Abstract
Glyphosate is a potent herbicide. It works by competitive inhibition of the enzyme 5-enol-pyruvyl shiki-mate-3-phosphate synthase (EPSPS), which catalyzes an essential step in the aromatic amino acid biosynthetic pathway. We report the genetic engineering of herbicide resistance by stable integration of the petunia EPSPS gene into the tobacco chloroplast genome using the tobacco or universal vector. Southern blot analysis confirms stable integration of the EPSPS gene into all of the chloroplast genomes (5000–10,000 copies per cell) of transgenic plants. Seeds obtained after the first self-cross of transgenic plants germinated and grew normally in the presence of the selectable marker, whereas the control seedlings were bleached. While control plants were extremely sensitive to glyphosate, transgenic plants survived sprays of high concentrations of glyphosate. Chloroplast transformation provides containment of foreign genes because plastid transgenes are not transmitted by pollen. The escape of foreign genes via pollen is a serious environmental concern in nuclear transgenic plants because of the high rates of gene flow from crops to wild weedy relatives.
Keywords: agricultural biotechnology, transformation, tobacco, glyphosate
Glyphosate is a potent, broad spectrum herbicide that is highly effective against a majority of grasses and broad leaf weeds. Glyphosate works by competitive inhibition of the enzyme 5-enol-pyruvyl shikimate-3-phosphate synthase (EPSPS) of the aromatic amino acid biosynthetic pathway. Synthesis of EPSP from shiki-mate-3-phosphate and inorganic phosphate is catalyzed by EPSPS. This particular reaction occurs only in plants and microorganisms, which explains why glyphosate is nontoxic to other living forms. Use of glyphosate is environmentally safe as it is inactivated rapidly in soil, has minimum soil mobility, and degrades to natural products, with little toxicity to non-plant life forms. However, glyphosate lacks selectivity and does not distinguish crops from weeds, thereby restricting its use. EPSPS-based glyphosate resistance has been genetically engineered via the nuclear genome either by the overproduction of the wild-type EPSPS1 or by the expression of a mutant gene (aroA) encoding glyphosate-resistant EPSPS2.
In all of the aforementioned examples, without exception, herbicide-resistant genes have been introduced into the nuclear genome. One common concern is the escape of a foreign gene through pollen dispersal from transgenic crop plants engineered for herbicide resistance to their weedy relatives, creating “superweeds.” Dispersal of pollen from a central test plot containing transgenic cotton plants to surrounding nontransgenic plants has been observed at varying distances in different directions3,4. The escape of foreign genes through pollen is a serious environmental concern, especially in the case of herbicide resistance genes, because of the high rates of gene flow from crops to wild relatives. For example, the frequencies of marker genes in wild sunflowers averaged about 28–38%; in wild strawberries growing within 50 m of a strawberry field, more than 50% of the wild plants contained marker genes from cultivated strawberries5. Similarly, transgenic oil seed rape, genetically engineered for herbicide resistance outcrossed with a weedy relative, Brassica campestris (field mustard) and conferred herbicide resistance even in the first back-cross generation under field conditions6.
Maternal inheritance of introduced genes prevents gene escape through pollen. Engineering foreign genes through chloroplast genomes (which are maternally inherited for most of the crops) might be a practical solution to this problem. The target enzymes or proteins for most herbicides (of the amino acid/fatty acid biosynthetic pathways or photosynthesis) are compartmentalized within the chloroplast. Another important advantage of chloroplast transformation is the higher levels of foreign gene expression due to a very high copy number (5000–10,000) of chloroplast genomes in plant cells. Because the transcriptional and translational machinery of the chloroplast is prokaryotic in nature, herbicide-resistant genes of bacterial origin can be expressed at extraordinarily high levels in chloroplasts.
