Abstract
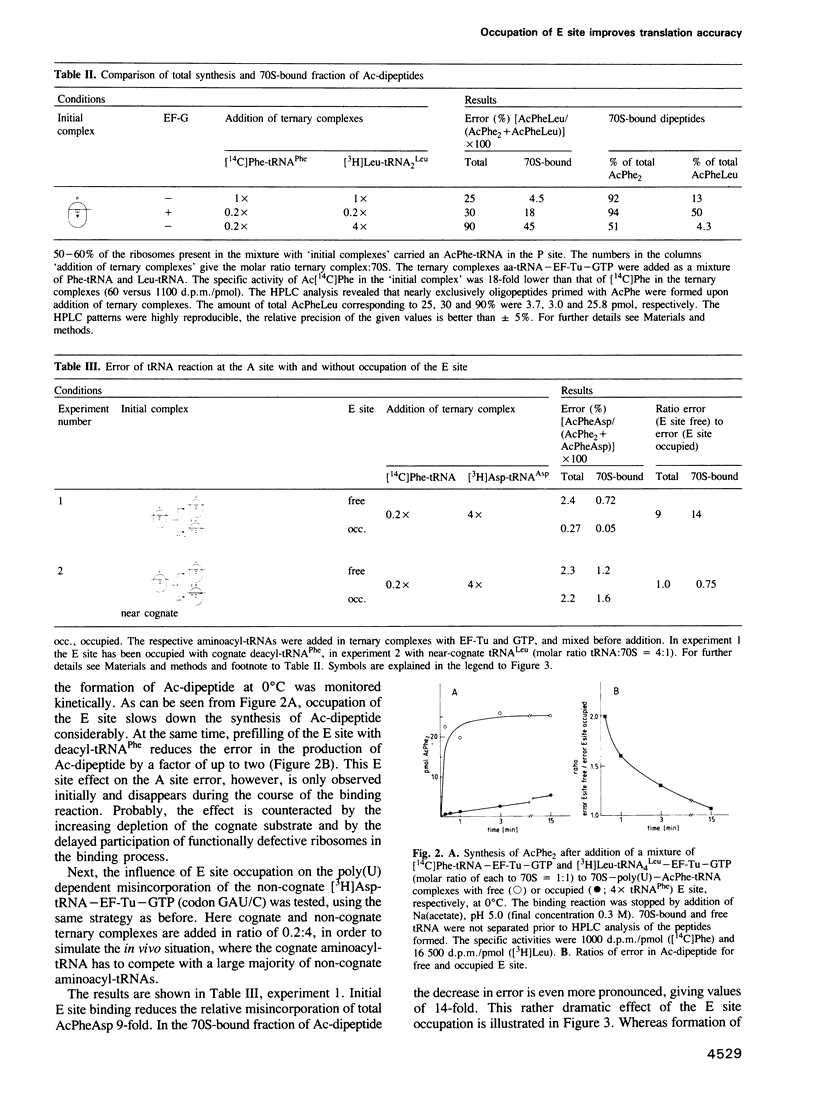
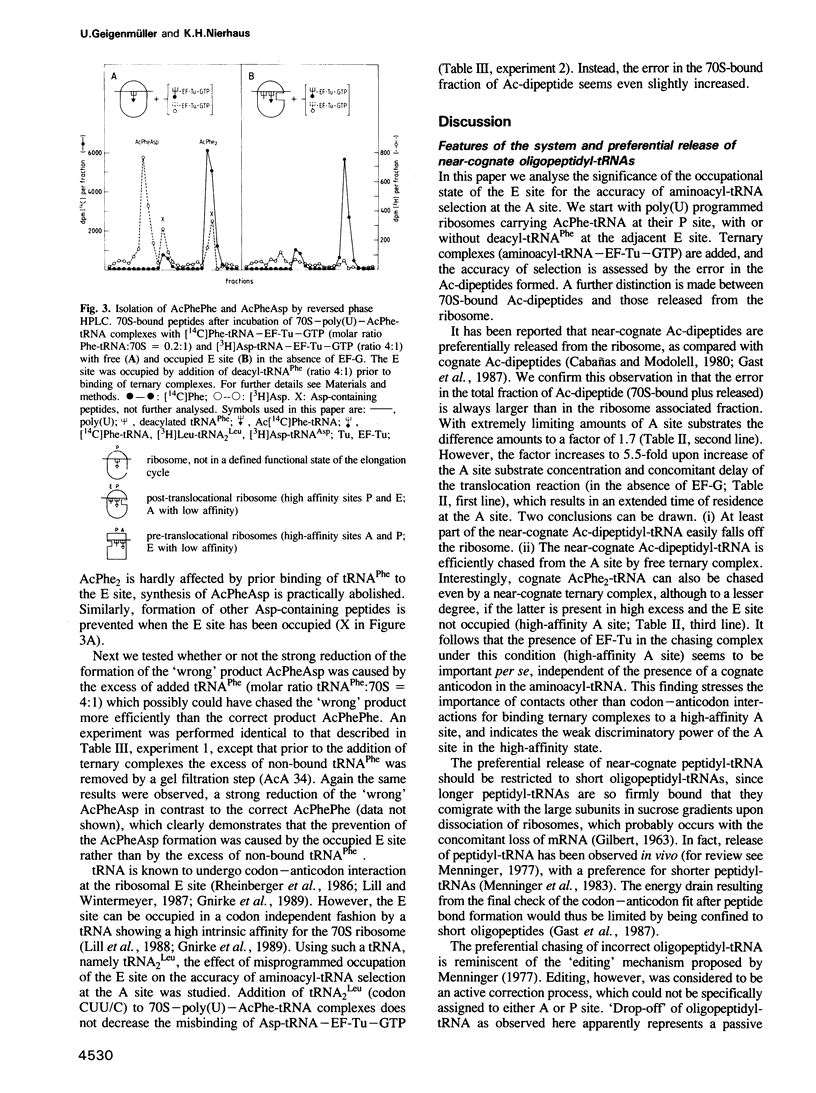
The E site (exit site for deacyl-tRNA) has been shown to be allosterically linked to the A site (aminoacyl-tRNA binding site), in that occupation of the E site reduces the affinity of the A site, and vice versa, whereas the intervening peptidyl-tRNA binding site (P site) keeps its high affinity. Here the question is analysed of whether or not the low affinity state of the A site caused by an occupied E site is of importance for the ribosomal accuracy of the aminoacyl-tRNA selection. In a poly(U) dependent system with high accuracy in poly(Phe) synthesis, the acceptance of the cognate ternary complex Phe-tRNA--EF-Tu--GTP (which has the correct anticodon with respect to the codon at the A site) was compared with the competing acceptance of ternary complexes with near-cognate Leu-tRNA(Leu) (which has a similar anticodon) or non-cognate Asp-tRNA(Asp) (which has a dissimilar anticodon), by monitoring the formation of AcPhePhe, AcPheLeu or AcPheAsp, respectively. Cognate (but not near-cognate) occupation of the E site reduced synthesis of the 'wrong' dipeptide AcPheLeu only marginally relative to that of the cognate AcPhe2, whereas the formation of AcPheAsp was decreased as much as 14-fold, thereby reducing it to the background level. It follows that the allosteric interplay between E and A sites, i.e. the low affinity of the A site induced by the occupation of the E site, excludes the interference of non-cognate complexes in the decoding process and thus reduces the number of aminoacyl-tRNA species competing for A site binding by an order of magnitude.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartetzko A., Nierhaus K. H. Mg2+/NH4+/polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol. 1988;164:650–658. doi: 10.1016/s0076-6879(88)64075-4. [DOI] [PubMed] [Google Scholar]
- Bergemann K., Nierhaus K. H. Spontaneous, elongation factor G independent translocation of Escherichia coli ribosomes. J Biol Chem. 1983 Dec 25;258(24):15105–15113. [PubMed] [Google Scholar]
