Summary
The meningococcal ST-11 complex (cc11) causes large invasive disease outbreaks with high case fatality rates, such as serogroup C (MenC) epidemics in industrialised nations in the 1990s and the serogroup W epidemic currently expanding globally. Glycoconjugate vaccines are available for serogroups A, C, W and Y. Broad coverage protein-based vaccines have recently been licensed against serogroup B meningococci (MenB), however, these do not afford universal MenB protection. Capsular switching from MenC to MenB among cc11 organisms is concerning because a large MenB cc11 (B:cc11) outbreak has the potential to cause significant morbidity and mortality. This study aimed to assess the potential for licensed and developmental non-capsular meningococcal vaccines to protect against B:cc11. The population structure and vaccine antigen distribution was determined for a panel of >800 geo-temporally diverse, predominantly MenC cc11 and B:cc11 genomes. The two licensed vaccines potentially protect against many but not all B:cc11 meningococci. Furthermore, strain coverage by these vaccines is often due to a single vaccine antigen and both vaccines are highly susceptible to vaccine escape owing to the apparent dispensability of key proteins used as vaccine antigens. cc11 strains with MenB and MenC capsules warrant special consideration when formulating future non-capsular meningococcal vaccines.
Keywords: Neisseria meningitidis, Meningococcal, Serogroup B, ST-11 complex, Vaccine
Highlights
-
•
The meningococcal ST-11 complex (cc11) is highly virulent and has caused large serogroup C and W outbreaks.
-
•
Serogroup C to B capsular switching is concerning owing to a lack of a universal vaccine against serogroup B meningococci.
-
•
Diverse serogroup B and C cc11 meningococci are predicted not to be covered by non-capsular vaccines targeting MenB.
-
•
Dispensability of multiple antigens raises the prospect of vaccine-escape by potentially covered outbreak strains.
-
•
Serogroup B and C cc11 meningococci merit special consideration when formulating future non-capsular meningococcal vaccines.
Introduction
Neisseria meningitidis (the meningococcus) is a leading cause of meningitis and septicaemia. The meningococcal ST-11 clonal complex (cc11) is diverse, comprised of two main lineages (lineage 11.1 and lineage 11.2; also known as sublineages 11.1 and 11.2), and includes serogroups C (MenC), W (MenW) and, to a lesser extent, B (MenB).1, 2 It is associated with relatively high case fatality rates and has been the cause of numerous large outbreaks including MenC cc11 (C:cc11) disease in the US military in the 1960s3 and educational institutions at a time of heightened endemic disease in the UK and elsewhere in the 1990s.4, 5 Outbreaks due to C:cc11 have recently been reported among men who have sex with men (MSM) in Europe and America.6 MenW cc11 (W:cc11) was the cause of a global outbreak among Hajj pilgrims in 2000 and has subsequently caused epidemics in the African meningitis belt and increased endemic disease in South Africa, South America, Europe and Australia.7
The UK and other countries responded to the increase in C:cc11 disease in the 1990s/2000s with highly effective glycoconjugate vaccination campaigns.8 More recently, Chile and the UK introduced quadrivalent meningococcal conjugate vaccination (MenACWY) to combat the increase in W:cc11 cases, an intervention being considered by other countries also experiencing a national W:cc11 outbreak.9, 10 There were concerns about capsular replacement from MenC to MenB during the MenC vaccine campaigns, especially given that the poor immunogenicity of the MenB polysaccharide precludes the development of MenB polysaccharide-based vaccines.11 Large modern MenB cc11 (B:cc11) outbreaks have yet to occur, however, an increase in cases was reported in France after 2002.12 Furthermore, a recent study indicated multiple capsular switching events within cc11, with MenB and MenC organisms highly interspersed within multiple lineage 11.1 and 11.2 sublineages.1
Several non-capsular vaccines have been developed to prevent MenB disease. Monovalent outer membrane vesicle (OMV) preparations are generally strain-specific due to the immunodominance of the diverse but poorly cross-protective porin A (PorA). Monovalent OMV preparations have targeted meningococci with PorA subtypes P1.7,16, P1.7-2,4, P1.19,15 and P1.3.13, 14 Multivalent OMV vaccines have also been proposed including a serogroup A + W OMV vaccine incorporating PorA subtypes P1.20,9 and P1.5,2,15 and a nine-valent formulation including PorA subtypes P1.7,16, P1.22,14, P1.5-1,2-2, P1.5-2,10, P1.7-1,1, P1.12-1,13, P1.19,15-1, P1.7-2,4 and P1.18-1,3.16
The 4CMenB vaccine includes an OMV (PorA P1.4) and three main protein antigens – factor H-binding protein (fHbp), Neisseria adhesin A (NadA) and neisserial heparin binding antigen (NHBA).17 The fHbp variant 1 component potentially protects against meningococci expressing fHbp variant 1, but not variant 2 or 3, peptides. The NadA variant 2/3 component potentially protects against meningococci with variant 1 or variant 2/3 NadA, but not variants 4, 5 or 6. Potential cross-protectivity afforded by the NHBA component is less well understood. Actual strain coverage by the vaccine depends on the antigenic similarity and/or expression level of the respective antigen within the infecting organism. The Meningococcal Antigen Typing System (MATS) is an ELISA that collectively measures both of these qualities to predict strain coverage afforded by the 4CMenB vaccine.18 Large scale studies incorporating MATS are contributing to our knowledge of potential 4CMenB coverage by virtue of specific peptide variants of the individual vaccine antigens within the study populations.19, 20
The bivalent fHbp vaccine, rLP2086, comprises variant 1 and variant 3 fHbp peptide components, the latter of which may be cross-protective against meningococci with variant 2, as well as those with variant 3, peptides.21 Strain coverage afforded by the bivalent fHbp vaccine is predicted using the Meningococcal Antigen Surface Expression (MeASurE) flow cytometric assay which quantifies fHbp surface expression.22
None of these vaccines are likely to protect against all MenB strains. The aim of this study was to assess genotypically the potential of these vaccines to protect against B:cc11 meningococci.