Early investigations on chloroplast transformation focused on the development of intact chloroplasts capable of efficient and prolonged transcription and translation7–9 and expression of foreign genes in isolated chloroplasts10. These experiments were done under the premise that it was possible to introduce isolated intact chloroplasts into protoplasts and regenerate transgenic plants11. The discovery of the gene gun as a transformation device opened the possibility of direct plastid transformation12. Transient expression of foreign genes in plastids of dicots13,14 and monocots15, prolonged foreign gene expression using autonomously replicating chloroplast expression vectors13, and stable integration of a selectable marker into the tobacco chloroplast genome16 were accomplished using the gene gun. Tobacco plants resistant to certain insects were obtained by integrating the cryIAc gene into the tobacco chloroplast genome17.
Results and discussion
Chloroplast integration and expression vectors with EPSPS
The chloroplast vector pZS-RD-EPSPS contains the 16S rRNA promoter (Prrn) driving the aadA (aminoglycoside adenyl transferase) and EPSPS genes with the psbA 3’ region (the terminator from a gene coding for photosystem II reaction center components) from the tobacco chloroplast genome. This construct integrates the EPSPS and aadA genes into the spacer region between the rbcL (the gene for the large subunit of RuBisCO) and orf512 genes (code for the accD gene) of the tobacco chloroplast genome. This vector, referred to as the tobacco vector, is useful for integrating foreign genes specifically into the tobacco chloroplast genome; this gene order is not conserved among other plant chloroplast genomes18. On the other hand, the universal chloroplast expression and integration vector pSBL-RD-EPSPS can be used to transform chloroplast genomes of several other plant species because the flanking sequences are highly conserved among higher plants; the universal vector uses trnA and trnI genes (chloroplast transfer RNA genes coding for alanine and isoleucine) from the inverted repeat region of the tobacco chloroplast genome as flanking sequences for homologous recombination.
Gene expression in Escherichia coli
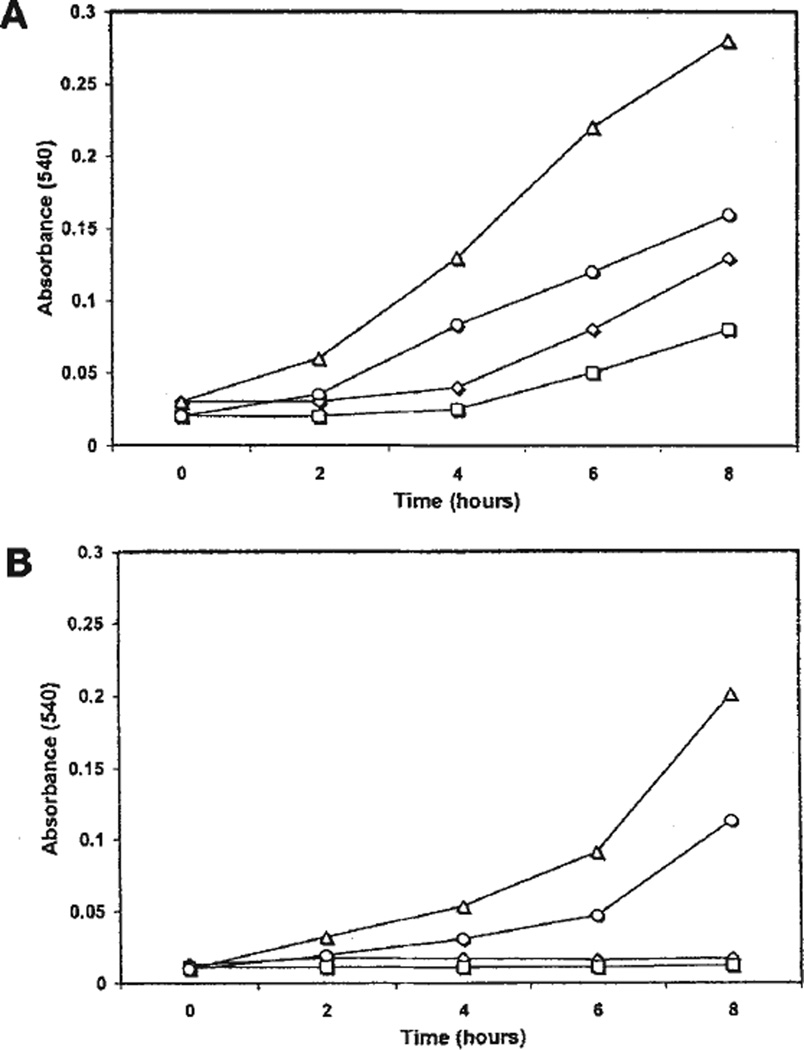
Because of the high similarity in the transcription and translation systems between E. coli and chloroplasts19, we first tested chloroplast expression vectors in E. coli before proceeding with transformation of higher plants. The higher growth rate of E. coli containing the tobacco vector compared with the control pZS197 (which is similar to pZS-RD-EPSPS but does not have the EPSPS gene) in the presence of 10 mM and 40 mM glyphosate (Fig. 1) indicates glyphosate tolerance of E. coli expressing the EPSPS gene. Glyphosate tolerance of E. coli is attributable to the expression of the EPSPS gene, present in both the tobacco and universal vectors.
