Abstract
Progressive, technological achievements in the quantitative fluorescence microscopy field are allowing researches from many different areas to start unraveling the dynamic intricacies of biological processes inside living cells. From super-resolution microscopy techniques to tracking of individual proteins, fluorescence microscopy is changing our perspective on how the cell works. Fortunately, a growing number of research groups are exploring single-molecule studies in living cells. However, no clear consensus exists on several key aspects of the technique such as image acquisition conditions, or analysis of the obtained data. Here, we describe a detailed approach to perform single-molecule tracking (SMT) of transcription factors in living cells to obtain key binding characteristics, namely their residence time and bound fractions. We discuss different types of fluorophores, labeling density, microscope, cameras, data acquisition, and data analysis. Using the glucocorticoid receptor as a model transcription factor, we compared alternate tags (GFP, mEOS, HaloTag, SNAP-tag, CLIP-tag) for potential multicolor applications. We also examine different methods to extract the dissociation rates and compare them with simulated data. Finally, we discuss several challenges that this exciting technique still faces.
Keywords: Transcription factor, glucocorticoid receptor, single-molecule tracking, dynamics, DNA binding, fluorescence microscopy
1. Introduction
One of the most fundamental tasks that occur inside the nucleus of eukaryotic cells is the process of transcription. The first step in transcription initiation involves the binding of transcription factors (TFs) to specific recognition sequences located at enhancers and/or promoters, which ultimately leads to the assembly of the pre-initiation complex [1]. Built upon biochemical and population-averaging studies, the classical view is that the subunits of the transcription machinery arrive in sequence to form a stable, functional end product. However, live-cell imaging studies has proven that the transcriptional regulatory complex is far more dynamic than originally anticipated, with subunits that quickly assemble and likely not always in a pre-defined order [1–4].
Groundbreaking developments in live-cell microscopy, fluorescence correlation spectroscopy (FCS), and fluorescent labeling have begun to open unique opportunities to study the dynamics of biological systems with high spatial and temporal resolution [5]. In particular, single-molecule tracking (SMT) approaches allow one to follow individual protein molecules in single live cells (Figure 1A). These technological advances now provide the means for the visualization and the measurement of the in vivo behavior of TF-binding events at chromatin targets such as enhancers and core promoters [6]. Several studies have now measured TF-binding events in both prokaryotes [7, 8] and eukaryotes [7, 9–21]. These experiments have shown that eukaryotic TFs spend most of their time freely diffusing, while only a small portion is specifically bound to chromatin at any given time [9–11, 13, 20]. In striking contrast, prokaryotic TFs appear to spend most of their time associated with DNA [7]. In all cases, binding events appear to be very fast, on the order of seconds at the most.
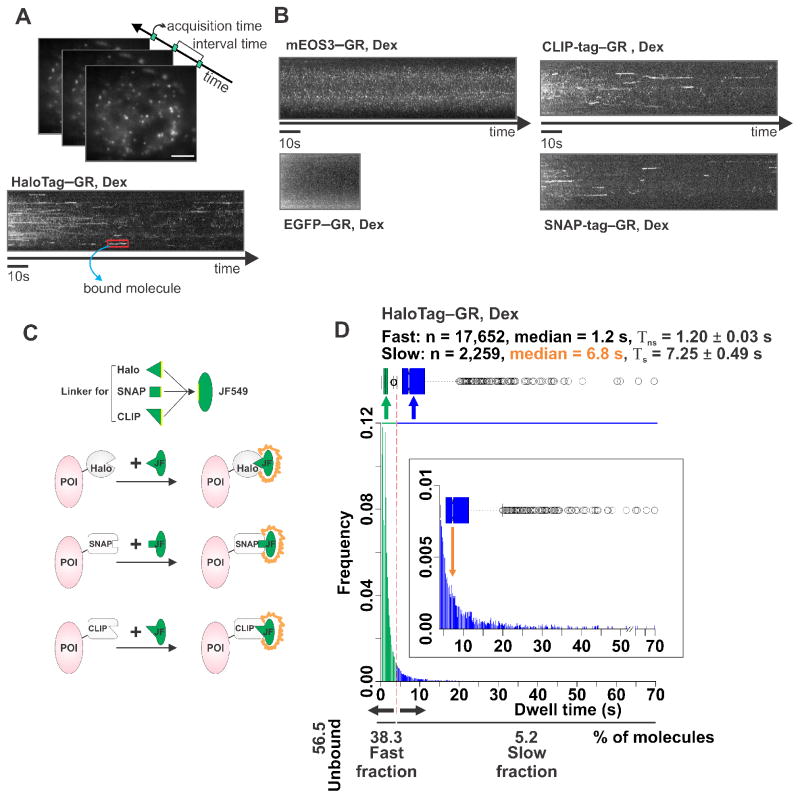
Figure 1. Single-molecule tracking (SMT) of GR molecules tagged with different labels.
