Abstract
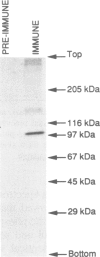
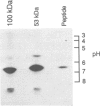
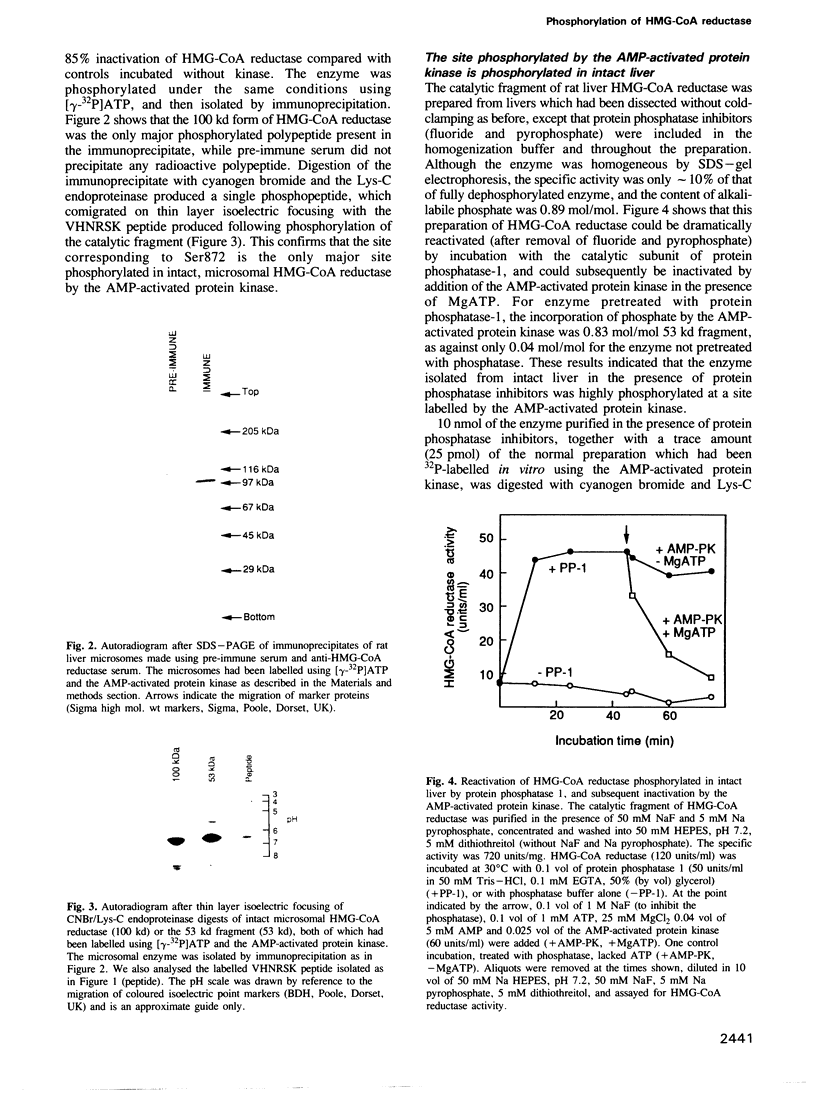
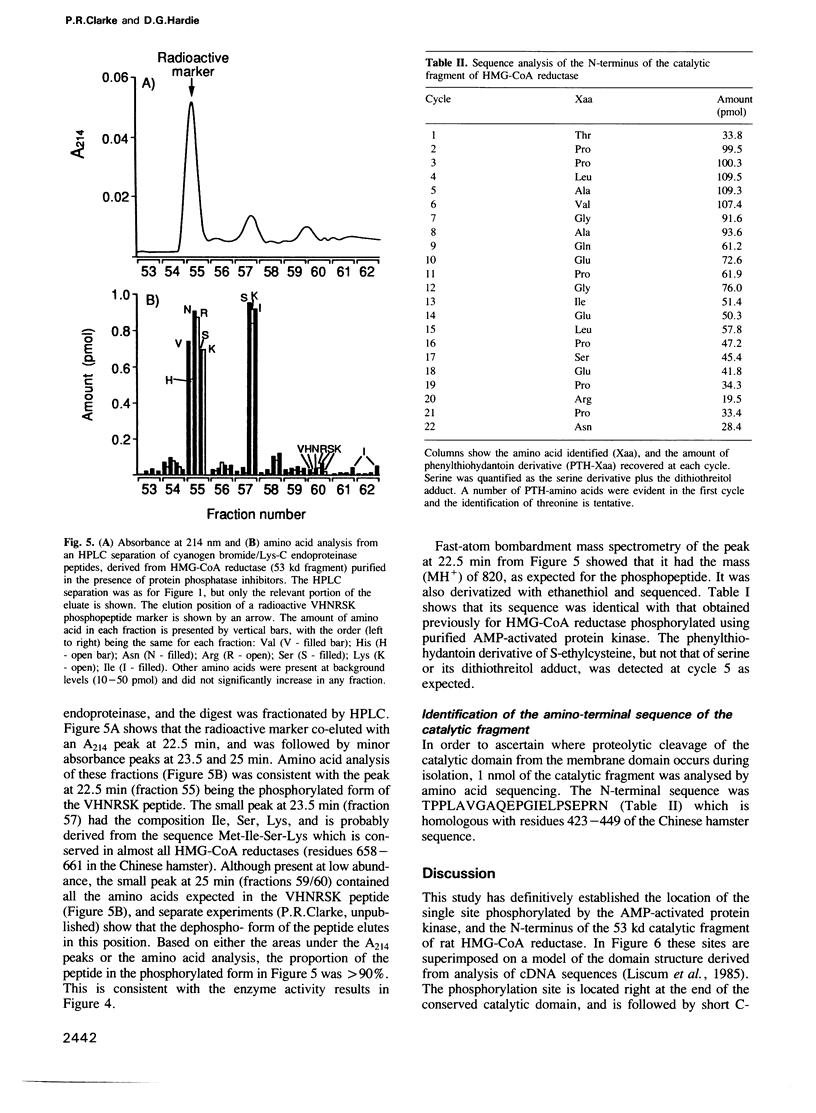
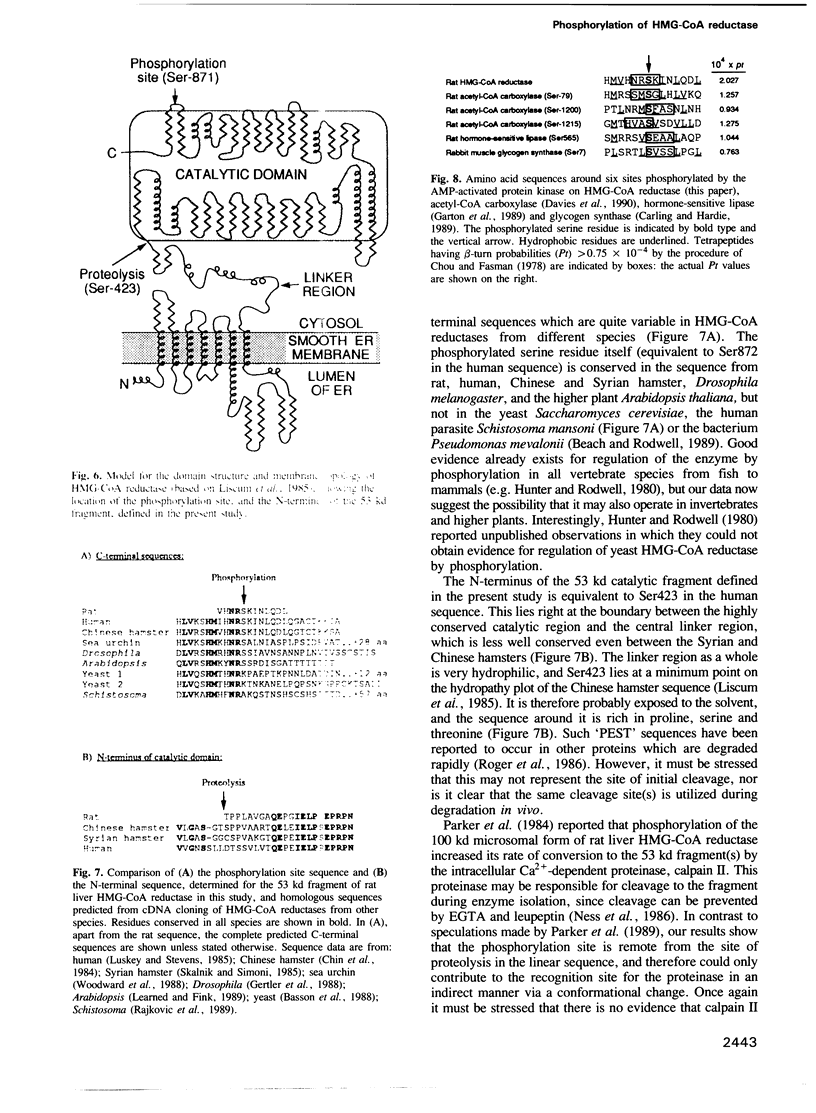
The intact, 100 kd microsomal enzyme and the 53 kd catalytic fragment of rat HMG-CoA reductase are both phosphorylated and inactivated by the AMP-activated protein kinase. Using the catalytic fragment, we have purified and sequenced peptides containing the single site of phosphorylation. Comparison with the amino acid sequence predicted from the cDNAs encoding other mammalian HMG-CoA reductases identifies this site as a serine residue close to the C-terminus (Ser872 in the human enzyme). Phosphopeptide mapping of native, 100 kd microsomal HMG-CoA reductase confirms that this C-terminal serine is the only major site phosphorylated in the intact enzyme by the AMP-activated protein kinase. The catalytic fragment of HMG-CoA reductase was also isolated from rat liver in the presence of protein phosphatase inhibitors under conditions where the enzyme is largely in the inactive form. HPLC, mass spectrometry and sequencing of the peptide containing Ser872 demonstrated that this site is highly phosphorylated in intact liver under these conditions. We have also identified by amino acid sequencing the N-terminus of the catalytic fragment, which corresponds to residue 423 of the human enzyme.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basson M. E., Thorsness M., Finer-Moore J., Stroud R. M., Rine J. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol. 1988 Sep;8(9):3797–3808. doi: 10.1128/mcb.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach M. J., Rodwell V. W. Cloning, sequencing, and overexpression of mvaA, which encodes Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Bacteriol. 1989 Jun;171(6):2994–3001. doi: 10.1128/jb.171.6.2994-3001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg Z. H., Stonik J. A., Brewer H. B., Jr Modulation of the enzymic activity of 3-hydroxy-3-methylglutaryl coenzyme A reductase by multiple kinase systems involving reversible phosphorylation: a review. Metabolism. 1987 Sep;36(9):900–917. doi: 10.1016/0026-0495(87)90101-6. [DOI] [PubMed] [Google Scholar]