Figure 1.
Growth curves of E. coli XL1 blue strain containing the plasmids (A) pZS-RD-EPSPS in the presence of 10 mM glyphosate (–Δ–) or 40 mM glyphosate (–○–) and pZS-197 (control) in the presence of 10 mM glyphosate (–◊–) or 40 mM glyphosate (–□–) or (B) pSBL-RD-EPSPS in the presence of 10 mM glyphosate (–Δ–) or 40 mM glyphosate (–○–) and pSBL-ctv2 (control) in the presence of 10 mM glyphosate (–◊–) or 40 mM glyphosate (–□–) in M9 minimal medium.
Characterization of transgenic plants
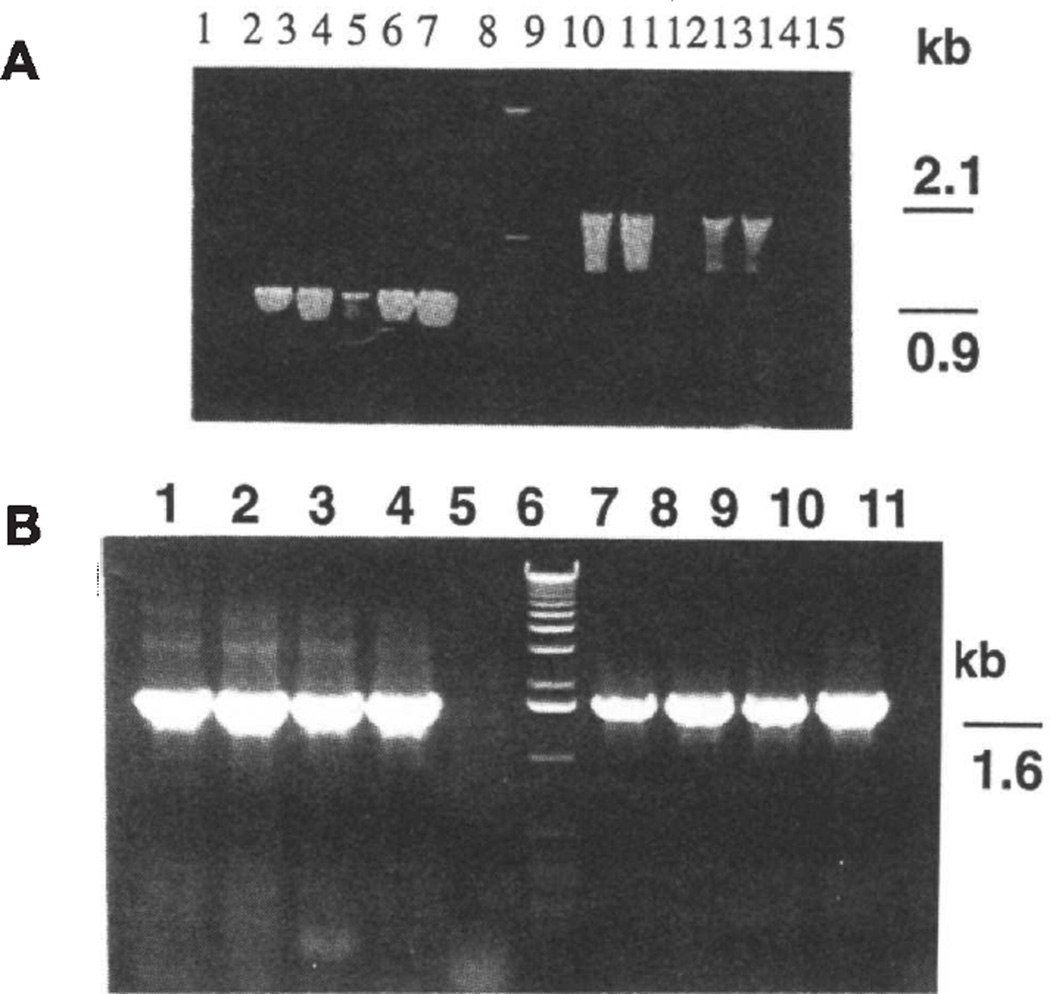
Transgenic plants were obtained within 3–5 months after bombardment. Typically, out of 16 bombarded leaves, 10 independently transformed shoots were identified. PCR analysis was performed with DNA isolated from the first- or second-generation shoots and also from the mature transgenic plants. Primers were used to confirm integration of the aadA gene into the plant genome from the tobacco vector as well as the universal vector. Lack of a product would indicate spontaneous mutants capable of growing on spectinomycin without the aadA gene. The expected PCR product (887 bp) was obtained from six lines (Fig. 2A) transformed with the tobacco vector. A PCR product of 1.57 kb was found in four lines (Fig. 2B) transformed with the universal vector. Under the selection conditions used, four mutants were detected out often lines transformed with the tobacco vector. On the other hand, all the transgenic lines transformed with the universal vector examined so far showed integration of the aadA gene.
Figure 2.
