Abstract
We have analyzed the CTF1 gene, identified in a screen for mutants with decreased chromosome transmission fidelity and shown to correspond to the previously identified chl1 mutation. Chl1 null mutants exhibited a 200-fold increase in the rate of chromosome III missegregation per cell division, and near wild-type rates of marker homozygosis on this chromosome by mitotic recombination. Analysis of the segregation of a marker chromosome indicated that sister chromatid loss (1:0 segregation) and sister chromatid non-disjunction (2:0 segregation) contributed equally to chromosome missegregation. A genomic clone of CHL1 was isolated and used to map its physical position on chromosome XVI. Nucleotide sequence analysis of CHL1 revealed a 2.6 kb open reading frame with a 99 kd predicted protein sequence that contained two PEST sequences and was 23% identical to the coding region of a nucleotide excision repair gene, RAD3. Domains of homology between these two predicted protein sequences included a helix-turn-helix motif and an ATP binding site containing a helicase consensus. Mutants lacking the CHL1 gene product are viable and display two striking, and perhaps interrelated, phenotypes: extreme chromosome instability and a delay in cell cycle progression in G2/M. This delay is independent of the cell cycle checkpoint that requires the function of the RAD9 gene.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bond J. F., Fridovich-Keil J. L., Pillus L., Mulligan R. C., Solomon F. A chicken-yeast chimeric beta-tubulin protein is incorporated into mouse microtubules in vivo. Cell. 1986 Feb 14;44(3):461–468. doi: 10.1016/0092-8674(86)90467-8. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Foury F., Lahaye A. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 1987 May;6(5):1441–1449. doi: 10.1002/j.1460-2075.1987.tb02385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1988 Mar;52(1):70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B., Carbon J. Toxic effects of excess cloned centromeres. Mol Cell Biol. 1986 Jun;6(6):2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring S. L., Connelly C., Hieter P. Positional mapping of genes by chromosome blotting and chromosome fragmentation. Methods Enzymol. 1991;194:57–77. doi: 10.1016/0076-6879(91)94007-y. [DOI] [PubMed] [Google Scholar]
- Haber J. E. Bisexual mating behavior in a diploid of Saccharomyces cerevisiae: evidence for genetically controlled non-random chromosome loss during vegetative growth. Genetics. 1974 Nov;78(3):843–858. doi: 10.1093/genetics/78.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harosh I., Naumovski L., Friedberg E. C. Purification and characterization of Rad3 ATPase/DNA helicase from Saccharomyces cerevisiae. J Biol Chem. 1989 Dec 5;264(34):20532–20539. [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
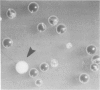
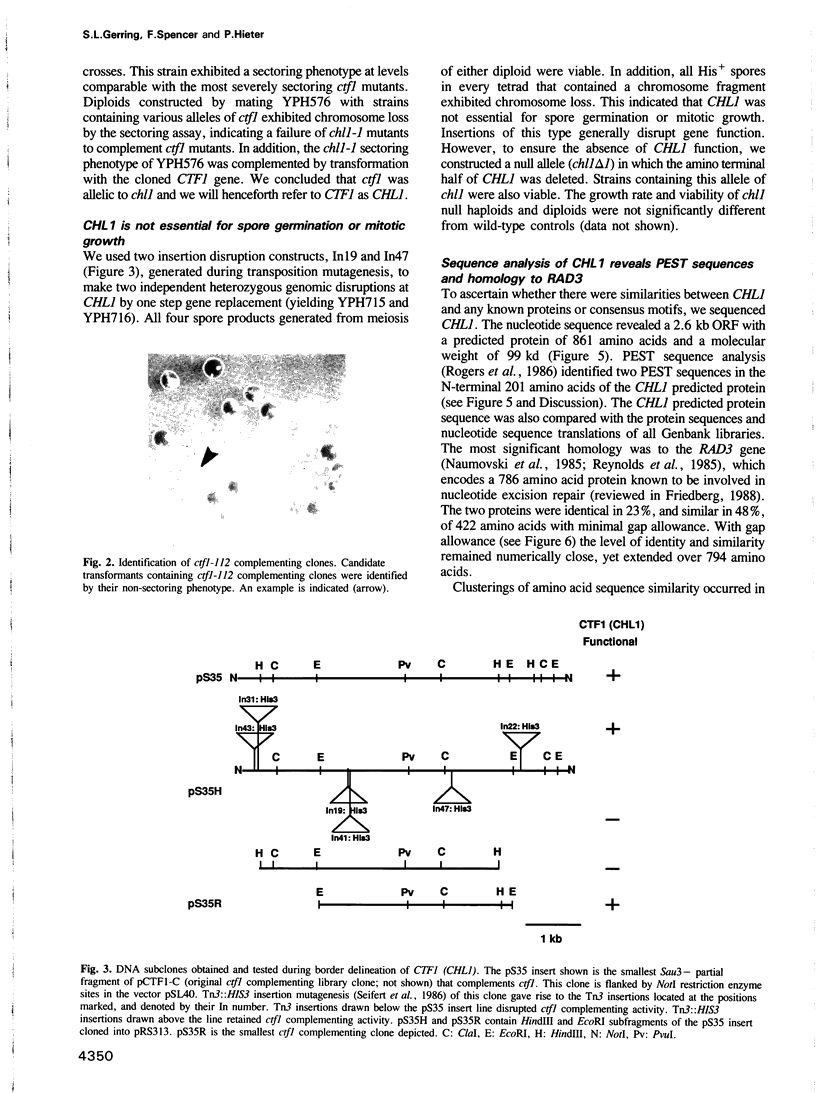
