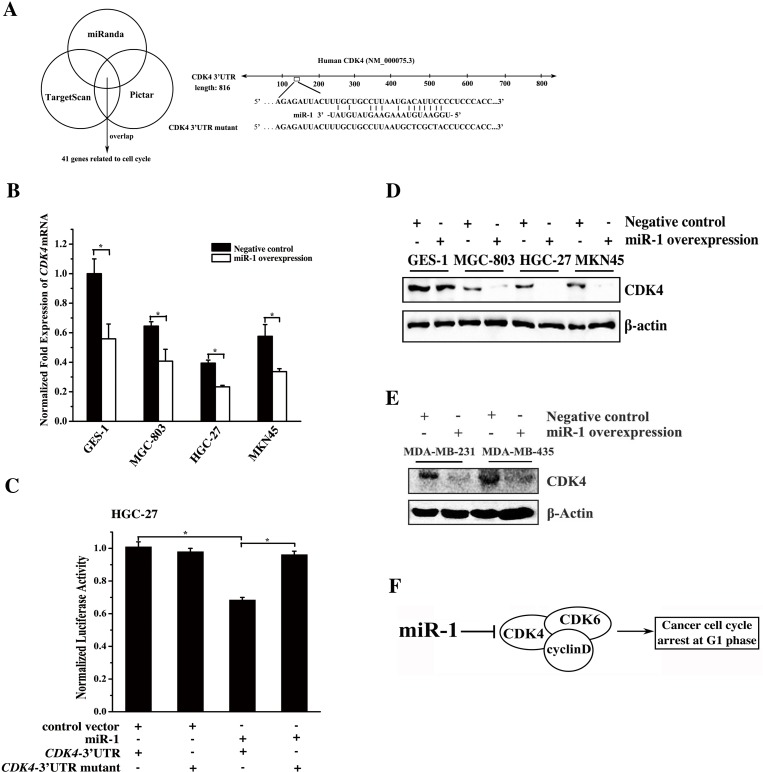
Figure 3. Mechanism of gastric and breast cancer cell cycle regulation mediated by miR-1.
A. Prediction of miR-1 target genes. The miR-1 targets related to the cell cycle were predicted using the miRanda, TargetScan and PicTar algorithms. CDK4 was predicted to be a target gene of miR-1. B. Expression levels of endogenous CDK4 in response to miR-1 overexpression in cells. Cells were transfected with the miR-1 precursor or the negative control. Twenty hours later, the level of CDK4 mRNA was detected using quantitative real-time PCR. C. Interaction between miR-1 and CDK4. The miR-1 construct or the vector only and the luciferase plasmid containing the 3′UTR of CDK4 mRNA or its mutant were co-transfected into HGC-27 cells. Then the luciferase activities were detected. The Renilla luciferase values were normalized to the firefly luciferase values. D. Western blot analysis of CDK4. Cells were treated with 30 nM of the miR-1 precursor or the negative control RNA for 48 h and whole-cell extracts were analyzed by Western blot analysis. β-actin was used for protein loading correction. E. Effects of miR-1 overexpression on its target gene expression in breast cancer cells. Breast cancer cells were transfected with 30 nM of the miR-1 precursor or the negative control miRNA. After 48 h, the expression of CDK4 in the cells was analyzed by Western blot. β-actin was used as a control. F. Model for the miR-1-mediated pathway in the cell cycle of cancer cells. Statistically significant differences are indicated with asterisks (*, p < 0.05; ** p < 0.01).