(A) The SMT technique visualizes individual molecules as bright diffraction-limited spots and tracks their movement or lack thereof over time. Hence, one can directly identify bound molecules as those that stop moving and remain stationary. HaloTag–GR labeled with Janelia Fluor 549 (JF549) HaloTag ligand can be visualized as such diffraction-limited spots under HILO microscopy. Scale bar 5 μm. A stack of images is taken from a single live cell with 10 ms acquisition time and 200 ms interval time. These parameters are used in order to capture most of the diffusing molecules as well as the stationary ones, while still being able to monitor longer binding events without excessive photobleaching. If molecules remain stationary, the time-projection stack will reveal a continuous signal that represents a bound GR molecule in a dexamethasone (Dex) exposed cell (red box). (B) Time-projection stacks from cells transfected with mEOS3-GR (upper left), EGFP-GR (lower left), CLIP-tag–GR labeled with JF549 (upper right), or SNAP-tag–GR labeled with JF549cp (lower right) in Dex treated conditions. (C) Schematic representation of HaloTag, SNAP-tag, and CLIP-tag post-translational fluorescent labeling system. Organic fluorescent dye (green oval), such as JF549, is made specific for different tags by changing the linker region (equilateral triangle, square, and isosceles right triangle). Each tag has different binding pocket specific for certain linker region. Protein-of-interest (POI) will become fluorescent when JF549 with right linker binds to the corresponding tag. (D) Distribution of residence times from individual GR (+Dex) stationary tracks, either in a histogram or in a Box-plot. A continuum of bi-exponentially distributed bound molecules is typically observed, based on the fitting of the survival distribution. The fast short-lived (Tns, non-specific) and slow long-lived (Ts, specific) fractions are color-coded (green and blue, respectively). In-set shows only the Ts population where the orange arrow points at the median value. The number (n) of tracks obtained, and the median dwell time in fast short-lived and slow long-lived fraction is shown above the histogram.
There has been intense interest in understanding how TFs find their way to their targets and several mechanisms have been proposed (reviewed in [22] and [6]). In this work, we will focus on how to performed SMT experiments to extract information regarding the binding characteristics of TFs, i.e. the residence time and bound fractions. We will describe key aspects of the technique: fluorophores, label density, microscope set-up, acquisition conditions and tracking analysis. Finally, we will discuss the advantages and disadvantages of this new methodology and perspectives for the future.
2. Single-molecule tracking for extracting binding characteristics
Single-molecule microscopy requires a bright and photostable fluorophore, low labeling density, a sensitive camera (capable of collecting photons originating from a single fluorophore), and a microscope set-up that will maximize the signal-to-noise ratio (SNR). Thanks to recent advances in all these areas, studying TF dynamics inside the nucleus of living cells at the single molecule level has become fairly feasible.
2.1. Fluorophores and labeling strategies
A key aspect of any fluorescent experiment is the selection of a suitable fluorophore. In particular, high molecular brightness (number of photons emitted per molecule per unit time), low photobleaching (inability to fluoresce after repeated cycles of absorption/emission), and high quantum yield (number of emitted photons per absorbed photons) are among the most valued commodities one needs to perform in vivo SMT experiments. When choosing a fluorescent label, the fluorophore’s blinking rate should also be considered, as blinking can easily be interpreted as one molecule dissociating from a site, followed by the binding of a new molecule to the same site. Live cell imaging was revolutionized by the discovery of genetically encoded fluorophores like GFP and its relatives (e.g. PA-GFP, Dendra2, mEOS3, etc) [23]. While there are some SMT studies performed on TFs using GFP-like proteins [7, 10, 24], their relatively poor brightness and photostability compared to organic dyes [25] makes them far from ideal to track potentially long chromatin binding events (Figure 1B). In this sense, organic dyes derived from rhodamine, coumarin, xanthene and cyanine such as Alexa Fluor, ATTO, and Cy present themselves as much better candidates for optimal single molecule detection [26]. Nevertheless, introduction of proteins conjugated with these dyes is very difficult and would require microinjection of individual proteins [16].