- Bilgin N., Ehrenberg M., Kurland C. Is translation inhibited by noncognate ternary complexes? FEBS Lett. 1988 Jun 6;233(1):95–99. doi: 10.1016/0014-5793(88)81362-0. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Kirsebom L. A., Ehrenberg M., Kurland C. G. Mutations in ribosomal proteins L7/L12 perturb EF-G and EF-Tu functions. Biochimie. 1988 May;70(5):611–618. doi: 10.1016/0300-9084(88)90244-1. [DOI] [PubMed] [Google Scholar]
- Bouadloun F., Donner D., Kurland C. G. Codon-specific missense errors in vivo. EMBO J. 1983;2(8):1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabañas M. J., Modolell J. Nonenzymic translocation and spontaneous release of noncognate peptidyl transfer ribonucleic acid from Escherichia coli ribosomes. Biochemistry. 1980 Nov 11;19(23):5411–5416. doi: 10.1021/bi00564a040. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M., Kurland C. G., Ruusala T. Counting cycles of EF-Tu to measure proofreading in translation. Biochimie. 1986 Mar;68(2):261–273. doi: 10.1016/s0300-9084(86)80023-2. [DOI] [PubMed] [Google Scholar]
- GILBERT W. Polypeptide synthesis in Escherichia coli. II. The polypeptide chain and S-RNA. J Mol Biol. 1963 May;6:389–403. doi: 10.1016/s0022-2836(63)80051-0. [DOI] [PubMed] [Google Scholar]
- Gast F. U., Peters F., Pingoud A. The role of translocation in ribosomal accuracy. Translocation rates for cognate and noncognate aminoacyl- and peptidyl-tRNAs on Escherichia coli ribosomes. J Biol Chem. 1987 Sep 5;262(25):11920–11926. [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Gnirke A., Geigenmüller U., Rheinberger H. J., Nierhaus L. H. The allosteric three-site model for the ribosomal elongation cycle. Analysis with a heteropolymeric mRNA. J Biol Chem. 1989 May 5;264(13):7291–7301. [PubMed] [Google Scholar]
- Haenni A. L., Chapeville F. The behaviour of acetylphenylalanyl soluble ribonucleic acid in polyphenylalanine synthesis. Biochim Biophys Acta. 1966 Jan 18;114(1):135–148. doi: 10.1016/0005-2787(66)90261-9. [DOI] [PubMed] [Google Scholar]
- Jelenc P. C., Kurland C. G. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Laughrea M., Latulippe J., Filion A. M., Boulet L. Mistranslation in twelve Escherichia coli ribosomal proteins. Cysteine misincorporation at neutral amino acid residues other than tryptophan. Eur J Biochem. 1987 Nov 16;169(1):59–64. doi: 10.1111/j.1432-1033.1987.tb13580.x. [DOI] [PubMed] [Google Scholar]
