Abstract
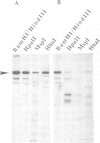
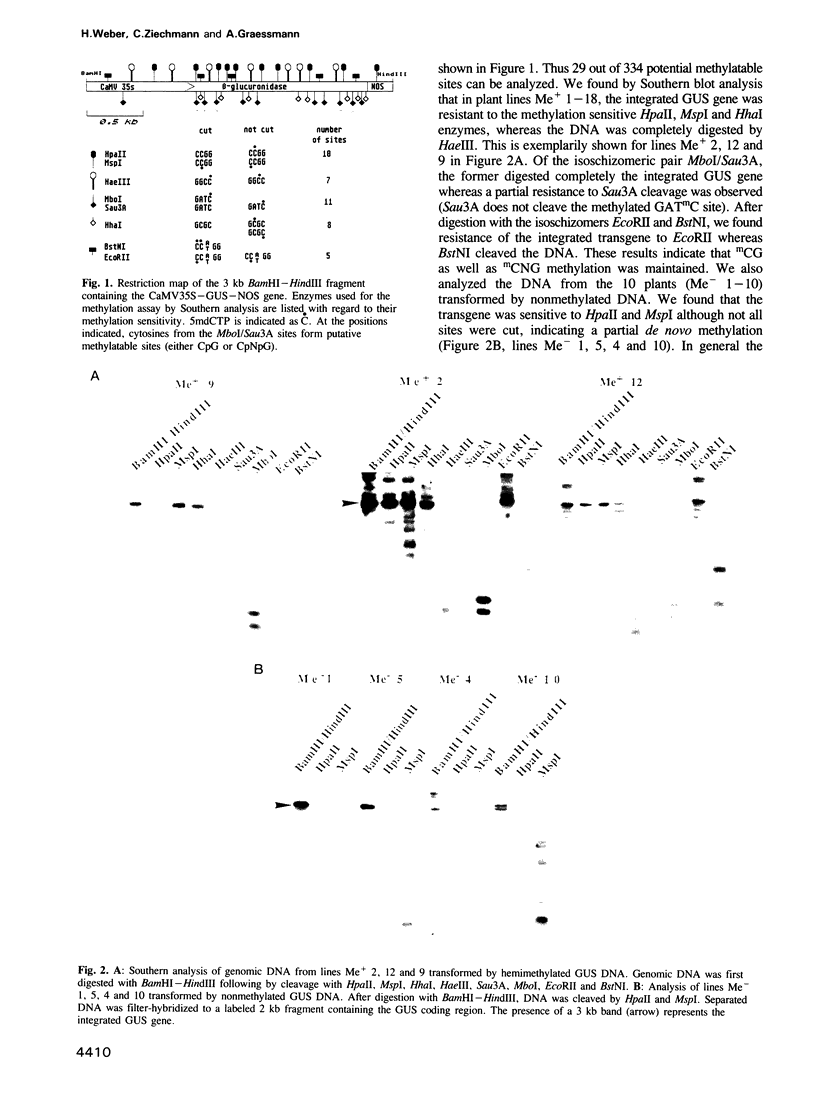
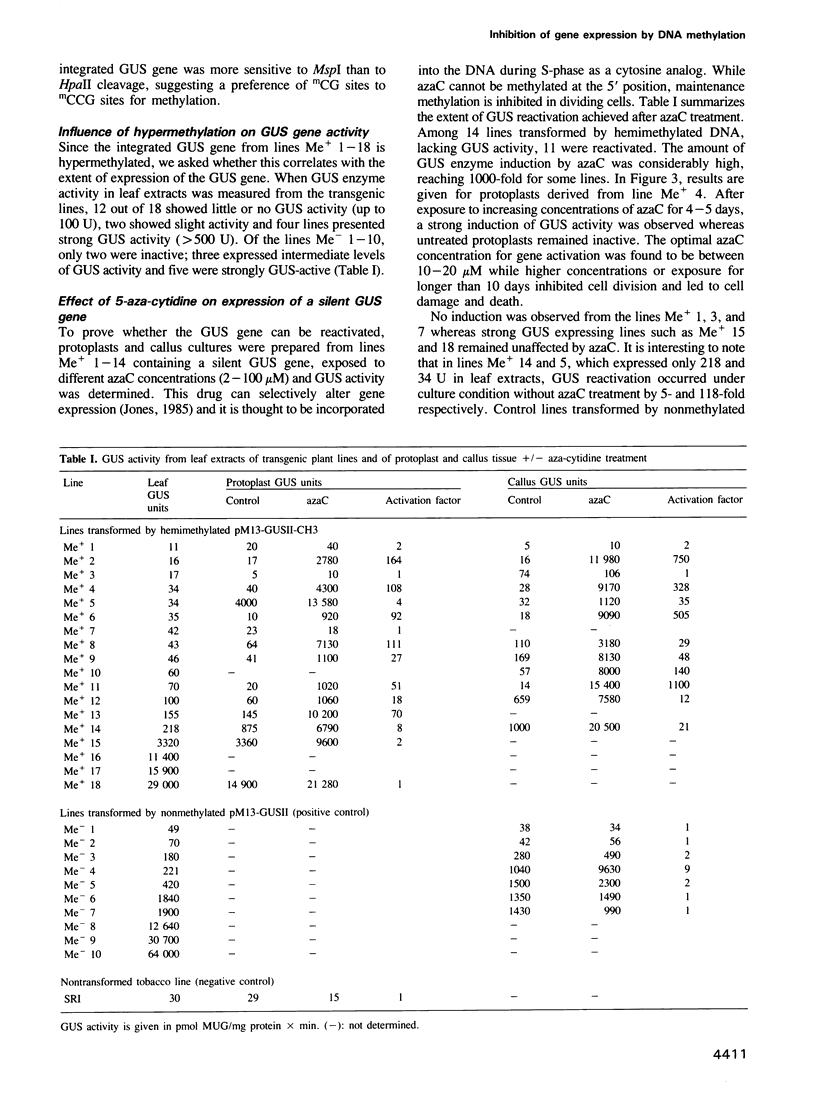
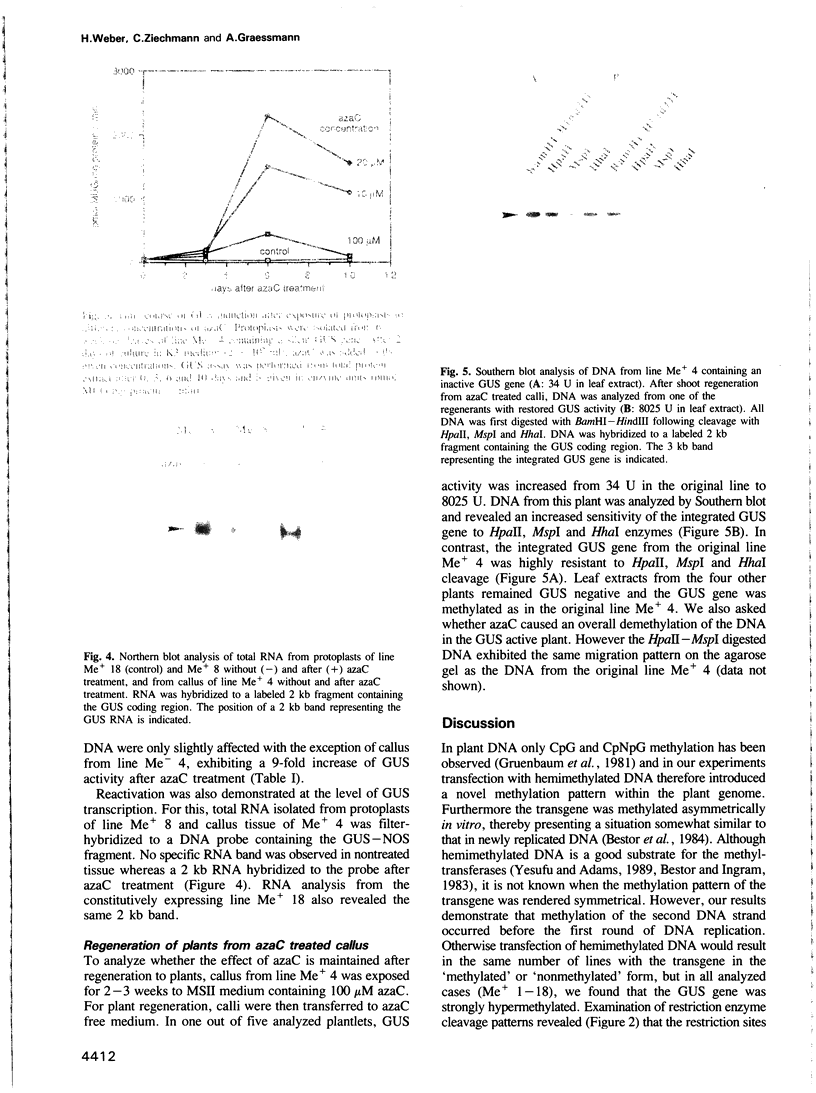
A hemimethylated chimeric gene, containing the cauliflower mosaic virus 35S promoter, the beta-glucuronidase coding region and the polyadenylation signal of nopaline synthase, was introduced into tobacco protoplasts by polyethylene glycol mediated transfection. Hemimethylation led to complete inhibition of transient gene expression. In regenerated transgenic plants the integrated gene was constitutively hypermethylated at the sequences CpG and CpNpG and this was correlated with an inactivation of beta-glucuronidase in 12 out of 18 analyzed plant lines whereas two showed slight and four strong activity. From 10 control lines transformed with nonmethylated DNA, only two were inactive; three showed slight and five strong activity. 5-aza-cytidine treatment of plant tissue from 'hypermethylated' lines led to induction of beta-glucuronidase in most cases. Shoots regenerated from azaC treated calli revealed stable enzyme restoration and demethylation of the integrated transgene.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L. DNA methylation. The effect of minor bases on DNA-protein interactions. Biochem J. 1990 Jan 15;265(2):309–320. doi: 10.1042/bj2650309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. H., Hellewell S. B., Ingram V. M. Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol Cell Biol. 1984 Sep;4(9):1800–1806. doi: 10.1128/mcb.4.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Chandler V. L., Walbot V. DNA modification of a maize transposable element correlates with loss of activity. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1767–1771. doi: 10.1073/pnas.83.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Gelvin S. B., Karcher S. J., DiRita V. J. Methylation of the T-DNA in Agrobacterium tumefaciens and in several crown gall tumors. Nucleic Acids Res. 1983 Jan 11;11(1):159–174. doi: 10.1093/nar/11.