to confirm that the detected DNA arose from living microbes.
A complementary paper by Thomas-White and co-workers9 used pre-existing samples collected in an NIH study designed to evaluate the treatment outcomes of well-characterized women enrolled in a multicentre prospective randomized trial (the Value of Urodynamic Evaluation (ValUE) study) to assess the role of preoperative urodynamic testing before surgery for stress urinary incontinence (SUI). Women with concomitant UUI were included, as clinically appropriate. The study relied on samples (mostly obtained through voiding (n = 174), but some obtained through catheterization (n=23)) obtained at baseline, before SUI treatment. Most (86%) of the 197 samples contained detectable levels of bacterial DNA. An important clinical message was that no association with SUI symptoms was observed, in agreement with the fact that SUI and UUI have different aetiologies. Similar to data from previous studies, the community characteristics — including diversity and organism predominance — of the urinary microbiota were associated with UUI symptoms. In addition, the large sample size meant that these investigators were also able to detect an association between hormonal status and BMI. Finally, they demonstrated that increased diversity of the urinary microbial community was associated with a concomitant lower frequency of Lactobacillus in clinically postmenopausal women who are not taking exogenous oestrogen.
A third paper by Price and co-workers5, addressed the clinical relevance of bacterial members of the urinary microbiota, focusing on the ability of the standard urine culture protocol to detect microorganisms. The standard protocol has been refined to grow certain uropathogens, especially E. coli. However, standard urine culture conditions are not ideal for many other known human uropathogens. Hilt and co-workers2 had previously described enhanced urine culture techniques that were established to grow organisms detected previously by sequencing. In 2016, Price et al.5 extended this work to a clinically relevant population of women based on their self-reported UTI. A key message of this paper was the low rate of uropathogen detection using standard urine culture techniques. Using baseline catheterized samples from 150 adults attending urogynaecology clinics, standard urine culture failed to detect 67% of uropathogens overall and 50% in participants with severe urinary symptoms. The investigators evaluated a variety of culture conditions to determine the optimal urine culture method that could be incorporated into any clinical laboratory for optimizing the detection of uropathogens. They reported that 100 μL of urine plated onto Blood (BAP), Colistin Naladixic Acid (CNA), and MacConkey agars in 5% CO2 for 48 h resulted in detection of 84% of all uropathogens, versus just 33% with the commonly used standard urine protocol (FIG 1).
Figure 1. Urine is not sterile.
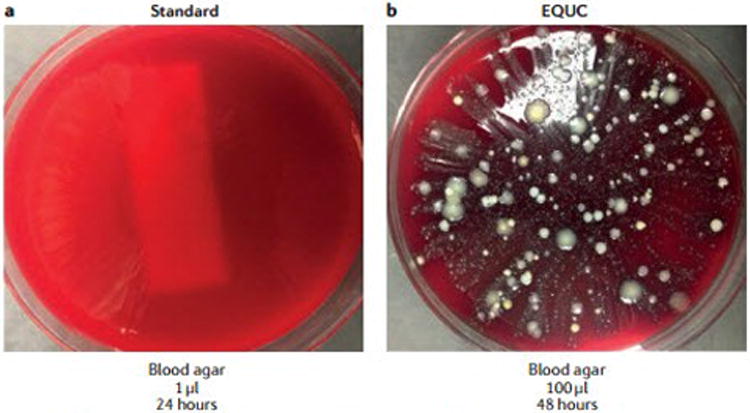
This urine sample was obtained by transurethral catheter from a woman seeking urogynaecology care, a | 1 μl urine was spread on a blood agar plate and incubated for 24 h at 35°C at ambient atmosphere. This technique is part of the standard urine culture protocol. b | 100 μl urine from the same patient was spread on a blood agar plate and incubated at 35 °C for 48h. Increased growth of bacterial colonies can be seen, demonstrating that the urine sample was not sterile, despite being obtained directly via a transurethral catheter. This technique is one of many growth conditions that comprise the enhanced quantitative urine culture (EQUC) protocol. Reprinted with permission from College of American Pathologists. CAP Today. Aug. 2016.