- Leberman R., Antonsson B., Giovanelli R., Guariguata R., Schumann R., Wittinghofer A. A simplified procedure for the isolation of bacterial polypeptide elongation factor EF-Tu. Anal Biochem. 1980 May 1;104(1):29–36. doi: 10.1016/0003-2697(80)90272-9. [DOI] [PubMed] [Google Scholar]
- Lill R., Lepier A., Schwägele F., Sprinzl M., Vogt H., Wintermeyer W. Specific recognition of the 3'-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J Mol Biol. 1988 Oct 5;203(3):699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- Lill R., Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J Mol Biol. 1987 Jul 5;196(1):137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- Menninger J. R., Caplan A. B., Gingrich P. K., Atherly A. G. Tests of the ribosome editor hypothesis. II. Relaxed (relA) and stringent (relA+) E. coli differ in rates of dissociation of peptidyl-tRNA from ribosomes. Mol Gen Genet. 1983;190(2):215–221. doi: 10.1007/BF00330642. [DOI] [PubMed] [Google Scholar]
- Menninger J. R. Ribosome editing and the error catastrophe hypothesis of cellular aging. Mech Ageing Dev. 1977 Mar-Apr;6(2):131–142. doi: 10.1016/0047-6374(77)90014-8. [DOI] [PubMed] [Google Scholar]
- Rheinberger H. J., Geigenmüller U., Wedde M., Nierhaus K. H. Parameters for the preparation of Escherichia coli ribosomes and ribosomal subunits active in tRNA binding. Methods Enzymol. 1988;164:658–670. doi: 10.1016/s0076-6879(88)64076-6. [DOI] [PubMed] [Google Scholar]
- Rheinberger H. J., Nierhaus K. H. Allosteric interactions between the ribosomal transfer RNA-binding sites A and E. J Biol Chem. 1986 Jul 15;261(20):9133–9139. [PubMed] [Google Scholar]
- Rheinberger H. J., Sternbach H., Nierhaus K. H. Codon-anticodon interaction at the ribosomal E site. J Biol Chem. 1986 Jul 15;261(20):9140–9143. [PubMed] [Google Scholar]
- Wurmbach P., Nierhaus K. H. Isolation of the protein synthesis elongation factors EF-Tu, EF-Ts, and EF-G from Escherichia coli. Methods Enzymol. 1979;60:593–606. doi: 10.1016/s0076-6879(79)60056-3. [DOI] [PubMed] [Google Scholar]
- Zaniewski R., Petkaitis E., Deutscher M. P. A multiple mutant of Escherichia coli lacking the exoribonucleases RNase II, RNase D, and RNase BN. J Biol Chem. 1984 Oct 10;259(19):11651–11653. [PubMed] [Google Scholar]



