Abstract
Background
Submicroscopic deletions in 14q12 spanning FOXG1 or intragenic mutations have been reported in patients with a developmental disorder described as a congenital variant of Rett syndrome. We aimed to further characterize and delineate the phenotype of FOXG1-mutation positive patients.
Method
We mapped the breakpoints of a 2;14 translocation by fluorescence in situ hybridization and analyzed three chromosome rearrangements in 14q12 by cytogenetic analysis and/or array CGH. We sequenced the FOXG1 gene in 210 patients, including 129 patients with unexplained developmental disorders and 81 MECP2-mutation negative individuals.
Results
We report one known mutation, seen in 2 patients, and 9 novel mutations of FOXG1 including 2 deletions, 2 chromosome rearrangements disrupting or displacing putative cis-regulatory elements from FOXG1, and 7 sequence changes. Analysis of our 11 patients, and further 15 patients reported in the literature, demonstrates a complex constellation of features including mild postnatal growth deficiency, severe postnatal microcephaly, severe mental retardation with absent language development, deficient social reciprocity resembling autism, combined stereotypies and frank dyskinesias, epilepsy, poor sleep patterns, irritability in infancy, unexplained episodes of crying, recurrent aspiration, and gastroesophageal reflux. Brain imaging studies reveal simplified gyral pattern and reduced white matter volume in the frontal lobes, corpus callosum hypogenesis, and variable mild frontal pachgyria.
Conclusions
We significantly expanded the number of FOXG1 mutations and identified two affecting possible cis-regulatory elements. While the phenotype of the patients overlaps both classic and congenital Rett syndrome, extensive clinical evaluation demonstrates a distinctive and clinically recognizable phenotype which we suggest to designate as the FOXG1 syndrome.
Keywords: Rett syndrome, FOXG1, mental retardation, microcephaly
INTRODUCTION
Recently, a series of reports have implicated deletions or sequence alterations of FOXG1 as the cause of a developmental disorder described as a congenital variant of Rett syndrome (RTT; MIM 312750). Small de novo interstitial deletions in 14q12 were detected by chromosome microarrays in four children1–4 and a complex chromosome rearrangement consisting of a translocation and adjacent small inversion in another.5 The common deleted region includes the telomeric breakpoint of the inversion in the latter patient and contains FOXG1, a strong candidate gene based on high and specific expression in developing brain and complex malformations of the telencephalon in both homozygous and heterozygous mouse mutants.6–8
Subsequently, sequencing of FOXG1 in five small cohorts of patients with overlapping phenotypes but negative results for MECP2 testing identified 11 patients with intragenic mutations including 5 nonsense, 4 frameshift and 2 missense mutations.9–13 Common clinical features in all these children included decelerating head circumference from 3 months that resulted in severe postnatal microcephaly, severe mental retardation with absent language, apraxia, jerky movements and generalized seizures. Brain imaging in several patients revealed hypogenesis of the corpus callosum.9,10,12 The phenotype was interpreted as resembling Rett syndrome except for no early period of normal development, which we view as one of several significant differences. Recently, duplications of FOXG1 have been associated with developmental epilepsy, mental retardation, and severe speech impairment in eight patients.14,15
After mapping a de novo chromosome translocation found in a severely mentally retarded girl to this region, we became interested in the types of mutations involving FOXG1 and the nature of the associated developmental disorder. Is the phenotype accurately described as a congenital variant of Rett syndrome, or is it a specific and recognizable syndrome in its own right? Here we report 2 microdeletions, 2 chromosome rearrangements most likely causing dysregulated FOXG1 expression, and 7 intragenic sequence changes among 210 patients tested. This cohort includes 129 patients referred to us directly because of unexplained developmental disorders, and 81 ascertained following negative testing for MECP2.
Our data and review of prior reports confirm mutations or dysregulation of FOXG1 as the cause of the developmental phenotype, which differs from classic Rett syndrome and other so-called Rett variants based on a specific and recognizable combination of developmental and brain imaging features. We prefer to designate this as the FOXG1 syndrome, which has overlapping features with a rapidly growing group of developmental encephalopathies that already includes Angelman (UBE3A-associated), Rett (MECP2-associated), Pitt-Hopkins (TCF4-associated), CNTNAP2-associated, NRXN1-associated and SLC9A6-associated syndromes.16–22
PATIENTS AND METHODS
Patients
We obtained clinical data, brain imaging studies and results of laboratory testing, as well as blood or DNA samples from 129 patients with mental retardation, microcephaly, frontal pachygyria or hypogenesis of the corpus callosum who were assessed by experienced clinical geneticists or neurologists. Two patients were recruited through the MRNET consortium (www.german-mrnet.de). We received 81 DNA samples from patients tested in a diagnostic lab for MECP2 and found to be negative. All available brain imaging studies from patients with a FOXG1 mutation were interpreted by one of the authors (W.B.D.) with selected images shown in Figures 1 and 2. The clinical data and samples were obtained with informed consent, including consent to use the photographs and videos in this report, under protocols approved by Institutional Review Boards at all participating institutions.
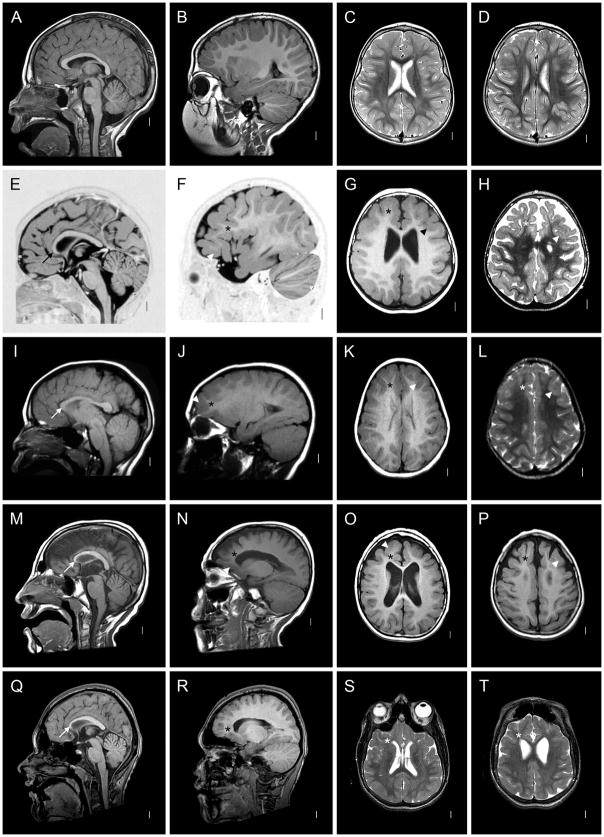
Figure 1.