- Beg Z. H., Stonik J. A., Brewer H. B., Jr Phosphorylation and modulation of the enzymic activity of native and protease-cleaved purified hepatic 3-hydroxy-3-methylglutaryl-coenzyme A reductase by a calcium/calmodulin-dependent protein kinase. J Biol Chem. 1987 Sep 25;262(27):13228–13240. [PubMed] [Google Scholar]
- Beg Z. H., Stonik J. A., Brewer H. B., Jr Phosphorylation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase and modulation of its enzymic activity by calcium-activated and phospholipid-dependent protein kinase. J Biol Chem. 1985 Feb 10;260(3):1682–1687. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L., Dietschy J. M. Active and inactive forms of 3-hydroxy-3-methylglutaryl coenzyme A reductase in the liver of the rat. Comparison with the rate of cholesterol synthesis in different physiological states. J Biol Chem. 1979 Jun 25;254(12):5144–5149. [PubMed] [Google Scholar]
- Carling D., Clarke P. R., Zammit V. A., Hardie D. G. Purification and characterization of the AMP-activated protein kinase. Copurification of acetyl-CoA carboxylase kinase and 3-hydroxy-3-methylglutaryl-CoA reductase kinase activities. Eur J Biochem. 1989 Dec 8;186(1-2):129–136. doi: 10.1111/j.1432-1033.1989.tb15186.x. [DOI] [PubMed] [Google Scholar]
- Carling D., Hardie D. G. The substrate and sequence specificity of the AMP-activated protein kinase. Phosphorylation of glycogen synthase and phosphorylase kinase. Biochim Biophys Acta. 1989 Jun 15;1012(1):81–86. doi: 10.1016/0167-4889(89)90014-1. [DOI] [PubMed] [Google Scholar]
- Carling D., Zammit V. A., Hardie D. G. A common bicyclic protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS Lett. 1987 Nov 2;223(2):217–222. doi: 10.1016/0014-5793(87)80292-2. [DOI] [PubMed] [Google Scholar]
- Chin D. J., Gil G., Russell D. W., Liscum L., Luskey K. L., Basu S. K., Okayama H., Berg P., Goldstein J. L., Brown M. S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature. 1984 Apr 12;308(5960):613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Davies S. P., Carling D., Hardie D. G. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur J Biochem. 1989 Dec 8;186(1-2):123–128. doi: 10.1111/j.1432-1033.1989.tb15185.x. [DOI] [PubMed] [Google Scholar]
- Davies S. P., Sim A. T., Hardie D. G. Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. Eur J Biochem. 1990 Jan 12;187(1):183–190. doi: 10.1111/j.1432-1033.1990.tb15293.x. [DOI] [PubMed] [Google Scholar]