A suitable alternative that combines the genetic specificity of fluorescent proteins with the favorable photophysics of chemical fluorophores is the “self-labeling tag” strategy such as the HaloTag system. The HaloTag protein is a bacterial dehalogenase enzyme variant that reacts rapidly and specifically with simple chloroalkane “ligands”. The protein has been engineered so that the catalytic cycle stops at the ester intermediate, allowing covalent labeling of a protein of interest [27]. Expression of a chimera between the HaloTag and the protein of interest, followed by labeling with a cell-permeable fluorophore–ligand molecule, allows rapid and specific formation of a fluorescent bioconjugate with an organic dye inside a living cell (Figure 1C). Extensive washes (see section 3.1.3) of the unbound dye must be implemented to guarantee specificity of the fluorescent signal and appropriate controls (e.g. addition of the dye in the absence of the Halo protein) should be tested for each new cell line/tissue. Other tags, based on the same principle, include the DNA-alkyl-transferase-based SNAP-tag or CLIP-tag [28], which are complementary to the HaloTag and therefore allow multicolor imaging [5].
Commercial panels of fluorophores were developed primarily for antibody labeling or single molecule localization of fixed samples, and typically their molecular structure (e.g. hydrophobicity, surface charges, polarity) limit their cell permeability. To overcome this limitation, most ligands for self-labeling tags are based on classic rhodamine structures, which are more permeable to cell membranes. For example, tetramethylrhodamine (TMR; circa 1887) has been successfully used as an organic probe in SMT inside living cells [9, 11, 13]. Although TMR is better than fluorescent proteins in terms of brightness, the dye exhibits a relatively low fluorescence quantum yield (Φ = 0.41) and poor photostability, leaving room for improvement. Inspired by computational experiments, the N,N-dimethyl groups in TMR were replaced with four-membered azetidine rings, which increased both the quantum yield (Φ = 0.88) and the photostability [25]. This new compound, named Janelia Fluor 549 (JF549) has proven more suitable for single-molecule tracking in live cells [12, 20, 21, 29]. This azetidine strategy is general and can be used to improve red-shifted rhodamine analogs, such as the silicon-containing JF646, allowing the possibility of multi-color imaging [5].
A longstanding question in SMT is whether the fluorophore label and/or tagging system has any effect on the obtained data and therefore can introduce bias into the biological results. To investigate this, we compared the JF549 dye behavior on HaloTag, SNAP-tag and CLIP-tag systems using the glucocorticoid receptor (GR) as a model TF. This ligand-induced TF has been previously characterized by SMT using either a mEOS2 tag [10], a TMR–HaloTag [11], or JF549–HaloTag [20]. We tracked individual GR molecules using HILO illumination (see section 2.2.2) by sub-optimal transient transfection of HaloTag, SNAP-tag, or CLIP-tag fused GR. Subsequently, the cells were treated with the synthetic ligand dexamethasone (Dex) prior to imaging. Dex binds to the ligand-binding pocket of the GR which induces the nuclear translocation of the receptor, and results in chromatin binding and subsequent regulation of transcription [30]. Using a custom made tracking software (see section 2.3), individual localizations of single molecules are classified as belonging to a bound track segment (i.e. moving less than a defined threshold rmax for more than Nmin consecutive frames) or unbound (see section 2.3). From the bound population, we then compute the duration of each binding event. Consistent with previous data [11, 20], this bound fraction presents a continuum of residence times bi-exponentially distributed (Figure 1D and 2G), ranging from 0.4 to 69.8s for JF549-HaloTag-GR. In marked contrast, GFP–GR or a mEOS3–GR produces only a one-component exponential distribution (Figure 2D–F and Movie S1), illustrating the significant effects of poor fluorophore photostability in residence time determination in SMT.