Brain imaging in a normal 3-year-old girl (A–D) and four patients with FOXG1 mutations (E–T). Here and in figure 2, each row shows four images from the same patient. The columns contain midline sagittal (left column) and parasagittal (2nd column) images, and axial images through the lateral ventricles (3rd column) and high convexity (right column). All four patients have a low forehead that reflects underlying microcephaly, foreshortened frontal lobes and reduced white matter volume that appears severe in the frontal lobes (asterisks in 12/16 images) and subtle in posterior regions. All four have hypogenesis of the anterior corpus callosum with a “pointed hook” appearance in three of them (arrows in E, M, Q) that consists of striking narrowing of the anterior body, genu and rostrum with the tiny rostrum forming a pointed tip, and relatively normal posterior callosum. In the remaining patient (arrow in I), the entire body of the corpus callosum is narrow and the genu small but less so than the others, leaving it dysmorphic but different from the “pointed hook” appearance. Three patients have subtle pachygyria over the frontal lobes only that consists of mildly short and wide gyri and mildly thick cortex (arrowheads in F–H, J–L, N–P), a subtle abnormality that may be accentuated by the reduced volume of the white matter. The increased cortical thickness was not seen in the remaining patients (R–T and Figure 2). Also, three patients had mildly enlarged lateral ventricles, with one having abnormally narrow anterior body and frontal horns (S). These images come from patients 1 (E–H), 5 (I–L), 7 (M–P) and 9 (Q–T), and a normal control (A–D).
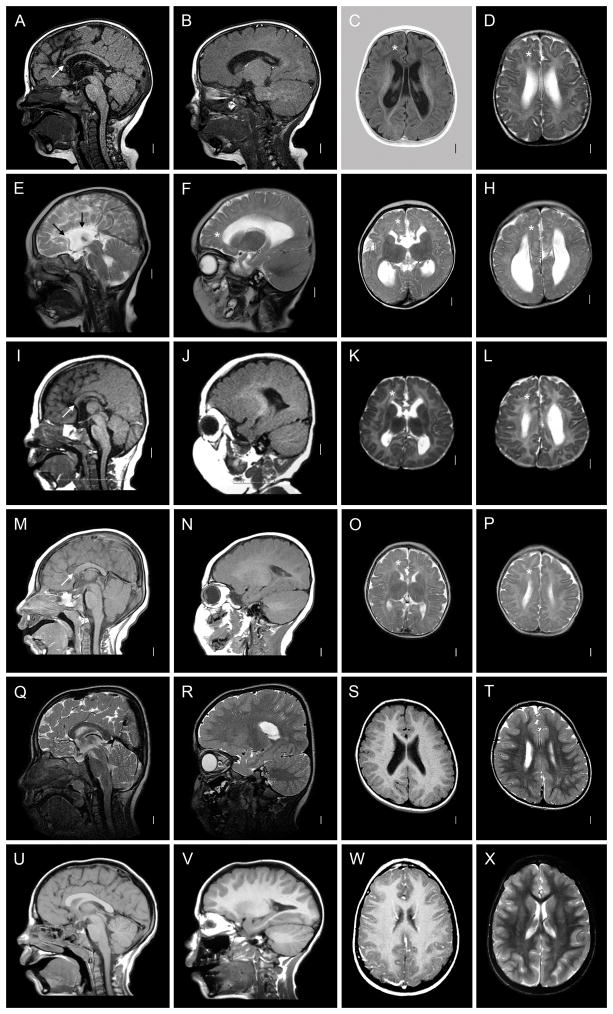
Figure 2.
Brain imaging in another six patients with FOXG1 mutations. Five have mildly enlarged lateral ventricles. The patients in the top four rows have changes similar to those seen in Figure 1, including microcephaly (E, I, M; not obvious in A, Q), short frontal lobes and reduced volume of frontal white matter (asterisks in 8/16 images). Hypogenesis of the corpus callosum was seen in 4/5, but with different patterns. The first patient (A) has a thin and dysmorphic corpus callosum similar to Figure 1I, the next total agenesis of the corpus callosum (E), and the third and fourth partial agenesis with a short and thin corpus callosum (I, M). No patients in this group have pachygyria. The last two patients have normal brain imaging except for subtle reduced volume of the frontal white matter and mildly simplified gyral pattern. These images come from patients 6 (A–D), 3 (E–H), 2 (I–L), 10 (M–P), 4 (Q–T), and 11 (U–X).
Fluorescence in situ hybridization (FISH)
Metaphase spreads from peripheral blood lymphocytes were prepared by standard procedure. FISH experiments for delineation of the 2q11.2 and 14q12 breakpoints were performed with BAC and fosmid clones. FISH with BAC clones was also performed to confirm array CGH and molecular karyotyping data. BACs (RP11 Human BAC Library) were obtained from the Resource Center for Genome Research at the Max-Planck-Institute for Molecular Genetics, Berlin, Germany, or the BACPAC Resource Center, Children's Hospital Oakland, CA, USA. Fosmid clones (WIBR-2 Human Fosmid Library [G248P8]) were received from the BACPAC Resource Center, Children's Hospital Oakland, CA, USA. BAC and fosmid DNA was prepared using the NucleoBond Xtra Midi kit (Machery-Nagel, Düren, Germany) or the Qiagen Large-Construct kit (Qiagen, Hilden, Germany). DNA was labeled by nick translation using the CGH Nick Translation Kit and Spectrum Green-dUTP (Vysis, Downers Grove, IL, USA) according to the protocol provided. Chromosomes were counterstained using 4',6-diamidino-2-phenylindole (DAPI) (Serva Feinbiochemica, Heidelberg, Germany) and mounted in antifading solution (Vector Labs, Burlingame, CA, USA). Slides were analyzed with a Leica Axioscope fluorescence microscope. Images were merged using a cooled CCD camera (Pieper, Schwerte, Germany) and Cyto Vision software (Applied Imaging, San Jose, CA, USA).
Molecular karyotyping and array comparative genomic hybridization (CGH)
Molecular karyotyping using the Affymetrix Mapping GeneChip 250 K Mapping SNP-Array (Nsp) (Affymetrix, Santa Clara, CA, USA) was performed in patient 3 according to the supplier’s instructions. Data were analyzed using Nexus and CNVF2.2 software.23 Array CGH in patient 4 was performed using a whole genome oligonucleotide array (244K, Agilent Technologies, Santa Clara, CA, USA). 244K image data were analyzed using Feature Extraction 9.5.3.1 and CGH Analytics 3.4.40 software (Agilent, Santa Clara, CA, USA) with the following analysis settings: aberration algorithm ADM-2; threshold: 6.0; window size: 0.2 Mb; filter: 5probes, log2ratio = 0.29.
Mutation analysis
We amplified the coding region of the FOXG1 gene (1 exon; GenBank accession no. NM_005249) from genomic DNA with five primer pairs that yielded overlapping amplicons. Primer sequences are available on request. Amplicons were directly sequenced using the ABI BigDye Terminator Sequencing Kit (Applied Biosystems, Darmstadt, Germany) and an automated capillary sequencer (ABI 3130; Applied Biosystems). Sequence electropherograms were analyzed using Sequence Pilot software (JSI medical systems, Kippenheim, Germany). Where mutations were shown to have arisen de novo, we verified self-reported relationships by genotyping both parents and the patient at ten microsatellite loci.
Results
Mapping of the chromosomal breakpoint in 14q12 by fluorescence in situ hybridization
Patient 1, a 12 year-old girl, was referred at 7 years because of severe mental retardation and postnatal microcephaly. She is the fourth child of healthy parents and her three older sisters are healthy. She was born at 40 weeks gestation by natural delivery with birth weight 3350 g (0 SD), length 51 cm (0 SD), and OFC 32.5 cm (−1 SD). At 6 months, she was able to turn, but she has never been able to sit or stand. She did not react to visual and acoustic stimuli in infancy, but this improved at later ages. Ophthalmologic examination revealed convergent strabismus. At 7 months she developed seizures. EEG revealed spikes and sharp waves over the left centroparietal and both centrotemporal regions. She could speak single words at 1 year, but lost this ability by 2 years. She had gastroesophageal reflux and constipation. Her brain MRI demonstrated striking reduced volume of white matter in the anterior frontal lobes with mildly thick cortex, striking hypoplasia and dysplasia of the anterior corpus callosum resembling a sharp hook, and probably dysplastic hippocampus (figure 1E–H).