Materials and methods
Genomes
Genomes and typing data were obtained from the PubMLST Neisseria database (https://pubmlst.org/neisseria, accessed 07/10/16)23 and the Meningitis Research Foundation Meningococcus Genome Library (http://www.meningitis.org/research/genome, accessed 07/10/16) the latter of which contains all English, Welsh and Northern Irish invasive disease isolates received by the Public Health England Meningococcal Reference Unit from July 2010 onwards. Owing to a paucity of MenB genomes among the MenW-associated lineage 11.1 sublineages, all corresponding genomes, irrespective of capsular group, were excluded from the analysis (Supplementary figure). With a single exception, relatively poorly sequenced/assembled genomes comprising more than an arbitrary cut-off of 500 contigs were also omitted from the study. The exception, M16 240144 (624 contigs), was included for completeness of the English, Welsh and Northern Irish data over the study period. The final panel (n = 900; supplementary information) included all remaining cc11-assigned genomes and those with incomplete MLST profiles that matched the ST-11 profile at more than four loci. It also included that of the recent English isolate M16 240518 (MenC cc11; submitted after 07/10/16) and two non cc11 ‘reference’ genomes (PubMLST IDs 27778 and 29645; cc41/44 and cc8, respectively).
The B:cc11 genomes (n = 89; of which 78 invasive, 3 carrier, and 8 other/unspecified) were from isolates collected in the UK (n = 50; 1970 to 2016), Czech Republic (n = 26; 1984 to 2011), France (n = 5; 2014 to 2016), Ireland (n = 2; 2014) and one each from Greece (1998), Norway (1969), Slovenia (2013), USA (1964), Tunisia (1987) and Saudi Arabia (1998). The remaining genomes included MenC isolates from 22 countries on five continents (n = 763, 1970 to 2016), MenW isolates from South Africa and the USA (n = 3, 2005 and 2012), a MenY isolate of unknown origin (n = 1) and isolates from various countries that were non-groupable or serogroup-unspecified (n = 42, 1993–2016).
Molecular typing and phylogenetic network analyses
Genomes were compared for 1546 core genes using the PubMLST genome comparator tool.23, 24, 25 Distance matrices were visualised using SplitsTree4.26 Untagged nadA or nhba alleles that were incompletely assembled due to the presence of an insertion sequence (IS) were identified by searching known interrupted alleles using the BLAST algorithm and aligning hits with the corresponding insertion sequence.
MATS analyses
Isolate-specific MATS data for English invasive B:cc11 isolates from 2007/8 were obtained from a previous study.19 An isolate was predicted to be covered by 4CMenB (MATS-positive) if one or more of fHbp, NadA or NHBA had a relative potency (RP) greater than the positive bactericidal threshold (PBT; 0.021 for fHbp, 0.009 for NadA, and 0.294 for NHBA), and/or the isolate possessed PorA P1.4.
Results
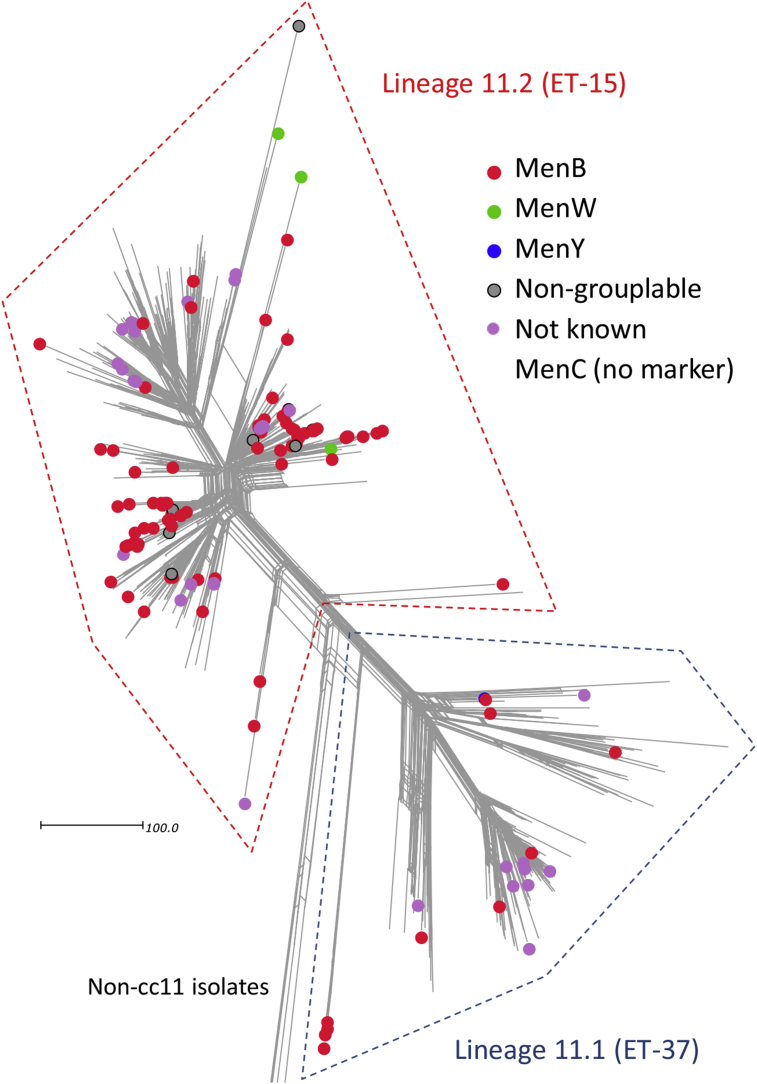
The MenB genomes from Tunisia and Saudi Arabia (PubMLST IDs 34577 and 34596) were found to cluster with cc8 isolates and were disregarded for the remainder of the study. The remaining MenB genomes were distributed between lineages 11.1 (n = 10; 1964–2014) and 11.2 (n = 77; 1970–2016) where they were broadly interspersed among the remaining predominantly MenC genomes (Fig. 1). Several predominantly MenB clusters were also observed.