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hutter K. J., Eipel H. E. Flow cytometric determinations of cellular substances in algae, bacteria, moulds and yeasts. Antonie Van Leeuwenhoek. 1978;44(3-4):269–282. doi: 10.1007/BF00394305. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- Koshland D., Rutledge L., Fitzgerald-Hayes M., Hartwell L. H. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987 Mar 13;48(5):801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Liras P., McCusker J., Mascioli S., Haber J. E. Characterization of a mutation in yeast causing nonrandom chromosome loss during mitosis. Genetics. 1978 Apr;88(4 Pt 1):651–671. [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash R., Tokiwa G., Anand S., Erickson K., Futcher A. B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988 Dec 20;7(13):4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Chu G., Berg P., Friedberg E. C. RAD3 gene of Saccharomyces cerevisiae: nucleotide sequence of wild-type and mutant alleles, transcript mapping, and aspects of gene regulation. Mol Cell Biol. 1985 Jan;5(1):17–26. doi: 10.1128/mcb.5.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. Analysis of the essential and excision repair functions of the RAD3 gene of Saccharomyces cerevisiae by mutagenesis. Mol Cell Biol. 1986 Apr;6(4):1218–1227. doi: 10.1128/mcb.6.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S. Yeast chromosome replication and segregation. Microbiol Rev. 1988 Dec;52(4):568–601. doi: 10.1128/mr.52.4.568-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Reed S. I. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980 Jul;95(3):561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S. I. Conservation of mitotic controls in fission and budding yeasts. Cell. 1989 Apr 21;57(2):295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Georges G. E., Solomon F., Botstein D. Insertions of up to 17 amino acids into a region of alpha-tubulin do not disrupt function in vivo. Mol Cell Biol. 1987 Oct;7(10):3799–3805. doi: 10.1128/mcb.7.10.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Reynolds P., Prakash S., Prakash L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol Cell Biol. 1989 May;9(5):1882–1896. doi: 10.1128/mcb.9.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H. S., Chen E. Y., So M., Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Feb;83(3):735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990 Feb;124(2):237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Matson S. W., Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Weber S., Prakash S. The RAD3 gene of Saccharomyces cerevisiae encodes a DNA-dependent ATPase. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6045–6049. doi: 10.1073/pnas.84.17.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath D., Davis R. W., Connelly C., Hieter P. Physical mapping of large DNA by chromosome fragmentation. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6027–6031. doi: 10.1073/pnas.85.16.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]