PCR analysis of the pZS-RD-EPSPS and pSBL-RD-EPSPS transgenic lines. (A) Lanes 2–7: primers for rbcL and aadA; lanes 10–15: external primers for rbcL and aadA. Lanes 1 and 9: unbombarded tissue; lane 8:1 kb ladder. (B) Lanes 1–4: internal primers for 16S rRNA and aadA. Lanes 7–10: external primers for 16S rRNA and aadA. Lanes 5 and 11: unbombarded tissue; lane 6:1 kb ladder.
Primers were also designed to determine whether the integration had occurred in the chloroplast genome. The strategy was to land one primer on the native chloroplast genome, adjacent to the point of integration of the vector, while landing the other on the aadA gene, for both chloroplast expression vectors. A primer was designed to land immediately outside the rbcL gene in the tobacco vector (2.08 kb PCR product). For the universal vector the primer on the native chloroplast genome landed in the 16S rRNA gene (1.60 kb PCR product). The expected products were observed for the transgenic lines obtained using the tobacco vector (Fig. 2A) as well as the universal vector (Fig. 2B). Unbombarded plants (controls) did not yield any PCR products, as expected (Fig. 2A and B). Thus, all transgenic plants examined are chloroplast, not nuclear, transformants. This may be due to the requirement of higher levels of accumulation of aminoglycoside adenyl transferase (AADA) in chloroplast transgenic plants under stringent selection conditions. Low levels of AADA present in the cytosol of nuclear transgenic plants should have eliminated nuclear transformants. The results of PCR analysis provide definitive evidence for chloroplast integration of foreign genes using both the tobacco and universal vectors.