Figure 1.
Population structure and capsular group distribution among meningococcal ST-11 complex genomes belonging to serogroup B/C-dominated lineage 11.1 and 11.2 sublineages. The scale bar indicates the number of differences among the 1546 core genome loci compared. NB lineage 11.1 and lineage 11.2 are also known as sublineage 11.1 and sublineage 11.2, respectively.1, 2
Lineage 11.1 MenB genomes
There were ten lineage 11.1 MenB genomes. Four of these (all P1.5-1,2-2:ST-3537) were closely related within a distinct sublineage and possessed alleles encoding variant 2 fHbp peptides and NHBA peptide 20, and were devoid of a nadA gene. The remaining six genomes (all P1.5,2:ST-11) were interspersed with MenC genomes within several distinct clusters and possessed alleles encoding variant 2 or 3 fHbp peptides, NHBA peptide 29, and NadA-2/3 peptides (Fig. 1).
Lineage 11.2 MenB genomes
The 77 lineage 11.2 MenB genomes were broadly interspersed with MenC genomes within multiple clusters and subclusters (Fig. 1). The main PorA subtypes were P1.5-1,10-8 (n = 41) and P1.5,2 (n = 27). The remaining PorA subtypes included P1.5-1,10-1 (n = 2), P1.7-1,1 (n = 2) and a single genome each for P1.17,16-3, P1.5,2-53, P1.5-1,10-4, P1.5-1,2-2 and P1.5-2,10. All of the lineage 11.2 MenB genomes possessed alleles encoding NHBA peptide 20. Based on their corresponding fHbp variant, these fell into the following three subgroups:
-
•
Sixty-two percent (48/77) possessed alleles encoding variant 1 fHbp peptides including peptides 10 (n = 8), 2 (n = 4), 453 (n = 4), 78 (n = 3) and a further 22 other peptides (n ≤ 2 isolates each). Of these, eight possessed alleles for NadA-2/3 peptides, 38 possessed insertionally inactivated or frameshifted nadA alleles, one possessed an incompletely assembled nadA allele (assembly artefact) and one was devoid of a nadA allele.
-
•
Twenty-five percent (19/77) possessed alleles for variant 2 or 3 fHbp peptides including peptides 18 (n = 9), 22 (n = 3), 92 (n = 2), 134 (n = 2) and three other peptides (n = 1 isolate each). Of these, two possessed alleles for NadA-2/3 peptides, 14 possessed insertionally inactivated nadA alleles, and three were devoid of nadA alleles.
-
•
Thirteen percent (10/77) possessed frameshifted fhbp alleles. Of these, three possessed alleles for NadA-2/3 peptides while the remaining seven possessed insertionally inactivated nadA alleles.
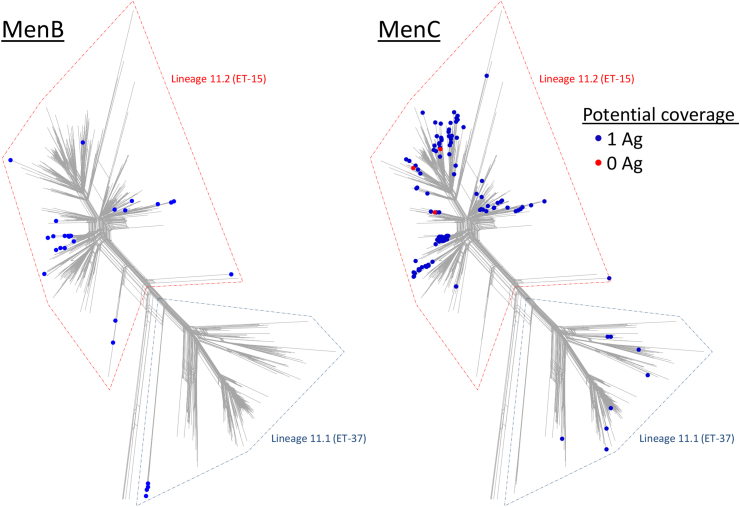
Distribution of minimally/non-vaccine-covered MenB and MenC cc11 isolates
Approximately one third (28/87) of all of the MenB isolates (from four countries between 1970 and 2014), and 17% (128/763) of all of the MenC isolates (from 14 different countries between 1970 and 2016) were potentially covered by virtue of a single 4CMenB antigen which was NHBA. With the exception of ten of the MenC isolates, these all possessed alleles for NHBA peptide 20. The remaining ten MenC isolates possessed alleles for NHBA peptides 29 (n = 8), 118 (n = 1) and 716 (n = 1). The MenB isolates were widespread within lineage 11.2, with some clustering, but were only observed in a single cluster within lineage 11.1 (Fig. 2). The MenC isolates were relatively widespread, including several lineage 11.1 clusters, but mainly belonged to lineage 11.2 (Fig. 2). Three diverse lineage 11.2 MenC isolates from the UK between 2000 and 2015 were predicted not to be covered by immunity against any of the 4CMenB antigens due, in part, to frameshifted or insertionally inactivated nhba alleles. A further four MenC isolates had similarly disrupted nhba alleles but were predicted to be covered by the immune response to other 4CMenB antigens (USA, 2006 and Canada, 2001/2).