- Easom R. A., Zammit V. A. A cold-clamping technique for the rapid sampling of rat liver for studies on enzymes in separate cell fractions. Suitability for the study of enzymes regulated by reversible phosphorylation-dephosphorylation. Biochem J. 1984 Jun 15;220(3):733–738. doi: 10.1042/bj2200733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easom R. A., Zammit V. A. Diurnal changes in the fraction of 3-hydroxy-3-methylglutaryl-CoA reductase in the active form in rat liver microsomal fractions. Biochem J. 1984 Jun 15;220(3):739–745. doi: 10.1042/bj2200739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in rat hepatocytes. J Biol Chem. 1983 Jun 25;258(12):7272–7275. [PubMed] [Google Scholar]
- Edwards P. A., Lemongello D., Fogelman A. M. Purification and properties of rat liver 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochim Biophys Acta. 1979 Jul 27;574(1):123–135. doi: 10.1016/0005-2760(79)90091-2. [DOI] [PubMed] [Google Scholar]
- Endo A., Hasumi K. Biochemical aspect of HMG CoA reductase inhibitors. Adv Enzyme Regul. 1989;28:53–64. doi: 10.1016/0065-2571(89)90063-0. [DOI] [PubMed] [Google Scholar]
- Garton A. J., Campbell D. G., Carling D., Hardie D. G., Colbran R. J., Yeaman S. J. Phosphorylation of bovine hormone-sensitive lipase by the AMP-activated protein kinase. A possible antilipolytic mechanism. Eur J Biochem. 1989 Jan 15;179(1):249–254. doi: 10.1111/j.1432-1033.1989.tb14548.x. [DOI] [PubMed] [Google Scholar]
- Gertler F. B., Chiu C. Y., Richter-Mann L., Chin D. J. Developmental and metabolic regulation of the Drosophila melanogaster 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Cell Biol. 1988 Jul;8(7):2713–2721. doi: 10.1128/mcb.8.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985 May;41(1):249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Guy P. S., Cohen P., Hardie D. G. Purification and physicochemical properties of ATP citrate (pro-3S) lyase from lactating rat mammary gland and studies of its reversible phosphorylation. Eur J Biochem. 1981 Feb;114(2):399–405. doi: 10.1111/j.1432-1033.1981.tb05160.x. [DOI] [PubMed] [Google Scholar]
- Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989 Jun 30;57(7):1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- Holmes C. F. A new method for the selective isolation of phosphoserine-containing peptides. FEBS Lett. 1987 May 4;215(1):21–24. doi: 10.1016/0014-5793(87)80106-0. [DOI] [PubMed] [Google Scholar]