Prenatal chromosome analysis revealed an apparently balanced chromosome translocation: 46,XX,t(2;14)(q13;q13). Chromosome analysis was normal in both parents, indicating that it occurred de novo (paternity confirmed). FISH analysis with fosmid F-1425A7 from chromosome 2 identified signals on 2q11.2 as well as the der(2) and der(14), indicating that this clone spanned the breakpoint (figure 3A and data not shown). No gene was directly disrupted by the breakpoint, although INPP4A is located 5–10 kb telomeric to the breakpoint (data not shown). A 1-bp deletion in Inpp4a encoding inositol polyphosphate 4-phosphatase type I occurred in the spontaneous weeble mouse mutant, which has severe locomotor instability and significant neuronal loss in the cerebellum and hippocampal CA1 sector.24 In addition, patient 1 is heterozygous for the breakpoint close to INPP4A, and further the weeble phenotype is a recessive condition that differs significantly from the phenotype in this girl.24
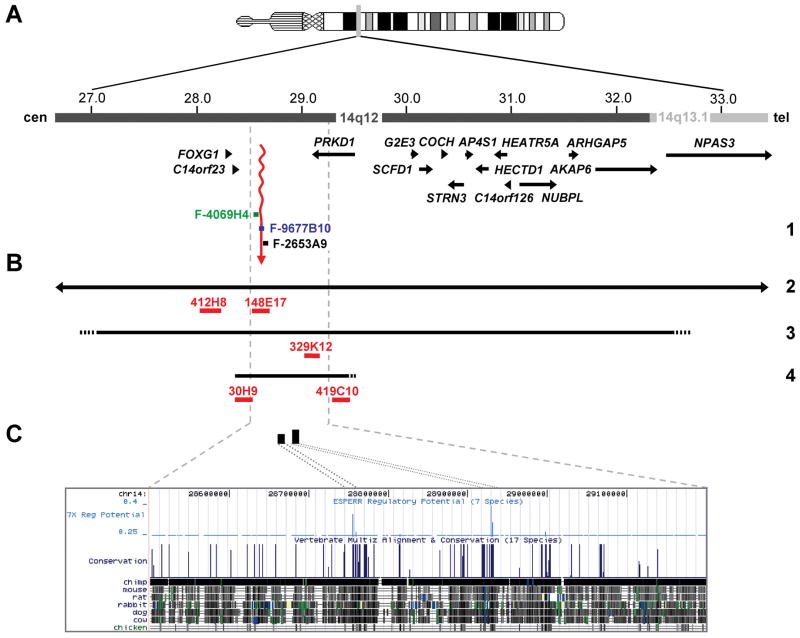
Figure 3.
Deletions including FOXG1 and regulatory mutations. (A) Physical map of the 14q12-q13.1 region. Genes in this region are represented by arrows indicating the 5’ 3’ orientation. Fosmid clones (F- [G248P8]; WIBR-2 Human Fosmid Library) used for mapping the 14q12 breakpoint of the 2;14 translocation in patient 1 (1) are indicated by colored bars and names are given. Color code of fosmids; black: mapped distal to the translocation breakpoint; blue: spanned the breakpoint; green: mapped proximal to breakpoint. The breakpoint region is indicated by a wavy vertical arrow in red. (B) Deletions in 14q12. Horizontal black lines depict the deletions identified in patients 2 – 4 (2, 3, 4). Arrowheads at the left and right side of the deletion in patient 2 show that this deletion extends to either side. Dotted lines indicate that the respective deletion breakpoints have not been fine-mapped. BAC clones (RP11 Human BAC Library) used for FISH analysis to confirm the presence of the deletions in the patients are indicated by red bars and names are given. (C) A detailed view of the region containing putative long-range cis-regulatory elements of FOXG1. In the upper part, the two black vertical bars represent potential regulatory elements (genomic positions chr14:28,754,647-28,756,495 bp and 28,930,280-28,932,099 bp) that have been identified with the ESPERR (evolutionary and sequence pattern extraction through reduced representations) computational method. In the lower part, a more detailed view of the region of high Regulatory Potential (RP) values, with the UCSC genome browser map of seven-way (human, chimpanzee, macaque, mouse, rat, dog, and cow; March 2006 assembly) RP analysis of the candidate cis-regulatory elements distal to FOXG1.
We therefore fine-mapped the 14q breakpoint, finding fosmid probes that hybridized proximal (F-4069H4) and distal (F-2653A9) to the breakpoint, as well as a single fosmid clone that spanned the breakpoint (F-9677B10, figure 3A). We narrowed the breakpoint to a ~10 kb gene-poor region in 14q12 located ~265 kb downstream of FOXG1 (figure 3A), suggesting that a position effect could cause transcriptional misregulation of this gene. In silico analysis using the Regulatory Potential score identified several candidate cis-regulatory elements that are evolutionary conserved throughout a 1-Mb region downstream of FOXG1 (figure 3C). Two of these noncoding elements located at genomic positions chr14:28,930,280-28,932,099 and chr14:28,754,647-28,756,495 (NCBI Build 36.1; figure 3C) have been analysed for gene enhancer activity as assessed in transgenic mice. Both DNA sequence elements are evolutionary highly conserved and were found to drive expression of a reporter gene (lacZ) in mouse forebrain (http://enhancer.lbl.gov/frnt_page.shtml). These data suggest that translocation of either or both of these putative cis-regulatory elements to chromosome 2 may have caused dysregulated expression of FOXG1 in the translocation patient.
Identification of deletions in 14q12 by cytogenetic and array CGH analysis
Patient 2 presented with developmental delay, slow head growth and severe feeding problems. Brain MRI showed a short corpus callosum (figure 2M–P). Routine chromosome analysis revealed a de novo deletion in chromosome 14 (46,XX,del(14)(q13.1q13.2)). We confirmed the deletion by FISH with two BAC clones, RP11-412H8 and RP11-148E7, flanking FOXG1 as well as BAC RP11-330O19 hybridizing proximal and RP11-908D14 and RP11-116N8 hybridizing distal to the deletion, respectively (data not shown and figure 3B). We estimate the maximum size of the deletion as ~8.4 Mb in 14q12-q13.1 including FOXG1.