Figure 2.
Diversity of MenB and MenC cc11 isolates potentially covered by zero or one 4CMenB antigen. MenB isolates potentially covered by zero or one 4CMenB antigens were diverse but mainly belonged to two large clusters in lineage 11.2. The minimally/non-covered MenC isolates were more widespread within the population but also predominantly belonged to lineage 11.2. NB lineage 11.1 and lineage 11.2 are also known as sublineage 11.1 and sublineage 11.2, respectively.1, 2
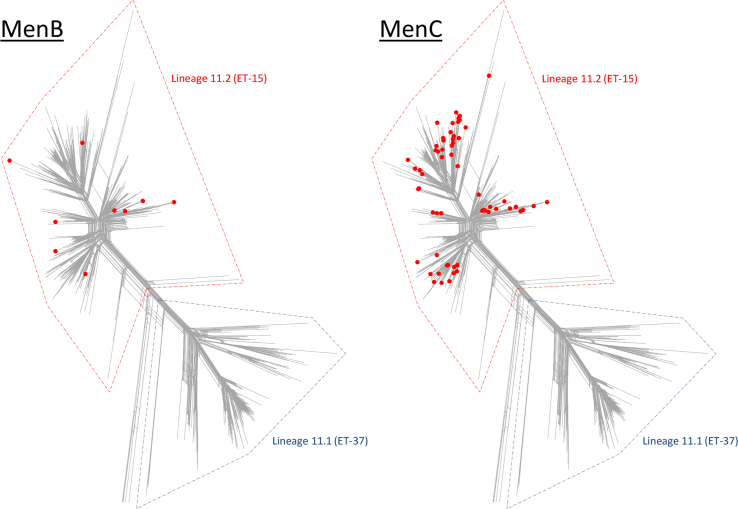
Thirteen percent (10/77) of the MenB lineage 11.2 isolates (from four countries between 1998 and 2014) and 12% (62/535) of the MenC lineage 11.2 isolates (from ten countries between 1996 and 2016) possessed frameshifted fhbp alleles and so were unlikely to be covered by fHbp-induced antibody. These isolates were widespread throughout the lineage 11.2 population structure (Fig. 3).
Figure 3.
Diversity of MenB and MenC cc11 isolates with frameshifted fhbp alleles. The MenB and MenC isolates with frameshifted fhbp alleles were widespread within lineage 11.2. NB lineage 11.1 and lineage 11.2 are also known as sublineage 11.1 and sublineage 11.2, respectively.1, 2
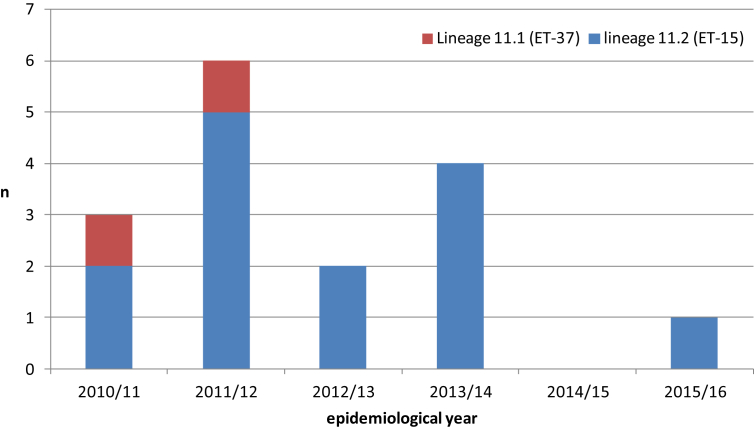
B:cc11 cases in England and Wales from 2010/11 to 2015/16
There were 16 B:cc11 cases in England and Wales between 2010/11 and 2015/16 with between zero and six cases annually (Fig. 4). Among these, 12.5% (2/16) were due to lineage 11.1 isolates, one with PorA subtype P1.5,2 and alleles for NHBA peptide 29, a variant 2 fHbp peptide and a NadA-2/3 peptide; and the other with PorA subtype P1.5-1,2-2 and alleles for NHBA peptide 20, a variant 2 fHbp peptide and no nadA gene.
Figure 4.
Distribution of B:cc11 lineage 11.1 and 11.2 genomes in England and Wales, 2010/11 onwards.
The remaining 87.5% (14/16) cases were due to lineage 11.2 organisms all of which had alleles for NHBA peptide 20. Of these 93% (13/14) were subtype P1.5,2 and 7% (1/14) were subtype P1.7-1,1. Seventy-one percent (10/14) of the lineage 11.2 isolates had alleles for variant 1 fHbp peptides (encoding nine different peptides in total), including four with alleles for NadA-2/3 peptides and six with insertionally inactivated nadA alleles. Seven percent (1/14) possessed an allele for a variant 2 fHbp peptide and had an insertionally inactivated nadA allele. The remaining 21% (3/14) of the lineage 11.2 isolates had frameshifted fhbp alleles, two with intact NadA2/3 alleles and one with insertionally inactivated nadA.
The case fatality rate among these cases was 31.3% (5/16) and included 0/3 infants, 2/6 1–5 year-olds, 2/6 adults (18–64 y) and 1/1 older adults.