- Holmes C. F., Campbell D. G., Caudwell F. B., Aitken A., Cohen P. The protein phosphatases involved in cellular regulation. Primary structure of inhibitor-2 from rabbit skeletal muscle. Eur J Biochem. 1986 Feb 17;155(1):173–182. doi: 10.1111/j.1432-1033.1986.tb09473.x. [DOI] [PubMed] [Google Scholar]
- Hunter C. F., Rodwell V. W. Regulation of vertebrate liver HMG-CoA reductase via reversible modulation of its catalytic activity. J Lipid Res. 1980 May;21(4):399–405. [PubMed] [Google Scholar]
- Ingebritsen T. S., Geelen M. J., Parker R. A., Evenson K. J., Gibson D. M. Modulation of hydroxymethylglutaryl-CoA reductase activity, reductase kinase activity, and cholesterol synthesis in rat hepatocytes in response to insulin and glucagon. J Biol Chem. 1979 Oct 25;254(20):9986–9989. [PubMed] [Google Scholar]
- Kleinsek D. A., Dugan R. E., Baker T. A., Porter J. W. 3-hydroxy-3-methylglutaryl-CoA reductase from rat liver. Methods Enzymol. 1981;71(Pt 100):462–479. doi: 10.1016/0076-6879(81)71057-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Fink G. R. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Arabidopsis thaliana is structurally distinct from the yeast and animal enzymes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2779–2783. doi: 10.1073/pnas.86.8.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., Cummings R. D., Anderson R. G., DeMartino G. N., Goldstein J. L., Brown M. S. 3-Hydroxy-3-methylglutaryl-CoA reductase: a transmembrane glycoprotein of the endoplasmic reticulum with N-linked "high-mannose" oligosaccharides. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7165–7169. doi: 10.1073/pnas.80.23.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy D. R., Willumsen B. M. Protein modification: new clue to Ras lipid glue. Nature. 1989 Oct 5;341(6241):384–385. doi: 10.1038/341384a0. [DOI] [PubMed] [Google Scholar]
- Luskey K. L., Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J Biol Chem. 1985 Aug 25;260(18):10271–10277. [PubMed] [Google Scholar]
- Meyer H. E., Hoffmann-Posorske E., Korte H., Heilmeyer L. M., Jr Sequence analysis of phosphoserine-containing peptides. Modification for picomolar sensitivity. FEBS Lett. 1986 Aug 11;204(1):61–66. doi: 10.1016/0014-5793(86)81388-6. [DOI] [PubMed] [Google Scholar]