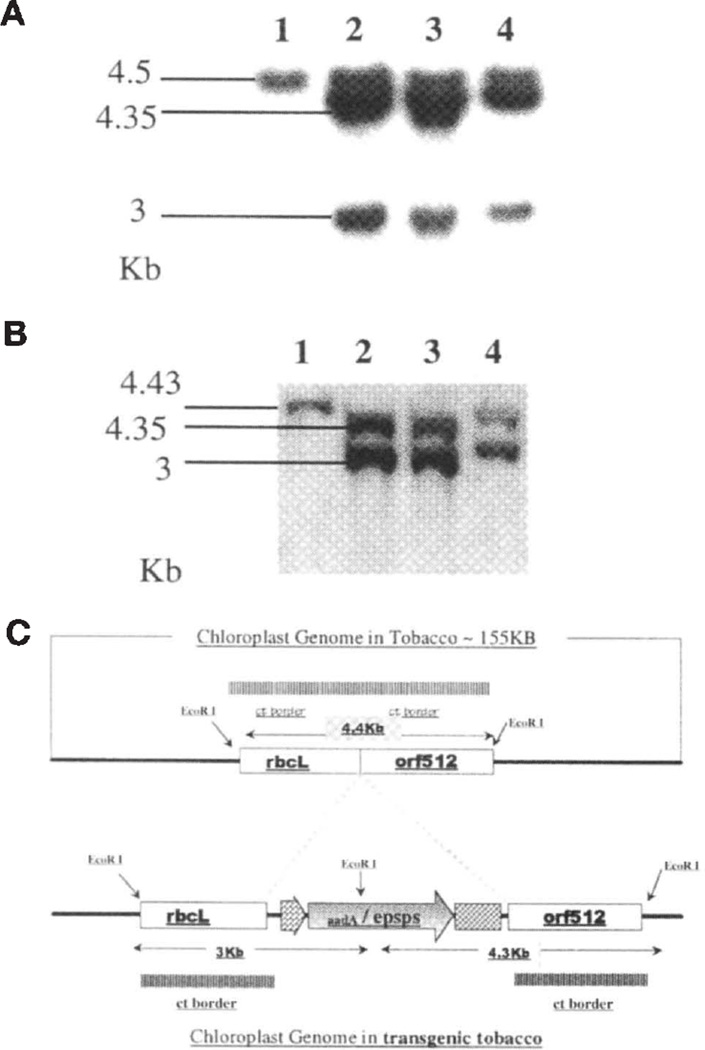
The integration of the EPSPS gene into the chloroplast was also confirmed by Southern blot analysis. To determine integration of the foreign gene into the chloroplast genome, a 654 bp fragment of the EPSPS gene was random prime labeled with 32P. The total DNA comprising both organellar and genomic DNA was digested with EcoRI. The presence of an EcoRI site 200 bp upstream of the integration site in the chloroplast genome was used to confirm integration of the EPSPS gene into the chloroplast genome. The probe hybridized to the native nuclear EPSPS gene (Fig. 3A). In addition, the probe hybridized to the digested chloroplast genomes of the transgenic tobacco plants (Fig. 3A). The probe did not hybridize to the digested chloroplast genome of the untransformed control plant (Fig. 3A). These data show the integration of the EPSPS gene into the chloroplast genome.
Figure 3.
Southern blot analysis of transgenic plants. (A) EcoRI digested total DNA probed with the EPSPS gene. (B) EcoRI digested total DNA probed with the chloroplast border fragment comprising rbcL-orf512. (A and B) Lane 1: untransformed tobacco plant; lanes 2–4: total DNA isolated from three transgenic lines (15A, 15-2, 15-5). (C) Structure of the chloroplast genome with the site of foreign gene integration represented by a shaded dotted line.
The copy number of the integrated gene was determined by establishing homoplasmy for the transgenic chloroplast genome. Tobacco chloroplasts contain 5000 to approximately 10,000 copies of their genome per cell17. If only a fraction of the genomes are actually transformed, the copy number must be less than 10,000. By establishing that the EPSPS transformed genome is the only one present in the transgenics, one could establish that the copy number is 5000 to approximately 10,000 per cell. This was done by digesting the total DNA with EcoRI and probing with the flanking sequences that enable homologous recombination into the chloroplast genome. The probe comprised a 2.9 kb fragment of the rbcL-orf512 sequences. A chloroplast genome transformed with the EPSPS gene incorporates an EcoRI site between the rbcL-orf512 region of the chloroplast genome, thereby generating an extra fragment when digested with this enzyme (Fig. 3C). Southern blot hybridization analysis revealed a 4.43 kb fragment for the untransformed control (Fig. 3B). Two fragments (4.35 and 3 kb) were generated as a result of the incorporation of the EPSPS gene cassette between the rbcL and orf512 regions (Fig. 3C). The 4.43 kb fragment present in the control is absent in the transgenics. Only the transgenic chloroplast genome is present in cells, and there is no native, untransformed, chloroplast genome without the EPSPS gene present. This establishes the homoplasmic nature of chloroplast transformants, simultaneously providing us with an estimate of 5000 to about 10,000 copies of the foreign EPSPS gene per cell.