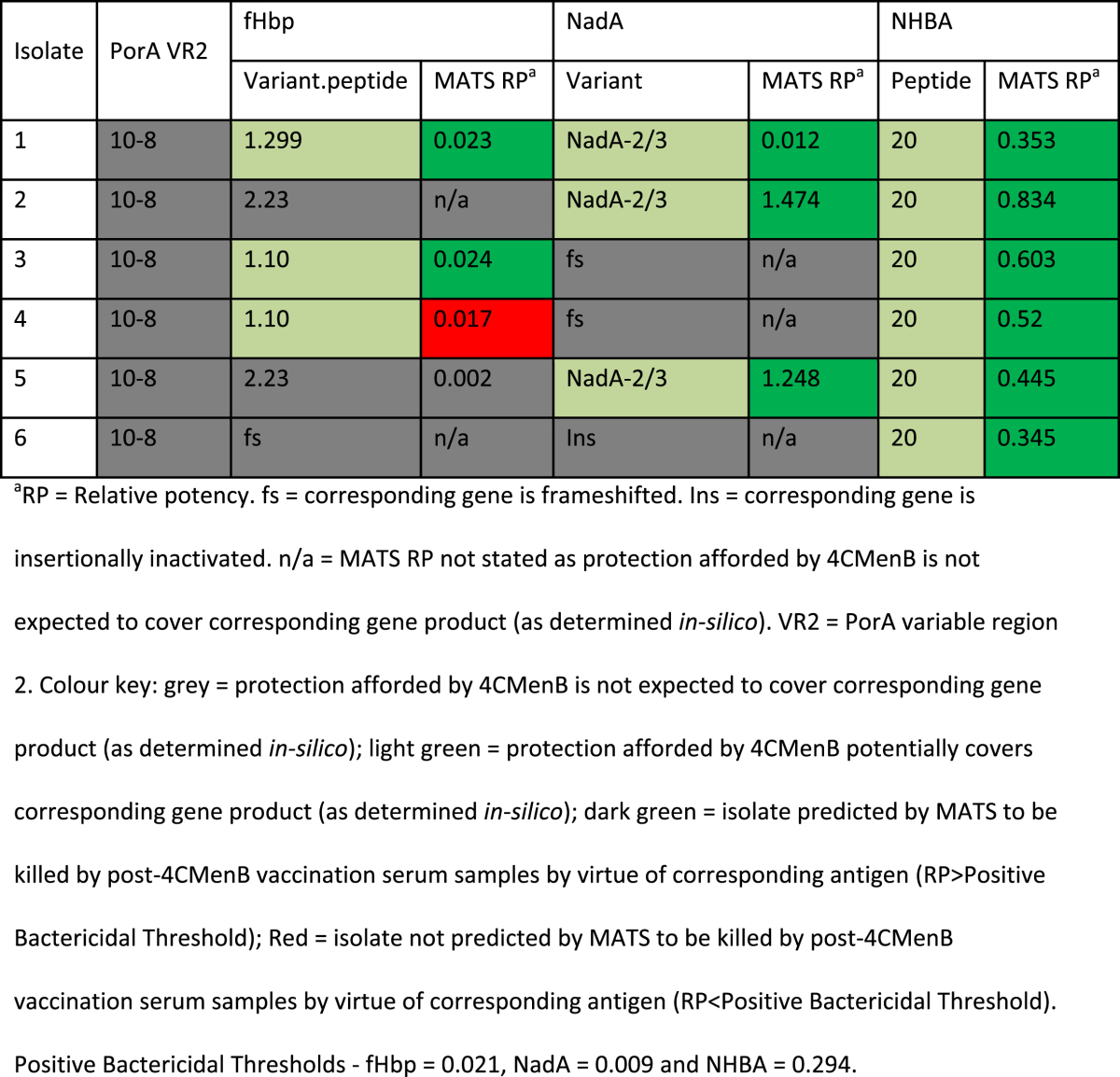
B:cc11 MATS analyses
All six B:cc11 isolates from England in 2007/8 were predicted by MATS to be killed by post-vaccination serum samples (MATS-covered). One had RP > PBT for fHbp, NadA and NHBA; one had RP > PBT for fHbp and NHBA, two had RP > PBT for NadA and NHBA; and two had RP > PBT for NHBA only. None of them possessed PorA P1.4 (Table 1).
Table 1.
MATS coverage among English B:cc11 isolates in 2007/8.
Discussion
Large-scale capsule replacement did not occur following the introduction of MenC glycoconjugate vaccination11; however, the recent W:cc11 outbreak affecting South America, Europe and elsewhere highlighted the unpredictable nature of meningococcal outbreaks. This also emphasises the propensity of members of cc11, expressing different serogroups, to cause outbreaks of aggressive invasive disease.7 Here, in agreement with previous work,27, 28 we found that B:cc11 in England and Wales was associated with high case fatality rates and we provide further evidence that diverse invasive B:cc11 organisms have been generated on multiple occasions through capsular switching of MenC organisms.1, 12 Furthermore, the existence of several persistent B:cc11 sublineages indicate that such populations are not necessarily transient. All major W:cc11 lineages are believed to have arisen from a common capsule switch event.29 Fortunately, conjugate vaccines were available to combat the recent W:cc11 outbreak, while the recently licensed 4CMenB vaccine was also likely to afford protection to infants against the outbreak strain.30 This poses the question, however, that - if one of these diverse MenB sublineages should expand to cause a large outbreak, would currently available non-capsular MenB vaccines afford protection?