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Faugeron G. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol. 1989 Jul;9(7):2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Burgess S. M., Hirsh D. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M. C., Amasino R. M. Extensive changes in DNA methylation patterns accompany activation of a silent T-DNA ipt gene in Agrobacterium tumefaciens-transformed plant cells. Mol Cell Biol. 1989 Oct;9(10):4298–4303. doi: 10.1128/mcb.9.10.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczek R. A., Siebert E. Optimization of northern analysis by vacuum-blotting, RNA-transfer visualization, and ultraviolet fixation. Anal Biochem. 1990 Jan;184(1):90–95. doi: 10.1016/0003-2697(90)90017-4. [DOI] [PubMed] [Google Scholar]
- Logemann J., Schell J., Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987 May 15;163(1):16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Maliga P., Sz-Breznovits A., Márton L. Streptomycin-resistant plants from callus culture of haploid tobacco. Nat New Biol. 1973 Jul 4;244(131):29–30. doi: 10.1038/newbio244029a0. [DOI] [PubMed] [Google Scholar]
- Martin C., Prescott A., Lister C., MacKay S. Activity of the transposon Tam3 in Antirrhinum and tobacco: possible role of DNA methylation. EMBO J. 1989 Apr;8(4):997–1004. doi: 10.1002/j.1460-2075.1989.tb03466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. A., Primig M., Trnovsky J., Matzke A. J. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989 Mar;8(3):643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngernprasirtsiri J., Kobayashi H., Akazawa T. Transcriptional regulation and DNA methylation of nuclear genes for photosynthesis in nongreen plant cells. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7919–7923. doi: 10.1073/pnas.86.20.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow D. W., Jacobs J. D., Howell S. H. Functional regions of the cauliflower mosaic virus 35S RNA promoter determined by use of the firefly luciferase gene as a reporter of promoter activity. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4870–4874. doi: 10.1073/pnas.84.14.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J., Baur M., Bogucki A., Potrykus I. Gene targeting in plants. EMBO J. 1988 Dec 20;7(13):4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J., Shillito R. D., Saul M., Mandák V., Hohn T., Hohn B., Potrykus I. Direct gene transfer to plants. EMBO J. 1984 Dec 1;3(12):2717–2722. doi: 10.1002/j.1460-2075.1984.tb02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. Gene-controlled cytosine demethylation in the promoter region of the Ac transposable element in maize. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2789–2793. doi: 10.1073/pnas.86.8.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990 Mar;15(3):103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spena A., Viotti A., Pirrotta V. Two adjacent genomic zein sequences: structure, organization and tissue-specific restriction pattern. J Mol Biol. 1983 Oct 5;169(4):799–811. doi: 10.1016/s0022-2836(83)80137-5. [DOI] [PubMed] [Google Scholar]
- Stein R., Razin A., Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I., Capesius I. Determination of 5-methylcytosine from plant DNA by high-performance liquid chromatography. Biochim Biophys Acta. 1981 Jun 26;654(1):52–56. doi: 10.1016/0005-2787(81)90135-0. [DOI] [PubMed] [Google Scholar]
- Watson J. C., Kaufman L. S., Thompson W. F. Developmental regulation of cytosine methylation in the nuclear ribosomal RNA genes of Pisum sativum. J Mol Biol. 1987 Jan 5;193(1):15–26. doi: 10.1016/0022-2836(87)90622-x. [DOI] [PubMed] [Google Scholar]
- Weissbach A., Ward C., Bolden A. Eukaryotic DNA methylation and gene expression. Curr Top Cell Regul. 1989;30:1–21. doi: 10.1016/b978-0-12-152830-0.50003-7. [DOI] [PubMed] [Google Scholar]
- Wigler M., Levy D., Perucho M. The somatic replication of DNA methylation. Cell. 1981 Apr;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]