Genome-wide array CGH was performed on a clinical basis on two patients with postnatal microcephaly and severe mental retardation. We detected a ~5.4-Mb heterozygous deletion in 14q12-q13.1 in patient 3 that encompasses at least 13 genes including FOXG1, and confirmed the deletion by FISH with BAC clone RP11-329K12 (figure 3B and data not shown). Neither parent carried the deletion (data not shown; paternity confirmed). We identified a ~1.09-Mb microdeletion downstream of FOXG1 in patient 4 that removes a large part of the next gene, PRKD1. PRKD1 encodes protein kinase D1 (also known as PKD or PKD1), a serine/threonine kinase that regulates a variety of cellular functions, including membrane receptor signalling, transport processes, protection from oxidative stress, transcriptional regulation, and F-actin reorganization.25,26 FISH with BAC clones RP11-30H9 and RP11-419C10 yielded only one signal on metaphase spreads, whereas two signals were obtained on metaphase spreads of his parents (figure 3B and data not shown; paternity confirmed). By quantitative PCR we confirmed the presence of two copies of the FOXG1 gene in patient 4 (data not shown), suggesting that similar to patient 1 a position effect might have caused altered expression of FOXG1. In line with this finding, the deletion spans the two putative long-range cis-regulatory elements for FOXG1 expression (figure 3C).
Sequence analysis of FOXG1
We next selected 125 patients with phenotypes overlapping patients 1–4, including mental retardation, postnatal microcephaly, mild frontal pachygyria, and callosal abnormalities. We sequenced FOXG1 in this cohort, and detected four intragenic mutations in the heterozygous state, including the transitions c.757A>G (p.N253D) in patient 5 and c.700T>C (p.S234P) in patient 11, the frameshift mutation c.460dupG (p.E154GfsX301) in patient 6, and the nonsense mutation c.256C>T (p.R86X) in patient 9. The missense mutations affect invariant amino acids, asparagine at position 253 and serine at position 234. Both residues are located in the DNA binding (forkhead) domain of FOXG1 suggesting that these amino acid exchanges may alter DNA binding properties of FOXG1. The c.757A>G mutation was not detected in the parents of patient 5 or in 204 control chromosomes. Similarly, the c.700T>C mutation was not found in the parents of patient 11. The 1-bp duplication was not identified in the parents of patient 6 or in 200 alleles, and the nonsense mutation was absent from the DNA sample of the mother of patient 9 (the father was deceased). Thus, three of four intragenic FOXG1 mutations occurred de novo (data not shown; all parental identities confirmed).
We also sequenced FOXG1 in a cohort of 81 patients (55 females and 26 males) tested for MECP2 mutations in a diagnostic lab with negative results. We detected the intragenic insertion-deletion mutation c.505_506delGGinsT (p.G168SfsX23) in patient 7 and the frameshift mutation c.263_278del16 (p.R88PfsX99) in patient 10, both in the heterozygous state. The parents of patients 7 and 10 did not carry the respective mutation (paternity confirmed), which was not detected in 200 control chromosomes. We also identified the same c.460dupG mutation in patient 8 as detected in patient 6. Sequencing of FOXG1 in patients 1–4 was negative.
Phenotype of the FOXG1-mutation positive patients
We reviewed clinical (table 1) and brain imaging (table 2) data in all eleven patients, and videotapes in four of the eleven patients (supplementary videos 1–4). Detailed results are listed in supplementary table 1 for all patients, and comparable information is collated for 16 previously reported patients in supplementary table 2. All of our patients had normal body measurements at birth with mildly slow growth thereafter leading to low weight (−1.5–2.5 SD) and low normal stature (0–2 SD) by 1–2 years. Birth head size was low normal (−0–2 SD) in 5/7 and borderline small (−2–3 SD) in 2/7 children. Severe postnatal microcephaly (MIC) was apparent by 2 years (−4–6 SD) in 6/8 patients.
Table 1.
Clinical data for patients with deletion 14q12 or FOXG1 mutations
| This report N=11 | Literature N=15 (ref. 1–5,9–13) | |
|---|---|---|
| FOXG1 mechanism | 4 CYTO, 7 intragenic | 5 CYTO, 10 intragenic |
| Subject demographics | ||
| Sex | 5F:6M | 14F:1M |
| Age last follow-up | 2y to 31y | 10mo to 22y |
| Growth | ||
| Weight birth (%) | 10/10 normal (10–90%) | 15/15 (10–90%) |
| Length birth (%) | 9/9 normal (10–75%) | 14/14 (25–90%) |
| OFC birth (SD) | 5/7 (−0–2 SD); 2/7 (−2.5 SD) | 4/14 (+1−0 SD); 10/13 (−0–2) |
| Weight age 1–2 years (SD), age | 5/6 (−1.5–2.6 SD); 1/6 25% | insufficient data |
| Length age 1–2 years (SD), age | 5/6 (−0–2 SD); 1/6 (−3.5 SD) | insufficient data |
| OFC age 1–2 years (SD), age | 8/8 (−2.5–6 SD) | insufficient data |
| Weight most recent (SD), age | 4/10 (−1–2 SD); 6/10 (−2–5.5 SD) | 6/9 (−1–2 SD); 3/9 (−3 SD) |
| Length most recent (SD), age | 2/8 (−1–2 SD); 5/8 (−2–5 SD) | 6/9 (−0–2 SD); 2/9 (−3.5–4) |
| OFC most recent (SD), age | 9/9 (−2.5–5.5 SD) | 12/12 (−2–5 SD) |
| Development | ||
| Developmental delay (DD) | 11/11 DD | |
| Mental retardation (MR) | 11/11 MR | 15/15 MR |
| Mental retardation (severity) | 11/11 severe | 5/5 severe |
| Speech-language development | 11/11 no language | 13/14 no language |
| Social interaction (eye contact) | 10/10 poor | 7/9 poor |
| Sitting (age) | 3/10 sit, all late | 3/13 sit |
| Walking (age) | 1/11 walk, late | 1/15 walk |
| Loss of psychomotor skills (age) | 2/11 vague history loss skills | 8–11 regression |
| Behavior, sleep, autonomic | ||
| Sleep pattern | 8/11 poor sleep pattern | 4/8 poor sleep pattern |
| Irritability | 5/10 excess irritability | insufficient data |
| Crying / weeping (inconsolable) | 6/10 excess crying | 3/6 excess crying |
| Laughing (inappropriate) | 0/11 excess laughing | insufficient data |
| Motor and dyskinesias | ||
| Hypotonia | 10/10 hypotonia | 6/6 hypotonia |
| Spasticity | 7/9 spasticity | 1/2 insufficient data |
| Functional hand use | 8/11 no hand use | 8/10 no hand use |
| Stereotypic movements | 8/11 stereotypies | 10/10 stereotypies |
| Dyskinesias (chorea, dystonia) | 8/10 dyskinesia (or all?) | 10/10 dyskinesia |
| Strabismus | insufficient data | 9/9 strabismus |
| Bruxism | 5/8 bruxism | 10/13 bruxism |
| Drooling (sialorhea) | no data | 7/9 drooling |
| Epilepsy | ||
| Seizures (age onset) | 8/10 seizures | 10/15 SZ (6mo to 14y) |
| Seizure types (specify) | CPS, FTCS, GTCS | CPS, GTCS, myoclonic |
| GI and respiratory systems | ||
| Feeding difficulties | 9/11 poor feeding | 5/6 poor feed |
| Aspiration | 6/10 aspiration | insufficient data |
| Gastroesophageal reflux (GER) | 9/10 GER often severe | 5/5 GER |
| Constipation | insufficient data | 6/6 constipation |
| Breathing abnormalities | non-specific | 3/5 abnormal respirations |
| Other abnormalities | ||
| Facial dysmorphism | 2/3 with deletions | 4/4 with deletions |
| Scoliosis | insufficient data | 4/11 scoliosis |
Abbreviations: CPS, complex partial seizures; CYTO, cytogenetic abnormality; FTCS, focal tonic clonic seizures; GER, gastroesophageal reflux; GTCS, generalized tonic clonic seizures; rec, recommended; SZ, seizures
Table 2.