- Munday M. R., Campbell D. G., Carling D., Hardie D. G. Identification by amino acid sequencing of three major regulatory phosphorylation sites on rat acetyl-CoA carboxylase. Eur J Biochem. 1988 Aug 1;175(2):331–338. doi: 10.1111/j.1432-1033.1988.tb14201.x. [DOI] [PubMed] [Google Scholar]
- Nakanishi M., Goldstein J. L., Brown M. S. Multivalent control of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mevalonate-derived product inhibits translation of mRNA and accelerates degradation of enzyme. J Biol Chem. 1988 Jun 25;263(18):8929–8937. [PubMed] [Google Scholar]
- Ness G. C., Sample C. E., Smith M., Pendleton L. C., Eichler D. C. Characteristics of rat liver microsomal 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Biochem J. 1986 Jan 1;233(1):167–172. doi: 10.1042/bj2330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness G. C., Way S. C., Wickham P. S. Proteinase involvement in the solubilization of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochem Biophys Res Commun. 1981 Sep 16;102(1):81–85. doi: 10.1016/0006-291x(81)91491-1. [DOI] [PubMed] [Google Scholar]
- Parker R. A., Miller S. J., Gibson D. M. Phosphorylation of native 97-kDa 3-hydroxy-3-methylglutaryl-coenzyme A reductase from rat liver. Impact on activity and degradation of the enzyme. J Biol Chem. 1989 Mar 25;264(9):4877–4887. [PubMed] [Google Scholar]
- Rajkovic A., Simonsen J. N., Davis R. E., Rottman F. M. Molecular cloning and sequence analysis of 3-hydroxy-3-methylglutaryl-coenzyme A reductase from the human parasite Schistosoma mansoni. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8217–8221. doi: 10.1073/pnas.86.21.8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resink T. J., Hemmings B. A., Tung H. Y., Cohen P. Characterisation of a reconstituted Mg-ATP-dependent protein phosphatase. Eur J Biochem. 1983 Jun 15;133(2):455–461. doi: 10.1111/j.1432-1033.1983.tb07485.x. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Skalnik D. G., Simoni R. D. The nucleotide sequence of Syrian hamster HMG-CoA reductase cDNA. DNA. 1985 Dec;4(6):439–444. doi: 10.1089/dna.1985.4.439. [DOI] [PubMed] [Google Scholar]
- Woodward H. D., Allen J. M., Lennarz W. J. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase of the sea urchin embryo. Deduced structure and regulatory properties. J Biol Chem. 1988 Dec 5;263(34):18411–18418. [PubMed] [Google Scholar]