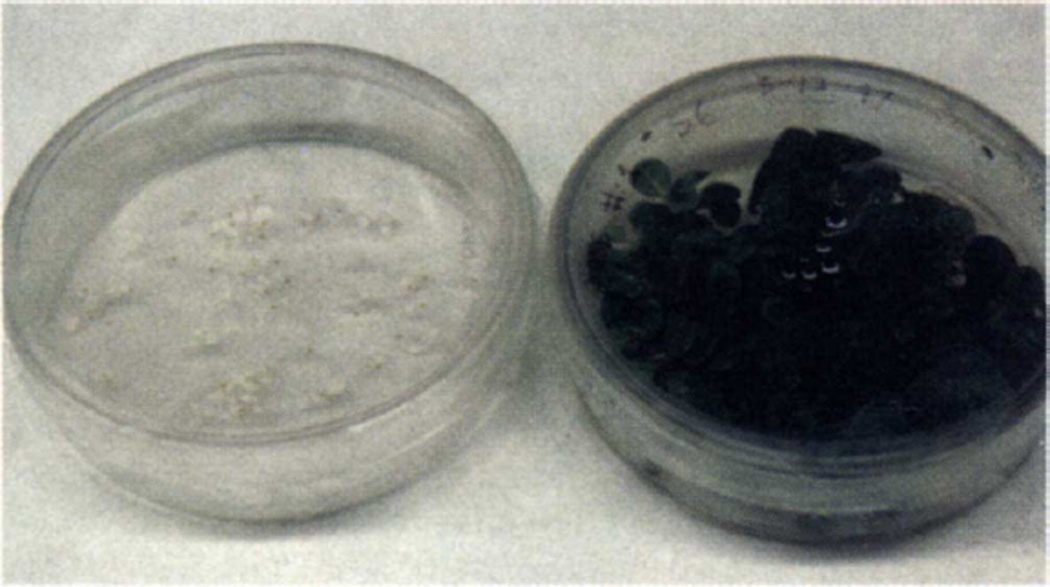
Seeds collected from transgenic plants after the first self-cross were germinated in the presence of spectinomycin. All of the seeds germinated, remained green, and grew normally (Fig. 4B). The 100% resistance to spectinomycin in all of the clones examined shows maternal inheritance of the introduced genes. Lack of pollen transmission of the aadA gene in chloroplast transgenic plants is expected in tobacco, a species with strict maternal inheritance of its plastids16. A heteroplasmic condition would have given rise to variegated progeny on spectinomycin16,20; lack of such variegated progeny also confirms homoplasmy as shown by Southern blot analysis (Fig. 3B). All of the untransformed seedlings were bleached and did not grow in the presence of spectinomycin (Fig. 4A). The lack of variation in chlorophyll pigmentation among the progeny also underscores the absence of position effect, an artifact of nuclear transformation (Fig. 4B).
Figure 4.
Analysis of maternal inheritance of spectinomycin resistance in the seed progeny of chloroplast transgenic plants. The control and transgenic seeds were germinated in MSO medium containing spectinomycin (500 µg/ml). (A) Control seedlings (B) Transgenic seedlings.