Brain imaging data for patients with deletion 14q12 or FOXG1 mutations
| This report N=11 | Literature N=15 (ref. 1–5,9–13) | |
|---|---|---|
| Simplified gyral pattern (SIMP) | 11/11 SIMP | 01/01 SIMP |
| Pachygyria, mild frontal | 03/11 frontal pachygyria | 00/02 frontal pachygyria |
| Reduced volume white matter (frontal) | 11/11 reduced WM | 04/04 reduced WM |
| Hypogenesis corpus callosum (frontal) | 09/11 hypogenesis CC | 09/09 hypogenesis CC |
| Agenesis corpus callosum (severe) | 01/11 ACC with deletion | 02/13 ACC |
Abbreviations: ACC, agenesis of corpus callosum; CC, corpus callosum; SIMP, simplified gyral pattern; WM, white matter
All eleven patients had early global developmental delay and later severe mental retardation. Only 3/10 learned to sit independently and only 1/11 learned to walk. None developed speech. All had deficient social interactions, especially poor eye contact. Several improved enough to allow use of pointing and assistive-communication devices (but not speech), and limited social interaction. Two children had a history of possible regression, but this was subtle and difficult to document. The remainder had no history suggesting regression.
Exams reported muscular hypotonia in 10/10 and spasticity in 7/9 patients. We found stereotypies especially of the hands in 8/11, and dyskinesias in 8/10 with features of chorea, athetosis and dystonia. However, we observed dyskinesia in 6/6 patients reviewed during the course of this study with videos available of four patients (see supplementary videos 1–4), which suggests that this may be seen in all patients.
Dysfunction in several vegetative functions was apparent in all children. Sleep was disrupted from infancy in at least 8/11 children, with frequent nighttime waking. This usually improved with age, or after gastrostomy placement in one child. Prominent irritability or crying was reported in 6/10 patients, especially in infancy. Gastroesophageal reflux was diagnosed in 9/10 children, and was often severe, becoming a prominent part of the overall phenotype. Aspiration was documented in 6/10. A gastrostomy tube was recommended for feeding in 2/7 children but only placed in one.
Seizures were documented in 8/10 patients with onset between 3 months and 6 years. They consisted of tonic, generalized tonic-clonic, and partial complex seizures. None had infantile spasms. EEG findings were variable from reports, but the original studies were not reviewed. Several other features are noted in supplementary table 1.
Brain MRI studies were reviewed in all eleven patients (figures 1 and 2). The forehead was usually low, although this was subjective. All eleven had a simplified gyral pattern defined as a reduced number of gyri separated by shallow sulci, and significantly reduced volume of white matter that appears most severe - by far - in the frontal lobes. We observed mild frontal pachygyria in 3/11 patients; our recognition of subtle frontal pachygyria in patient 1 led directly to testing and detection of an intragenic mutation in patient 5. We found hypogenesis of the corpus callosum in 9/11 patients including three with a novel defect in which the frontal portion of the corpus callosum narrows to a sharp point or hook, with an absent rostrum and very small genu (figure 1E, M and Q). One child with a large 14q12 deletion had complete agenesis of the corpus callosum (figure 2I). The remaining patients had a short or thin corpus callosum with dysmorphic or absent rostrum and narrow or absent splenium. The two patients with a normal corpus callosum were those with a 14q12 deletion 35 kb distant from FOXG1 (figure 2Q) and the patient with the missense mutation p.S234P (figure 2U).
The two children with large deletions (patients 2 and 3) had mild dysmorphic facial features in addition to the MIC, including wide and round face, flat midface, large ears and small jaw. The remaining children had obvious MIC with subtle and non-specific dysmorphic features (figure 4).
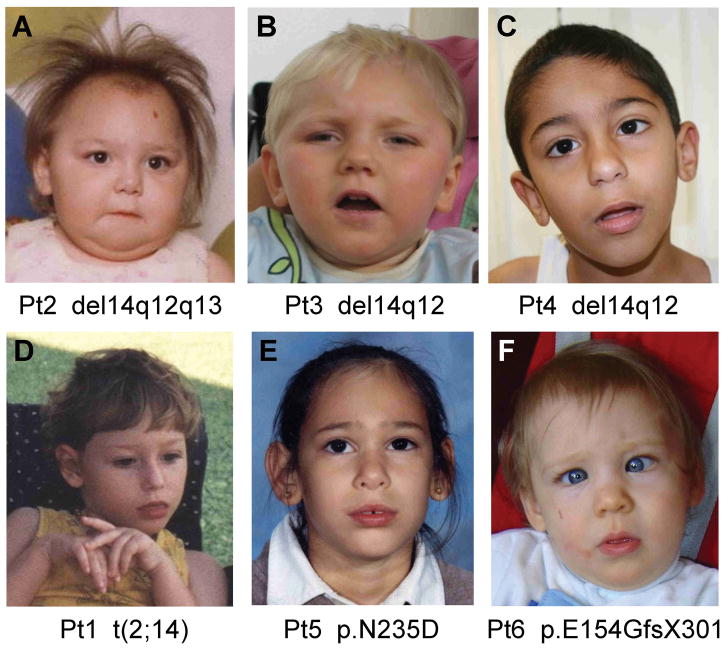
Figure 4.
Patients with FOXG1 mutations. Two patients with 14q12 deletions (top row, A–B) demonstrate mild facial dysmorphism consisting of round face with flat midface, low nasal bridge, bulbous nasal tip, thin upper lip and prominent ears (A–B). Four children with regulatory (top row, C and bottom row, D) or intragenic mutations (bottom row, E–F) have normal facial appearance except for mildly prominent ears. One photo shows hand stereotypy (D), and another shows striking esotropia (F). These patient numbers and mutations are shown at the bottom of each photo.
DISCUSSION
Foxg1 is a transcription factor that regulates development of the telencephalon from early embryonic to adult stages by multiple diverse mechanisms.6,7,27 Not surprisingly, it was recognized as a strong candidate gene to explain the developmental disorder associated with deletion 14q12 and as a single gene cause of human developmental disorders.5,9 Our study extends several previous reports, demonstrating that heterozygous loss of FOXG1 results in a consistent and severe developmental phenotype with emerging genotype-phenotype correlations.
Foxg1 function
Foxg1 is a conserved transcriptional repressor that is strongly expressed in progenitors of the ventricular zone and early postmitotic neurons in the developing neuroepithelium of the telencephalon and visual structures.28–31 It is involved in the timing of neuronal differentiation and in the specification of subdomain identity, regulating dorsal-ventral and subsequently rostral-lateral regional differentiation and later neural progenitor versus Cajal-Retzius cell fate, expansion of the neural progenitor pool, and progenitor cell cycle length.8,30,32–35
Homozygous Foxg1−/− mutants die at birth with a dramatic reduction in the size of the cerebral hemispheres affecting ventral more severely than dorsal structures, and depletion of neural progenitor pools.8,30,32–35 The cortical plate is poorly organized with loss of layer-specific markers, and appears relatively thick in lateral cortical regions as a result of accelerated neuronal differentiation (see figure 1 in Hanashima et al 2004).