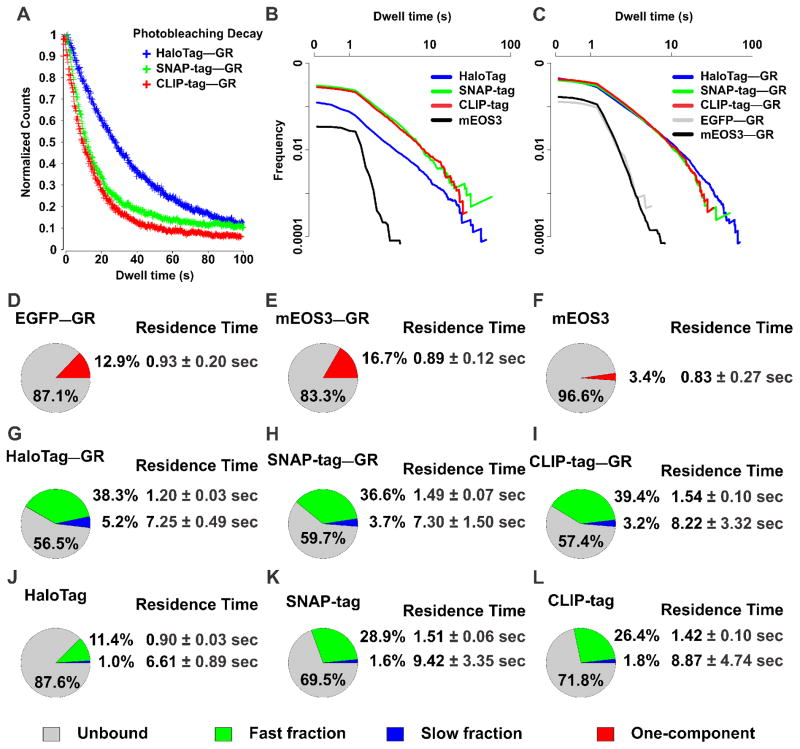
Figure 2. HaloTag is more resistant to photobleaching and enables tracking of longer binding events compated to SNAP-tag and CLIP-tag.
(A) Photobleaching decay of HaloTag–GR (blue), SNAP-tag–GR (green), and CLIP-tag–GR (red) labeled with JF549 was plotted by comparing the detected particles as a function of time. Total count of molecules was normalized to 1. (B) Comparison of dwell time distribution between HaloTag (+JF549) (blue line), SNAP-tag (+JF549) (green line), CLIP-tag (+JF549) (red line), and mEOS3 (black line). Both axes are plotted as a log10. (C) Comparison of dwell time distribution between HaloTag–GR (+JF549) (blue line), SNAP-tag–GR (+JF549) (green line), CLIP-tag–GR (+JF549) (red line), EGFP-GR (grey line), and mEOS3-GR (black line). Both axes are plotted as a log10. (D–F) Pie-charts represent percentage of molecules unbound (grey), and bound at a single-exponential component fraction (one component, red) of EGFP-GR (D), mEOS3-GR (E), or mEOS3 (F). The average residence time of one-component fraction is presented next to chart. (G–L) Pie-charts represent percentage of molecules unbound (grey), bound at the fast short-lived fraction (green), and bound at the slow long-lived fraction (blue) of tag with GR (G–I); HaloTag–GR (G), SNAP-tag–GR (H), CLIP-tag–GR (I) labeled with JF549, or tag alone (J–L); HaloTag (J), SNAP-tag (K), and CLIP-tag (L) labeled with JF549. The average residence time of fast short-lived and slow long-lived fraction is presented next to their representative fractions. For each condition, the data has been corrected for photobleaching. The average number of tracks captured per cell: EGFP-GR, 47; mEOS3, 30; mEOS3-GR, 183; HaloTag, 48; HaloTag–GR, 160; SNAP-tag, 50; SNAP-tag–GR, 112; CLIP-tag, 47; CLIP-tag–GR, 81. Exposure time 10 ms; interval time 200 ms.
Interestingly, we observed clear differences in the photostability of the JF549 dye whether it is bound to HaloTag–GR, SNAP-tag–GR, or CLIP-tag–GR under the same acquisition conditions (Figure 2A–C and Movie S2). In general, judging by the photobleaching curves (Figure 2A), the HaloTag conjugate appears more photostable than the SNAP-tag or CLIP-tag conjugates when labeled with JF549, at least by a factor of 2. This difference could stem from the different fluorophore environments generated by the protein tags. Although crystal structures of dye-bound adducts of the HaloTag and SNAP-tag have not been reported, models of the labeled HaloTag protein suggest a tighter dye-protein interface when compared to labeled SNAP-tag [31]. Nevertheless, after photobleaching correction (see section 3.3), residence times for the slow component are very similar between the alternate tags, although the uncertainty is bigger for SNAP-tag and CLIP-tag (Figure 2G–I, note the bigger error bars). This is likely due to the smaller number of molecules that survive for extended periods of time. Differences in behavior between the tags can be observed even in the absence of a TF. All Halo/SNAP/CLIP tags present a small proportion of slow-component binding inside the nucleus (Figure 2J–L), but the HaloTag presents the smallest amount of unspecific binding. Moreover, the SNAP/CLIP tags show a relatively high amount of fast-stops events compared to HaloTag (Figure 2J–L). Collectively, these results indicate that the HaloTag-JF549 behaves better for SMT experiments than the SNAP-tag- or CLIP-tag-JF549, showing the highest photostability and lowest unspecific binding.