The B:cc11 isolates were all potentially covered by immunity to the NHBA component of 4CMenB, but none were potentially covered by immunity to the PorA P1.4 component. None of the Lineage 11.1 isolates were potentially covered by the 4CMenB fHbp component, but some were potentially covered by immunity to the NadA component. Collectively, the lineage 11.2 MenB isolates possessed diverse fhbp alleles encoding peptides of all three main variants, as well as some isolates with frameshifted fhbp alleles, as has been described previously.31, 32 Similarly, the presence of intact alleles for potentially covered NadA-2/3 peptides was sporadic among lineage 11.2 MenB isolates. Thus, from a genotypic standpoint, 4CMenB had the potential to protect against all of the MenB isolates by virtue of at least one vaccine antigen and this was supported by MATS results for the seemingly typical panel of six lineage 11.2 isolates from 2007/8.19 MATS is believed to provide a conservative estimate of strain coverage.19 In a recent study of ∼500 students, however, a third did not exhibit an hSBA response against a MATS-positive outbreak strain eight weeks following their second dose of 4CMenB.33
In the present study, potential 4CMenB coverage among many MenB isolates was limited to that afforded by the NHBA component. These isolates possessed alleles for NHBA peptide 20 which exhibited relatively high levels of MATS coverage in two large studies.19, 20 It is, however, of concern that there were seven MenC genomes with disrupted nhba alleles, three of which also had frameshifted fhbp alleles and insertionally inactivated nadA alleles. The gene encoding the NHBA peptide may, therefore, be dispensable for lineage 11.2 organisms, as appears to also be the case for NadA and fHbp. Furthermore, the diversity of fHbp among lineage 11.2 isolates suggests a lack of immunological constraints, as has been proposed for other antigens,34 which adds to the variant 1 fHbp vaccine component's vulnerability to vaccine escape.
In terms of other non-capsular vaccines, the bivalent fHbp vaccine, rLP2086, would potentially cover the majority of the B:cc11 meningococci but not those with a frameshifted fhbp gene. It would also be particularly vulnerable to vaccine escape owing to the apparent dispensability of fhbp among diverse lineage 11.2 meningococci, given that this is the sole antigen in the vaccine. None of the monovalent OMV vaccines or the nine-valent vaccine included exact matches to either of the predominant PorA subtypes (P1.5,2 or P1.5-1,10-8).13 The protective potential of matching or closely-related VR1s (P1.5-1 or P1.5-2), or the closely-related VR2 (P1.2-2), of the nine-valent vaccine against these isolates is not known. A previous study has shown VR2 to be the main target for the immune response against PorA P1.7-2,4.35 Cross-protectivity of the immune response against PorA, meanwhile, may be dependent on the number of doses and age of the vaccinee.36 Four PorAs contained in the nine-valent vaccine did, however, constitute rare PorAs among the B:cc11 isolates and may be useful if an outbreak were caused by a rare strain.16 The investigational serogroup A + W OMV vaccine did include an exact match to one of the predominant PorA subtypes (P1.5,2) and so may offer protection depending on the emergent strain.15 The extent of possible protection afforded by non-PorA OMV components of these vaccines,13 or indeed that of Bexsero, among these isolates is not known.
With the exception of the recent English, Welsh and Northern Irish isolates, the dataset analysed here was largely opportunistic and not globally comprehensive and, as such, percentages provided in the results should be used with caution. The important thing to note, however, is that a number of geo-temporally and evolutionarily diverse B:cc11 and C:cc11 isolates are minimally/not covered and/or are able to dispense with genes for key antigens. The number of MenB isolates, their overall geo-temporal diversity, and their diverse antigenic repertoires, adds to the strength of the conclusions. Future genomic characterisation of large carriage strain panels37 will provide further information on the underlying diversity of cc11 and supplement these data. The data for England and Wales from 2010/11 onwards were comprehensive in terms of disease isolates submitted to the MRU. There was no trend for increase of B:cc11 cases and the majority of these (n = 12/16) were potentially covered by virtue of two 4CMenB antigens. On the other hand, it was noteworthy that the potential coverage of the remainder of the isolates was by virtue of just one antigen. Furthermore, three out of 16 isolates had fhbp alleles that were inactivated by frameshift mutations and one of these was an isolate with an interrupted nadA allele.
In conclusion, in the event of the public health emergency that a large B:cc11 outbreak would represent, it may not be possible to intervene as rapidly as with the W:cc11 outbreak, due to the absence of a suitable vaccine, although there may be other vaccines in development that were not discussed in the present manuscript. Any protection afforded by the MenB vaccines available at the time of writing may be vulnerable to vaccine escape. Several previously employed or investigational OMV vaccines could provide a contingency response but rapid availability would be unlikely, especially for investigational vaccines that require further evaluation. Therefore it would be prudent to investigate the inclusion of relatively stable B:cc11 surface antigen variants in future MenB vaccine formulations. Such candidates would include PorA subtypes P1.5,2 and P1.5-1,10-8.
Conflict of interests
The PHE Immunisation Department has provided marketing authorisation holders with post-marketing surveillance reports on invasive meningococcal disease which the companies are required to submit to the UK Licensing authority in compliance with their Risk Management Strategy. A cost recovery charge is made for these reports.