Heterozygous Foxg1+/− (especially Foxg1+/cre) mutants have less severe developmental defects including reduced volumes of the cerebral cortex, striatum and hippocampus, and thin cortex due to reduced thickness of the superficial layers (II–III) of the cortex. The latter results from loss of neuron-generating intermediate progenitor cells during mid- and late corticogenesis and, to a lesser extent, from a prolonged cell cycle in late corticogenesis.6,7 These data predict that humans with FOXG1 haploinsufficiency should have microcephaly, thin cortex with abnormal cortical cytoarchitecture (but with a thicker cortex possible in lateral cortical regions), and cognitive plus behavioral defects.
FOXG1 mutations
Combining our series and prior reports, 18 intragenic sequence changes of FOXG1 have been reported including 14 null − 6 nonsense and 8 frameshift − and 4 missense mutations (supplementary tables 1 and 2). The missense mutations (p.F215L, p.N227K, p.S234P, and p.N253D) all affect evolutionary highly conserved amino acid residues in the forkhead domain involved in DNA binding. One group hypothesized that amino acid substitutions in functional domains of FOXG1 and late-truncating mutations may lead to a milder phenotype including typical and atypical Rett syndrome because the resulting FOXG1 proteins may retain some functions, especially their ability to bind corepressors.13 However, the p.Y400X and p.S323fsX325 mutations, identified in patients with a severe phenotype leave one FOXG1 domain intact which interacts with the Groucho corepressor family.36 Thus, missense as well as clear loss-of-function mutations can have a severe impact on FOXG1 function suggesting that other factors may modulate the phenotypic outcome.
The recurrent c.460dupG mutation causing duplication of a guanine after 7 subsequent guanines in FOXG1 was identified twice in our patient cohort and in one reported patient.10 These findings suggest that this guanine stretch may be prone to replication errors and represent a mutational hot spot in FOXG1.
We identified the 14q12 breakpoint in patient 1 about 265 kb downstream of the FOXG1 gene. The finding of de novo FOXG1 mutations in patients with a developmental encephalopathy overlapping Rett syndrome strongly suggests that altered expression of FOXG1 due to displacement of cis-regulatory elements important for its spatial and temporal expression caused the disease phenotype in patient 1. The 14q12 breakpoint of a different de novo 2;14 translocation was mapped ~5 kb downstream of the FOXG1 gene.5 The authors postulated that the breakpoint disrupted novel FOXG1 splice variants between exons 3 and 4 with a brain-specific expression pattern, represented by the EST sequence BX248251. However, the non-coding RNA gene C14orf23 maps distal to FOXG1 (figure 3A), and the reference sequence of this gene was derived from BX248251, which suggests that the putative FOXG1 exons 2–5 more likely represent exons of C14orf23. In line with this, no mutations in these exons have yet been reported.13 Thus, we suggest that both 2;14 translocations resulted in removal of long-range regulatory elements of FOXG1, located in the gene desert between FOXG1 and PRKD1.
Further evidence supporting the existence of sequences regulating FOXG1 expression comes from our identification of an ~1-Mb microdeletion not covering FOXG1 in patient 4. The phenotypes of both patients 1 and 4 are very similar to FOXG1-mutation positive individuals, suggesting that dysregulated FOXG1 expression may be responsible for their clinical features. In addition, two more patients with small overlapping microdeletions of 0.14 − 1.8 Mb that do not seem to include FOXG1 have been reported and are associated with a phenotype fitting with that of FOXG1-mutation positive patients.3 Our in silico analysis revealed two highly conserved non-coding elements of 1819 bp and 1848 bp between FOXG1 and PRKD1 that drive expression of a reporter gene specifically in the forebrain of transgenic mice (http://enhancer.lbl.gov/frnt_page.shtml). Thus, we propose that regulatory mutations, i.e. displacement and deletion of the two putative cis-regulatory elements by the translocation and the ~1-Mb microdeletion, respectively, cause a phenotype similar to that caused by mutations directly affecting FOXG1. Nonetheless, the phenotype of patient 4 is less severe than our other patients. This suggests disrupted function of a spatiotemporally specific subset of the normal expression pattern.37
The FOXG1 syndrome
We report the largest series of patients with FOXG1 mutations to date. Analysis of our eleven patients and 16 patients from the literature1–5,9,10,12,13 reveals a complex and consistent developmental phenotype that affects many areas of neurologic and behavioural function (thus classified as a developmental encephalopathy). By recognizing classic dyskinesias and a novel and specific pattern of brain malformations in our cohort of patients we also expand the phenotype.
The characteristic features of the FOXG1 syndrome include (1) moderate postnatal growth deficiency, (2) severe postnatal microcephaly, (3) severe mental retardation with (4) absent or minimal language development, (5) deficient social interactions including poor eye contact denoting a syndromic form of autism, (6) combined stereotypies and frank dyskinesias with mixed features of athetosis, chorea and dystonia, (7) epilepsy, (8) poor sleep patterns, (9) irritability especially in infancy, (10) excessive episodes of crying, (11) recurrent aspiration, and (12) frequent gastroesophageal reflux. While a few prior reports described developmental regression in some children, the history was vague as it was in two of our patients, leading us to exclude this as a diagnostic feature. Several other clinical problems were reported in a few patients, such as clubfeet, scoliosis, bruxism, and constipation.
Brain imaging studies reveal frontal predominant simplified gyral pattern, reduced white matter volume and callosal hypogenesis, with variable appearance of mild frontal pachygyria. Complete agenesis of the corpus callosum was observed in only 3/24 patients overall, but in 2/7 patients with deletion 14q12, which suggests modifying genes in the region. Considering the thin cortex observed in the Foxg1+/− mouse, it seems possible that the mild pachygyria seen in a few patients is an artifact resulting from image averaging of the cortical ribbon due to the very thin frontal white matter. However, the Foxg1−/− mouse has a dysplastic cortex, so we prefer to reserve judgment on this issue until higher resolution brain scans or human brain tissue become available.
FOXG1 genotype-phenotype analysis
The number of reported mutations is now large enough to begin genotype-phenotype correlation. Combining our data and several literature reports, the core phenotype is seen in individuals with mutations causing protein truncation and in some with missense and other mutations. Patients with large deletions of 14q12 that include FOXG1 and other genes may have more severe phenotypes, adding complete agenesis of the corpus callosum and mild facial dysmorphism to the core phenotype.
Also, some patients with missense or putative regulatory mutations appear to have less severe phenotypes. Combining our series and literature reports, only 6/23 patients were able to sit, and 4 of the 6 had less severe mutations including a late truncation (p.S323fsX325), a regulatory mutation (deletion 34 kb downstream of FOXG1 in patient 4), and two missense mutations (p.F215L, and p.N253D in patient 5). Only 2/26 patients learned to walk and both had less severe mutations including one late truncation (p.Y400X) and the same missense mutation (patient 5 with p.N253D).