Eighteen-week-old control and transgenic plants were sprayed with equal volumes of different concentrations (0.5–5 mM) of glyphosate. Untransformed control tobacco plants were extremely sensitive to glyphosate; they died within 7 days even at 0.5 mM glyphosate (Fig. 5B). On the other hand, the chloroplast transgenic plants survived concentrations as high as 5 mM glyphosate (Fig. 5A). These results are intriguing, considering the fact that the EPSPS gene from petunia used in these chloroplast vectors has a low level of tolerance to glyphosate. Sensitivity to glyphosate by EPSPS should have been compensated by overproduction of the enzyme by thousands of copies of the EPSPS gene present in each cell of the transgenic plants. Pre-EPSPS is catalytically active and has a similar sensitivity to glyphosate as the mature enzyme21.
Figure 5.
Herbicide resistance in the progeny of chloroplast transgenic plants. Eighteen-week-old plants sprayed with 5 mM glyphosate. (A) Transgenic plants. (B) Control plants.
Codon preference is significantly different between the prokaryotic chloroplast compartment and the eukaryotic nuclear compartment. Ideally, a mutant aroA gene from a prokaryotic system (which does not bind glyphosate) should be expressed in the chloroplast compartment; such genes are now available and exhibit a thousand-fold higher level of resistance to glyphosate than the petunia gene used in this investigation. In light of these observations, it is possible that integration of prokaryotic herbicide resistance genes into the chloroplast genome could result in incredibly high levels of resistance to herbicides while still maintaining the efficacy of biological containment.
Experimental protocol
Construction of chloroplast vectors
The tobacco chloroplast expression vector pZS-RD-EPSPS was constructed by inserting the BglII–BamHI fragment of pMON-894, containing the petunia pre-EPSPS gene, into the Spel blunt ended site of pZS-197. The universal chloroplast vector pSBL-RD-EPSPS was constructed by inserting a BglII–BamHI pre-EPSPS fragment from pMON-894 into the unique XbaI site of pSBL-ctv2. Standard protocols for vector construction, including Klenow filling and dephosphorylation, were used. Both plasmids were amplified in the XL1 Blue strain of E. coli. Growth curves were recorded in M-9 minimal medium22.
Plant transformation and analysis
Generation of chloroplast transgenic plants was carried out as described23. Primers were designed for PCR analysis using PC Gene (Oxford Molecular Group, Campbell, CA) and Primer 3 v 0.5 (Whitehead Institute for Biomedical Research, Cambridge, MA). Primer synthesis was done by Genosys, Woodlands, TX). DNA was isolated from leaves using the protocol of Edwards et al.24
Acknowledgments
The authors are grateful to P. Maliga (Rutgers University) for providing pZS-197, and G. Kishore (Monsanto) for pMON-894, and P.J. Brixey for technical assistance. This study was supported in part by the USDA-NRICGP grants 93-37311 and 95-02770 to H.D.
References
- 1.Shah DM, Horch RB, Klee HJ, Kishore GM, Winter JA, E.Tumer N, et al. Engineering herbicides tolerance in transgenic plants. Science. 1986;233:478–481. doi: 10.1126/science.233.4762.478. [DOI] [PubMed] [Google Scholar]
- 2.Cioppa GD, Baner SC, Tayler ML, Roshester DE, Klein BK, Shah DM, et al. Targeting a herbicide resistant enzyme from Escherichia coli to chloroplasts of higher plants. Bio/Technology. 1987;5:579–584. [Google Scholar]
- 3.Llewellyn D, Fitt G. Pollen dispersal from two field trials of transgenic cotton in the Namoi valley, Australia. Molecular Breeding. 1996;2:157–166. [Google Scholar]
- 4.Umbeck PF, Barton KA, Nordheim EV, McCarty JC, Parrot WL, Jenkins JN. Degree of pollen dispersal by insects from a field test of genetically engineered cotton. J. Econ. Entomol. 1991;84:1943–1950. [Google Scholar]
- 5.King J. Could transgenic supercrops one day breed superweeds? Science. 1996;274:180–181. [Google Scholar]