Clinicians face a number of challenges when caring for women with lower urinary tract symptoms. The simple dichotomy of ‘infection’ or ‘no infection’ does not incorporate evidence documenting the existence of the female urinary microbiota. The spectrum of health, dysbiosis, and disease likely relates to the urinary microbial community, as it does to other human microbial niches. Reliance on standard urine culture alone might limit the information available to physicians; the additional information available from enhanced urine culture techniques helps inform clinicians of the status of the urinary microbiota, including the presence of uropathogens that might be missed using standard urine culture protocols. The robust information generated by enhanced urine culture techniques provide an opportunity to advance clinical care and refine best practices in antibiotic stewardship and avoid use of interventions that wipe out bacterial communities that have a favourable biological function.
Key advances.
Most adult women have a detectable community of bacteria in their urine1
The characteristics of the female urinary microbiota relate to certain common lower urinary tract conditions, notably urgency urinary incontinence8
The standard urine culture is considerably limited in detection of organisms that make up the female urinary microbiota5
Enhanced quantitative urine culture techniques confirm that the DNA of the organisms detected by sequencing technology are living and cultivatable5
The female urinary microbiota seems to be associated with probability of successful treatment in certain women with urgency urinary incontinence9
Acknowledgments
The authors would like to thank the members of the Loyola Urinary Education and Research Collaborative for their contributions to the work described, in particular Dr Paul Schreckenberger from the Loyola Department of Pathology, for help in putting together this manuscript. A.J.W. and L.B. have been supported by NIH grants 2U10 HD41250, U01 DK58229, R21 DK097435, R56 DK104718, P20 DK108268, R01 DK104718, a translational grant from the Falk Foundation and by RFC LU206998 from Loyola University Chicago. A.J.W. has received funding for an Investigator Initiated Studies VESI-12D01 and MYRB-15A01 from Astellas Scientific and Medical Affairs.
Our funding sources have had no role in design or conduct of our studies; collection, management, analysis, and interpretation of our data, or in preparation, review, or approval of this or any other manuscript.
Competing interests statement: Dr Brubaker has received editorial honoraria from UpToDate.
Dr Wolfe has received research support from Astellas Scientific and Medical Affairs for urinary microbiome research. Both authors have received funding from the NIH and Loyola University Chicago for urinary microbiome research.
Footnotes
Author contributions Both authors researched data for article, made substantial contributions to discussions of content, wrote the article, and reviewed and edited the manuscript before submission.
Contributor Information
Linda Brubaker, Departments of Obstetrics & Gynecology and Urology, Loyola University Chicago Stritch School of Medicine, 2160S 1st Ave, Maywood, Illinois 60153, USA.
Alan J. Wolfe, Department of Microbiology and Immunology, Loyola University Chicago Stritch School of Medicine, 2160S 1st Ave, Maywood, Illinois 60153, USA
References
- 1.Wolfe AJ, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 2012;50:1376–1383. doi: 10.1128/JCM.05852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilt EE, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khasriya R, et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol. 2013;51:2054–2062. doi: 10.1128/JCM.03314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearce MM, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio. 2014;5:e01283–14. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price TK, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54:1216–1222. doi: 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas-White KJ, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J. 2016;27:723–733. doi: 10.1007/s00192-015-2847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tadic SD, Griffiths D, Schaefer W, Cheng CI, Resnick NM. Brain activity measured by functional magnetic resonance imaging is related to patient reported urgency urinary incontinence severity. J Urol. 2010;183:221–228. doi: 10.1016/j.juro.2009.08.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karstens L, et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol. 2016;6:78. doi: 10.3389/fcimb.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas-White KJ, et al. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am J Obstet Gynecol. 2016 doi: 10.1016/j.ajog.2016.07.049. http://dx.doi.org/10.1016/j.ajog.2016.07.049. [DOI] [PMC free article] [PubMed]