The association between FOXG1 and developmental abnormalities in humans is further supported by recent reports of duplication 14q12 including the FOXG1 in 8 patients.14,15 The phenotype resembles the FOXG1 haploinsufficiency syndrome described above, including severe developmental delay, mental retardation and absent or minimal language in 8/8, seizures in 5/8, hypotonia in 2/8, stereotyped behaviour especially perseverative hand movements in at least 4/8, and inappropriate laughing and sleep disturbance in at least 1/8 patients. However, the seizures were more severe, with typical infantile spasms reported in 4/8 patients. Other differentiating features include normal head circumference by age 1–3 years in 7/8 patients, and walking in at least 3/8 patients. Based on limited data, brain imaging features appear to be similar but less severe including subtle hypogenesis of the corpus callosum in at least 3 patients, and reduced white matter volume in at least 2 patients (this interpretation is based partly on our interpretation of Figure 2 in14).
The FOXG1 syndrome or congenital Rett syndrome variant
This disorder was first described as a congenital variant of Rett syndrome. While many features do overlap with Rett syndrome, several features beyond the congenital onset differ as well. For example, the presence of true dyskinesias and brain imaging abnormalities, as well as the lack of regression and lack of respiratory arrhythmia differ from classic Rett syndrome. We believe that the combination of developmental and brain imaging features observed in individuals with FOXG1 mutations are sufficiently distinct to allow clinical recognition of this disorder, which we therefore prefer to designate simply as the FOXG1 syndrome.
Further, the phenotypic overlap with other complex developmental syndromes extends far beyond similarity to Rett syndrome to include at least nine different syndromes and genes: Angelman syndrome, classic Rett and other MECP2-associated syndromes, the ARX-, CDKL5-, CNTNAP2-, NRXN1-, SLC9A6-, and TCF4-associated syndromes, and maternal isodicentric and duplication 15q11.2q13 syndromes.16,18,21,22,38–42 Most are single gene disorders, and all present with a wide spectrum of cognitive, social, affective, autonomic and motor deficits, as well as epilepsy. Considering the increasing number and phenotypic heterogeneity of these disorders, we have begun to use the term complex or mixed “developmental encephalopathies” to refer to the group as a whole.
In conclusion, we present the first evidence for mutations of cis-regulatory elements required for spatially, temporally, and quantitatively correct activity of FOXG1 in patients with the same phenotype as individuals with intragenic FOXG1 lesions, as well as several novel intragenic mutations. The specific pattern of developmental features and brain malformations observed in individuals with FOXG1 mutations should allow clinical recognition of the FOXG1 syndrome and prompt genetic testing of the gene.
Supplementary Material
Acknowledgments
We are grateful to the patients and their families who contributed to this study. Some of the results summarized here form part of the diploma work of Fanny Kortüm and the MD thesis of Max Flindt at the University of Hamburg. We are grateful to Birgit Schulze (Sozialpädiatrisches Zentrum, Frankfurt, Germany) and Christoph Korenke (Zentrum für Kinder- und Jugendmedizin, Klinikum Oldenburg, Germany) for their clinical assessment of patient 6, and Rainer Gabriel (Gemeinschaftspraxis Neuropädiatrie, Lüneburg, Germany) for clinical assessment of patient 11. We thank Inka Jantke, Stefanie Dora, and Fabienne Trotier for skillful technical assistance.
Funding
This work was supported by grants from the Werner Otto-Stiftung [WOS 3/76 to G.U.]; the Deutsche Forschungsgemeinschaft (DFG) [GRK1459 to K.K.]; and the National Institutes of Health (NIH) [1R01-NS058721 to W.B.D]. This work was part of the German Mental Retardation Network (MRNET) funded through a grant from the German Ministry of Research and Education to A. Rauch [01GS08160] and D.W. [01GS08164].
Footnotes
Competing interest
None.
Patient consent
Parental consents obtained.
Licence for Publication statement:
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd to permit this article (if accepted) to be published in JMG and any other BMJPGL products and sublicences such use and exploit all subsidiary rights, as set out in our licence (http://group.bmj.com/products/journals/instructions-for-authors/licence-forms).
References
- 1.Bisgaard AM, Kirchhoff M, Tumer Z, Jepsen B, Brondum-Nielsen K, Cohen M, Hamborg-Petersen B, Bryndorf T, Tommerup N, Skovby F. Additional chromosomal abnormalities in patients with a previously detected abnormal karyotype, mental retardation, and dysmorphic features. Am J Med Genet A. 2006;140:2180–7. doi: 10.1002/ajmg.a.31425. [DOI] [PubMed] [Google Scholar]
- 2.Jacob FD, Ramaswamy V, Andersen J, Bolduc FV. Atypical Rett syndrome with selective FOXG1 deletion detected by comparative genomic hybridization: case report and review of literature. Eur J Hum Genet. 2009;17:1577–81. doi: 10.1038/ejhg.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mencarelli MA, Kleefstra T, Katzaki E, Papa FT, Cohen M, Pfundt R, Ariani F, Meloni I, Mari F, Renieri A. 14q12 Microdeletion syndrome and congenital variant of Rett syndrome. Eur J Med Genet. 2009;52:148–52. doi: 10.1016/j.ejmg.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Papa FT, Mencarelli MA, Caselli R, Katzaki E, Sampieri K, Meloni I, Ariani F, Longo I, Maggio A, Balestri P, Grosso S, Farnetani MA, Berardi R, Mari F, Renieri A. A 3 Mb deletion in 14q12 causes severe mental retardation, mild facial dysmorphisms and Rett-like features. Am J Med Genet A. 2008;146A:1994–8. doi: 10.1002/ajmg.a.32413. [DOI] [PubMed] [Google Scholar]
- 5.Shoichet SA, Kunde SA, Viertel P, Schell-Apacik C, von Voss H, Tommerup N, Ropers HH, Kalscheuer VM. Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations and microcephaly. Hum Genet. 2005;117:536–44. doi: 10.1007/s00439-005-1310-3. [DOI] [PubMed] [Google Scholar]
- 6.Eagleson KL, Schlueter McFadyen-Ketchum LJ, Ahrens ET, Mills PH, Does MD, Nickols J, Levitt P. Disruption of Foxg1 expression by knock-in of cre recombinase: effects on the development of the mouse telencephalon. Neuroscience. 2007;148:385–99. doi: 10.1016/j.neuroscience.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex. 2008;18:1865–75. doi: 10.1093/cercor/bhm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xuan S, Baptista CA, Balas G, Tao W, Soares VC, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–52. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 9.Ariani F, Hayek G, Rondinella D, Artuso R, Mencarelli MA, Spanhol-Rosseto A, Pollazzon M, Buoni S, Spiga O, Ricciardi S, Meloni I, Longo I, Mari F, Broccoli V, Zappella M, Renieri A. FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet. 2008;83:89–93. doi: 10.1016/j.ajhg.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahi-Buisson N, Nectoux J, Girard B, Van Esch H, De Ravel T, Boddaert N, Plouin P, Rio M, Fichou Y, Chelly J, Bienvenu T. Revisiting the phenotype associated with FOXG1 mutations: two novel cases of congenital Rett variant. Neurogenetics. 2010;11:241–9. doi: 10.1007/s10048-009-0220-2. [DOI] [PubMed] [Google Scholar]