Acknowledgements
This work was supported by a Wellcome Trust grant awarded to Martin Maiden [grant number 087622]. The publication made use of the Neisseria Multi Locus Sequence Typing website (https://pubmlst.org/neisseria/) developed by Keith Jolley and sited at the University of Oxford.23 The development of this site has been funded by the Wellcome Trust and European Union. The publication also made use of the Meningitis Research Foundation Meningococcus Genome Library (http://www.meningitis.org/research/genome) developed by Public Health England, the Wellcome Trust Sanger Institute and the University of Oxford as a collaboration. The project is funded by the Meningitis Research Foundation.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jinf.2017.05.015.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Lucidarme J., Hill D.M., Bratcher H.B., Gray S.J., du Plessis M., Tsang R.S. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect. 2015 Nov;71(5):544–552. doi: 10.1016/j.jinf.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucidarme J., Scott K.J., Ure R., Smith A., Lindsay D., Stenmark B. An international invasive meningococcal disease outbreak due to a novel and rapidly expanding serogroup W strain, Scotland and Sweden, July to August 2015. Euro Surveill. 2016 Nov 10;21(45) doi: 10.2807/1560-7917.ES.2016.21.45.30395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brundage J.F., Zollinger W.D. Evolution of meningococcal disease epidemiology in the US army. In: Vedros N.A., editor. Evolution of meningococcal disease. CRC Press; Boca Raton: 1987. [Google Scholar]
- 4.Gray S.J., Trotter C.L., Ramsay M.E., Guiver M., Fox A.J., Borrow R. Epidemiology of meningococcal disease in England and Wales 1993/94 to 2003/04: contribution and experiences of the Meningococcal Reference Unit. J Med Microbiol. 2006 Jul;55(Pt 7):887–896. doi: 10.1099/jmm.0.46288-0. [DOI] [PubMed] [Google Scholar]
- 5.Jelfs J., Munro R., Ashto F.E., Caugant D.A. Genetic characterization of a new variant within the ET-37 complex of Neisseria meningitidis associated with outbreaks in various parts of the world. Epidemiol Infect. 2000 Oct;125(2):285–298. doi: 10.1017/s0950268899004471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kupferschmidt K. Infectious diseases. Bacterial meningitis finds new niche in gay communities. Science. 2013 Jul 26;341(6144):328. doi: 10.1126/science.341.6144.328. [DOI] [PubMed] [Google Scholar]
- 7.Mustapha M.M., Marsh J.W., Harrison L.H. Global epidemiology of capsular group W meningococcal disease (1970–2015): multifocal emergence and persistence of hypervirulent sequence type (ST)-11 clonal complex. Vaccine. 2016 Mar 18;34(13):1515–1523. doi: 10.1016/j.vaccine.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Borrow R., Abad R., Trotter C., van der Klis F.R., Vazquez J.A. Effectiveness of meningococcal serogroup C vaccine programmes. Vaccine. 2013 Sep 23;31(41):4477–4486. doi: 10.1016/j.vaccine.2013.07.083. [DOI] [PubMed] [Google Scholar]
- 9.Ladhani S.N., Ramsay M., Borrow R., Riordan A., Watson J.M., Pollard A.J. Enter B and W: two new meningococcal vaccine programmes launched. Arch Dis Child. 2016 Jan;101(1):91–95. doi: 10.1136/archdischild-2015-308928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safadi M.A., O'Ryan M., Valenzuela Bravo M.T., Brandileone M.C., Gorla M.C., de Lemos A.P. The current situation of meningococcal disease in Latin America and updated Global Meningococcal Initiative (GMI) recommendations. Vaccine. 2015 Nov 27;33(48):6529–6536. doi: 10.1016/j.vaccine.2015.10.055. [DOI] [PubMed] [Google Scholar]
- 11.Maiden M.C., Ibarz-Pavon A.B., Urwin R., Gray S.J., Andrews N.J., Clarke S.C. Impact of meningococcal serogroup C conjugate vaccines on carriage and herd immunity. J Infect Dis. 2008 Mar 01;197(5):737–743. doi: 10.1086/527401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lancellotti M., Guiyoule A., Ruckly C., Hong E., Alonso J.M., Taha M.K. Conserved virulence of C to B capsule switched Neisseria meningitidis clinical isolates belonging to ET-37/ST-11 clonal complex. Microbes Infect. 2006 Jan;8(1):191–196. doi: 10.1016/j.micinf.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Holst J., Oster P., Arnold R., Tatley M.V., Naess L.M., Aaberge I.S. Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum Vaccin Immunother. 2013 Jun;9(6):1241–1253. doi: 10.4161/hv.24129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boslego J., Garcia J., Cruz C., Zollinger W., Brandt B., Ruiz S. Efficacy, safety, and immunogenicity of a meningococcal group B (15:P1.3) outer membrane protein vaccine in Iquique, Chile. Chilean National Committee for Meningococcal Disease. Vaccine. 1995 Jun;13(9):821–829. doi: 10.1016/0264-410x(94)00037-n. [DOI] [PubMed] [Google Scholar]
- 15.Norheim G., Tunheim G., Naess L.M., Kristiansen P.A., Caugant D.A., Rosenqvist E. An outer membrane vesicle vaccine for prevention of serogroup A and W-135 meningococcal disease in the African meningitis belt. Scand J Immunol. 2012 Aug;76(2):99–107. doi: 10.1111/j.1365-3083.2012.02709.x. [DOI] [PubMed] [Google Scholar]
- 16.Kaaijk P., van Straaten I., van de Waterbeemd B., Boot E.P., Levels L.M., van Dijken H.H. Preclinical safety and immunogenicity evaluation of a nonavalent PorA native outer membrane vesicle vaccine against serogroup B meningococcal disease. Vaccine. 2013 Feb 04;31(7):1065–1071. doi: 10.1016/j.vaccine.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 17.Bai X., Findlow J., Borrow R. Recombinant protein meningococcal serogroup B vaccine combined with outer membrane vesicles. Expert Opin Biol Ther. 2011 Jul;11(7):969–985. doi: 10.1517/14712598.2011.585965. [DOI] [PubMed] [Google Scholar]
