Figure 1. DLL4 and JAG1 activated Notch signalling and affected sprouting angiogenesis.
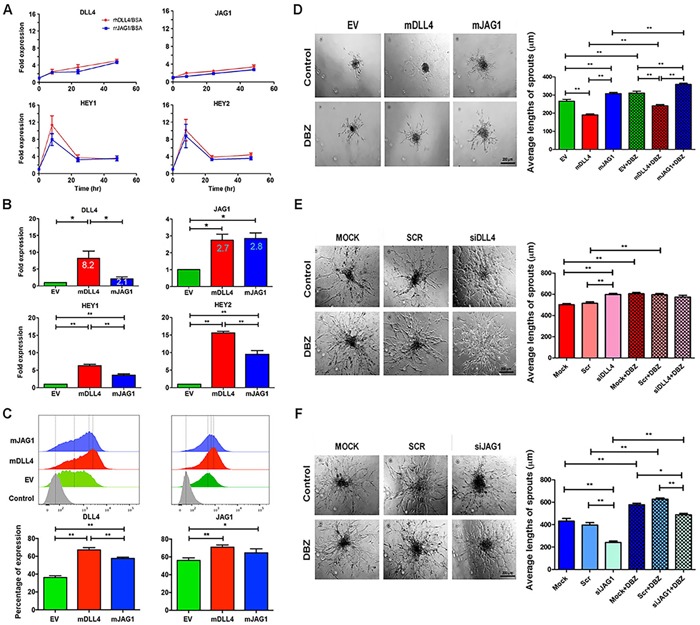
in vitro. A. Expression profile of Notch target genes in HMEC-1 stimulated with rhDLL4 or rrJAG1 over a time course. QPCR was used to determine mRNA levels of DLL4, JAG1, HEY1 and HEY2. Fold changes were obtained by normalizing against the EV control.B. Expression profile of Notch target genes in HMEC-1 over-expressing mDLL4 or mJAG1 by retrovirus transductions. QPCR was used to determine mRNA levels of DLL4, JAG1, HEY1 and HEY2 (ANOVA with Bonferroni's post-test). C. Expression of endogenous hDLL4 and hJAG1 proteins in parental HMEC-1 cells (GFP-negative) sorted from co-culture of EV-, mDLL4- or mJAG1-overexpressing HMEC-1 with an equal amount of parental HMEC-1 cells by FACS analysis (ANOVA with Bonferroni's post-test). D. Effect of mDLL4 and mJAG1 expressed in HMEC-1 cells on sprouting in HMEC-1 spheroids and treated with DBZ. Average lengths of three longest sprouts were calculated for statistical test (ANOVA with Bonferroni's post-test). Graphs are means of 3 independent experiments. E. Effect of knockdown of DLL4 in HMEC-1 cells by specific siDLL4 on sprouting in HMEC-1 spheroids. Average lengths of three longest sprouts were calculated for statistical test (ANOVA with Bonferroni's post-test). Graphs are means of 3 independent experiments. F. Effect of knockdown of JAG1 in HMEC-1 cells by specific siJAG1 on sprouting in HMEC-1 spheroids. Average lengths of three longest sprouts were calculated for statistical test (ANOVA with Bonferroni's post-test). Graphs are means of 3 independent experiments. *P<0.05, ** P<0.01.