- 11.Le Guen T, Bahi-Buisson N, Nectoux J, Boddaert N, Fichou Y, Diebold B, Desguerre I, Raqbi F, Daire VC, Chelly J, Bienvenu T. A FOXG1 mutation in a boy with congenital variant of Rett syndrome. Neurogenetics. 2011;12:1–8. doi: 10.1007/s10048-010-0255-4. [DOI] [PubMed] [Google Scholar]
- 12.Mencarelli MA, Spanhol-Rosseto A, Artuso R, Rondinella D, De Filippis R, Bahi-Buisson N, Nectoux J, Rubinsztajn R, Bienvenu T, Moncla A, Chabrol B, Villard L, Krumina Z, Armstrong J, Roche A, Pineda M, Gak E, Mari F, Ariani F, Renieri A. Novel FOXG1 mutations associated with the congenital variant of Rett syndrome. J Med Genet. 2010;47:49–53. doi: 10.1136/jmg.2009.067884. [DOI] [PubMed] [Google Scholar]
- 13.Philippe C, Amsallem D, Francannet C, Lambert L, Saunier A, Verneau F, Jonveaux P. Phenotypic variability in Rett syndrome associated with FOXG1 mutations in females. J Med Genet. 2010;47:59–65. doi: 10.1136/jmg.2009.067355. [DOI] [PubMed] [Google Scholar]
- 14.Brunetti-Pierri N, Paciorkowski AR, Ciccone R, Mina ED, Bonaglia MC, Borgatti R, Schaaf CP, Sutton VR, Xia Z, Jelluma N, Ruivenkamp C, Bertrand M, de Ravel TJ, Jayakar P, Belli S, Rocchetti K, Pantaleoni C, D'Arrigo S, Hughes J, Cheung SW, Zuffardi O, Stankiewicz P. Duplications of FOXG1 in 14q12 are associated with developmental epilepsy, mental retardation, and severe speech impairment. Eur J Hum Genet. 2011;19:102–7. doi: 10.1038/ejhg.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung A, Bruno D, Scheffer IE, Carranza D, Burgess T, Slater HR, Amor DJ. 4.45 Mb microduplication in chromosome band 14q12 including FOXG1 in a girl with refractory epilepsy and intellectual impairment. Eur J Med Genet. 2009;52:440–2. doi: 10.1016/j.ejmg.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Amiel J, Rio M, de Pontual L, Redon R, Malan V, Boddaert N, Plouin P, Carter NP, Lyonnet S, Munnich A, Colleaux L. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet. 2007;80:988–93. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 18.Gilfillan GD, Selmer KK, Roxrud I, Smith R, Kyllerman M, Eiklid K, Kroken M, Mattingsdal M, Egeland T, Stenmark H, Sjoholm H, Server A, Samuelsson L, Christianson A, Tarpey P, Whibley A, Stratton MR, Futreal PA, Teague J, Edkins S, Gecz J, Turner G, Raymond FL, Schwartz C, Stevenson RE, Undlien DE, Stromme P. SLC9A6 mutations cause X-linked mental retardation, microcephaly, epilepsy, and ataxia, a phenotype mimicking Angelman syndrome. Am J Hum Genet. 2008;82:1003–10. doi: 10.1016/j.ajhg.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–3. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 20.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–7. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 21.Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, Bijlsma EK, Oortveld MA, Ekici AB, Reis A, Schenck A, Rauch A. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–66. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, Clayton-Smith J, Reardon W, Saraiva J, Cabral A, Gohring I, Devriendt K, de Ravel T, Bijlsma EK, Hennekam RC, Orrico A, Cohen M, Dreweke A, Reis A, Nurnberg P, Rauch A. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am J Hum Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyer J, Dreweke A, Becker C, Gohring I, Thiel CT, Peippo MM, Rauch R, Hofbeck M, Trautmann U, Zweier C, Zenker M, Huffmeier U, Kraus C, Ekici AB, Ruschendorf F, Nurnberg P, Reis A, Rauch A. Molecular karyotyping in patients with mental retardation using 100K single-nucleotide polymorphism arrays. J Med Genet. 2007;44:629–36. doi: 10.1136/jmg.2007.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nystuen A, Legare ME, Shultz LD, Frankel WN. A null mutation in inositol polyphosphate 4-phosphatase type I causes selective neuronal loss in weeble mutant mice. Neuron. 2001;32:203–12. doi: 10.1016/s0896-6273(01)00468-8. [DOI] [PubMed] [Google Scholar]
- 25.Jaggi M, Du C, Zhang W, Balaji KC. Protein kinase D1: a protein of emerging translational interest. Front Biosci. 2007;12:3757–67. doi: 10.2741/2349. [DOI] [PubMed] [Google Scholar]
- 26.Storz P. Protein kinase D1: a novel regulator of actin-driven directed cell migration. Cell Cycle. 2009;8:1975–6. [PubMed] [Google Scholar]
- 27.Roth M, Bonev B, Lindsay J, Lea R, Panagiotaki N, Houart C, Papalopulu N. FoxG1 and TLE2 act cooperatively to regulate ventral telencephalon formation. Development. 2010;137:1553–62. doi: 10.1242/dev.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourguignon C, Li J, Papalopulu N. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development. 1998;125:4889–900. doi: 10.1242/dev.125.24.4889. [DOI] [PubMed] [Google Scholar]
- 29.Murphy DB, Wiese S, Burfeind P, Schmundt D, Mattei MG, Schulz-Schaeffer W, Thies U. Human brain factor 1, a new member of the fork head gene family. Genomics. 1994;21:551–7. doi: 10.1006/geno.1994.1313. [DOI] [PubMed] [Google Scholar]
- 30.Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–66. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhao XF, Suh CS, Prat CR, Ellingsen S, Fjose A. Distinct expression of two foxg1 paralogues in zebrafish. Gene Expr Patterns. 2009;9:266–72. doi: 10.1016/j.gep.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Dou CL, Li S, Lai E. Dual role of brain factor-1 in regulating growth and patterning of the cerebral hemispheres. Cereb Cortex. 1999;9:543–50. doi: 10.1093/cercor/9.6.543. [DOI] [PubMed] [Google Scholar]
- 33.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–9. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 34.Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–27. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Muzio L, Mallamaci A. Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J Neurosci. 2005;25:4435–41. doi: 10.1523/JNEUROSCI.4804-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao J, Lai E, Stifani S. The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol Cell Biol. 2001;21:1962–72. doi: 10.1128/MCB.21.6.1962-1972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinjan DA, Lettice LA. Long-range gene control and genetic disease. Adv Genet. 2008;61:339–88. doi: 10.1016/S0065-2660(07)00013-2. [DOI] [PubMed] [Google Scholar]
- 38.Battaglia A. The inv dup (15) or idic (15) syndrome (Tetrasomy 15q) Orphanet J Rare Dis. 2008;3:30. doi: 10.1186/1750-1172-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–37. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Jedele KB. The overlapping spectrum of rett and angelman syndromes: a clinical review. Semin Pediatr Neurol. 2007;14:108–17. doi: 10.1016/j.spen.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Kato M. A new paradigm for West syndrome based on molecular and cell biology. Epilepsy Res. 2006;70(Suppl 1):S87–95. doi: 10.1016/j.eplepsyres.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Russo S, Marchi M, Cogliati F, Bonati MT, Pintaudi M, Veneselli E, Saletti V, Balestrini M, Ben-Zeev B, Larizza L. Novel mutations in the CDKL5 gene, predicted effects and associated phenotypes. Neurogenetics. 2009;10:241–50. doi: 10.1007/s10048-009-0177-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.