- 18.Donnelly J., Medini D., Boccadifuoco G., Biolchi A., Ward J., Frasch C. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci U S A. 2010 Nov 09;107(45):19490–19495. doi: 10.1073/pnas.1013758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel U., Taha M.K., Vazquez J.A., Findlow J., Claus H., Stefanelli P. Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: a qualitative and quantitative assessment. Lancet Infect Dis. 2013 May;13(5):416–425. doi: 10.1016/S1473-3099(13)70006-9. [DOI] [PubMed] [Google Scholar]
- 20.Parikh S.R., Newbold L., Slater S., Stella M., Moschioni M., Lucidarme J. Meningococcal serogroup B strain coverage of the multicomponent 4CMenB vaccine with corresponding regional distribution and clinical characteristics in England, Wales, and Northern Ireland, 2007–08 and 2014–15: a qualitative and quantitative assessment. Lancet Infect Dis. 2017 Mar 30 doi: 10.1016/S1473-3099(17)30170-6. http://dx.doi.org/10.1016/S1473-3099(17)30170-6, pii: S1473-3099(17)30170-6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Gandhi A., Balmer P., York L.J. Characteristics of a new meningococcal serogroup B vaccine, bivalent rLP2086 (MenB-FHbp; Trumenba(R)) Postgrad Med. 2016 Aug;128(6):548–556. doi: 10.1080/00325481.2016.1203238. [DOI] [PubMed] [Google Scholar]
- 22.McNeil L.K., Zagursky R.J., Lin S.L., Murphy E., Zlotnick G.W., Hoiseth S.K. Role of factor H binding protein in Neisseria meningitidis virulence and its potential as a vaccine candidate to broadly protect against meningococcal disease. Microbiol Mol Biol Rev. 2013 Jun;77(2):234–252. doi: 10.1128/MMBR.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolley K.A., Maiden M.C. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinforma. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladhani S.N., Beebeejaun K., Lucidarme J., Campbell H., Gray S., Kaczmarski E. Increase in endemic Neisseria meningitidis capsular group W ST-11 complex associated with severe invasive disease in England and Wales. Clin Infect Dis. 2014 Nov 10;60(4):578–585. doi: 10.1093/cid/ciu881. [DOI] [PubMed] [Google Scholar]
- 25.Bratcher H.B., Corton C., Jolley K.A., Parkhill J., Maiden M.C. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics. 2014;15:1138. doi: 10.1186/1471-2164-15-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huson D.H. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14(1):68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 27.Zarantonelli M.L., Lancellotti M., Deghmane A.E., Giorgini D., Hong E., Ruckly C. Hyperinvasive genotypes of Neisseria meningitidis in France. Clin Microbiol Infect. 2008 May;14(5):467–472. doi: 10.1111/j.1469-0691.2008.01955.x. [DOI] [PubMed] [Google Scholar]
- 28.Trotter C.L., Fox A.J., Ramsay M.E., Sadler F., Gray S.J., Mallard R. Fatal outcome from meningococcal disease–an association with meningococcal phenotype but not with reduced susceptibility to benzylpenicillin. J Med Microbiol. 2002 Oct;51(10):855–860. doi: 10.1099/0022-1317-51-10-855. [DOI] [PubMed] [Google Scholar]
- 29.Mustapha M.M., Marsh J.W., Krauland M.G., Fernandez J.O., de Lemos A.P., Dunning Hotopp J.C. Genomic investigation reveals highly conserved, mosaic, recombination events associated with capsular switching among invasive Neisseria meningitidis serogroup W sequence type (ST)-11 strains. Genome Biol Evol. 2016 Jul 03;8(6):2065–2075. doi: 10.1093/gbe/evw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladhani S.N., Giuliani M.M., Biolchi A., Pizza M., Beebeejaun K., Lucidarme J. Effectiveness of meningococcal B vaccine against endemic hypervirulent Neisseria meningitidis W Strain, England. Emerg Infect Dis. 2016 Feb;22(2):309–311. doi: 10.3201/eid2202.150369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang R.S., Hoang L., Tyrrell G., Horsman G., Wylie J., Jamieson F.B. Genetic and antigenic characterization of Canadian invasive Neisseria meningitidis serogroup C (MenC) case isolates in the post-MenC conjugate vaccine era, 2009–2013. J Med Microbiol. 2015 Feb;64(Pt 2):174–179. doi: 10.1099/jmm.0.000006-0. [DOI] [PubMed] [Google Scholar]
- 32.Lucidarme J., Tan L., Exley R.M., Findlow J., Borrow R., Tang C.M. Characterization of Neisseria meningitidis isolates that do not express the virulence factor and vaccine antigen factor H binding protein. Clin Vaccine Immunol. 2011 Jun;18(6):1002–1014. doi: 10.1128/CVI.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basta N.E., Mahmoud A.A., Wolfson J., Ploss A., Heller B.L., Hanna S. Immunogenicity of a meningococcal B vaccine during a University outbreak. N Engl J Med. 2016 Jul 21;375(3):220–228. doi: 10.1056/NEJMoa1514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckee C.O., Gupta S., Kriz P., Maiden M.C., Jolley K.A. Long-term evolution of antigen repertoires among carried meningococci. Proc Biol Sci. 2010 Jun 7;277(1688):1635–1641. doi: 10.1098/rspb.2009.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D.R., Ruijne N., McCallum L., O'Hallahan J., Oster P. The VR2 epitope on the PorA P1.7-2,4 protein is the major target for the immune response elicited by the strain-specific group B meningococcal vaccine MeNZB. Clin Vaccine Immunol. 2006 Apr;13(4):486–491. doi: 10.1128/CVI.13.4.486-491.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tappero J.W., Lagos R., Ballesteros A.M., Plikaytis B., Williams D., Dykes J. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA. 1999 Apr 28;281(16):1520–1527. doi: 10.1001/jama.281.16.1520. [DOI] [PubMed] [Google Scholar]
- 37.Read R.C., Baxter D., Chadwick D.R., Faust S.N., Finn A., Gordon S.B. Effect of a quadrivalent meningococcal ACWY glycoconjugate or a serogroup B meningococcal vaccine on meningococcal carriage: an observer-blind, phase 3 randomised clinical trial. Lancet. 2014 Aug 18;384(9960):2123–2131. doi: 10.1016/S0140-6736(14)60842-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.