- 6.Mikkelsen TR, Anderson B, Jörgensen RB. The risk of crop transgene spread. Nature. 1996;380:31. [Google Scholar]
- 7.Daniell H, Rebeiz CA. Chloroplast culture IX: chlorphyll(ide) A biosynthesis in vitro at rates higher than in vivo. Biochem. Biophys. Res. Commun. 1982;106:466–471. doi: 10.1016/0006-291x(82)91133-0. [DOI] [PubMed] [Google Scholar]
- 8.Daniell H, Ramanujan P, Krishnan M, Gnanam A, Rebeiz CA. In vitro synthesis of photosynthetic membranes: I. Development of photosystem I activity and cyclic phosphorylation. Biochem. Biophys. Res. Commun. 1983;111:740–749. doi: 10.1016/0006-291x(83)90367-4. [DOI] [PubMed] [Google Scholar]
- 9.Daniell H, Krishnan M, Umabal U, Gnanam A. An efficient and prolonged in vitro translational system from cucumber etioplasts. Biochem. Biophys. Res. Commun. 1986;135:248–255. doi: 10.1016/0006-291x(86)90969-1. [DOI] [PubMed] [Google Scholar]
- 10.Daniell H, McFadden BA. Uptake and expression of bacterial and cyanobacterial genes by isolated cucumber etioplasts. Proc. Natl. Acad. Sci. USA. 1987;84:6349–6353. doi: 10.1073/pnas.84.18.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson PS. The use of protoplasts for genetic research. Proc. Natl. Acad. Sci. USA. 1973;70:598–602. doi: 10.1073/pnas.70.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniell H. Foreign gene expression in chloroplasts of higher plants mediated by tungsten particle bombardment. Methods Enzymol. 1993;217:536–556. doi: 10.1016/0076-6879(93)17088-m. [DOI] [PubMed] [Google Scholar]
- 13.Daniell H, Vivekananda J, Nielsen BL, Ye GN, Tewari KK, Sanford JC. Transient foreign gene expression in chloroplast of cultured tobacco cells following biolistic delivery of chloroplast vectors. Proc. Natl. Acad. Sci. USA. 1990;87:88–92. doi: 10.1073/pnas.87.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye GN, Daniell H, Sanford JC. Optimization of delivery of foreign DNA into higher plant chloroplasts. Plant Mol. Biol. 1990;15:809–819. doi: 10.1007/BF00039421. [DOI] [PubMed] [Google Scholar]
- 15.Daniell H, Krishnan M, McFadden BA. Expression of β-glucuronidase gene in different cellular compartments following biolistic delivery of foreign DNA into wheat leaves and calli. Plant Cell Reports. 1991;9:615–619. doi: 10.1007/BF00231800. [DOI] [PubMed] [Google Scholar]
- 16.Svab Z, Maliga P. High frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride KE, Svab Z, Schaaf DJ, Hogen PS, Stalker DM, Maliga P. Amplification of a chimeric Bacillus gene in chloroplasts leads to extraordinary level of an insecticidal protein in tobacco. Bio/Technology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- 18.Maier RM, Neckermann K, Igloi GL, Kössel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 19.Brixey PJ, Guda C, Daniell H. The chloroplast psbA promoter is more efficient in E. coli than the T7 promoter for hyper expression of a foreign protein. Biotechnology Letters. 1997;19:395–399. [Google Scholar]
- 20.Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Della-Cioppa G, Bauer SC, Klein BK, Shaw DM, Fraley RT, Kishore GM. Translocation of the precursor of 5-enolpyruvylshikimate-3-phosphate synthase into chloroplasts of higher plants in vitro. Proc. Natl. Acad. Sci. USA. 1986;83:6873–6877. doi: 10.1073/pnas.83.18.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers SG, Brand LA, Holder SB, Sharps ES, Brackin MJ. Amplification of the aro A gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl. Environ. Microbiol. 1983;46:37–43. doi: 10.1128/aem.46.1.37-43.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniell H. Transformation and foreign gene expression in plants mediated by microprojectile bombardment. Methods Mol. Biol. 1997;62:463–489. doi: 10.1385/0-89603-480-1:463. [DOI] [PubMed] [Google Scholar]
- 24.Edwards K, Johnstone C, Thompson C. A simple and rapid method for preparation of plant genomic DNA for PCR analysis. Nucl. Acids. Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]