2.2. Data Acquisition
2.2.1 Labeling density
The image of a single point object with an optical microscope appears in the detector as a diffraction limited spot of ~250 nm in the x–y plane. This diffraction pattern, known as the point spread function (PSF), is created by the action of interference in the image plane [5]. Therefore, no matter how small an object really is, its image will never appear smaller than the PSF. The resolution, defined as the minimal distance where two objects can be resolved as separate entities, is limited by the Rayleigh equation d=0.61λ/NA, where d is the minimal distance, λ is the emission wavelength and NA is the numerical aperture of the objective. Interestingly, single molecule localization “breaks” the diffraction barrier (i.e. increases resolution) by fitting the intensity profile of the PSF with a 2D-Gaussian function down to a few nanometers. However, this can only be achieved with the use of sparse labeling conditions to guarantee that individual intensity spots are always well separated in space [32], as illustrated in Movie S3.
There are basically two strategies to achieve low density conditions. Either one can introduce a small number of molecules, all fluorescent; or alternatively, to have large number of molecules but only label a small fraction of them. Microinjection of the ex-vivo labeled ATTO647N-STAT1 transcription factor [16] is a clear example of the “small number of molecules” strategy. For the latter approach, the HaloTag system allows control over labeling density by varying the amount and incubation time of the small-molecule label [11, 20]. Even transient transfection is appropriate as it yields a wide-range of expression levels, ensuring there will be many cells in the sample with an appropriate molecular density (Movie S4). A more physiologically relevant methodology is to stably integrate the HaloTag–protein of interest fusion, which is now easily achieved through CRISPR/Cas9 technology [33]. However, endogenous expression levels may exceed the appropriate amounts for single molecule microscopy (Movie S5). Therefore, in the case of the HaloTag labeling system, decreasing the amount of ligand will achieve the expected outcome, as only a small fraction of the molecules will be labeled with the fluorescent dye (Movie S6). Alternatively, optimal label density can also be obtained by utilizing photoactivatable dyes, such as PA-JF549, and activating a small fraction of the population with 405 nm light [34].
2.2.2 The microscope set-up
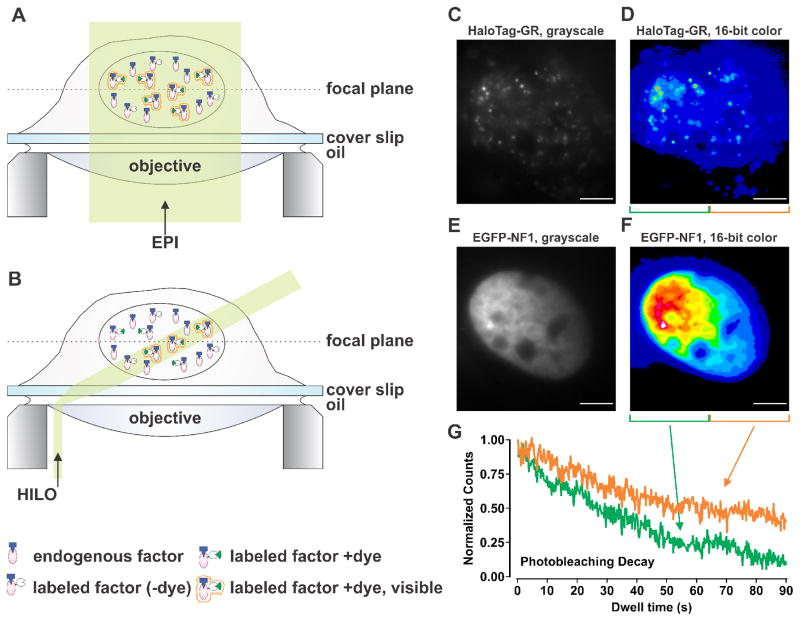
There are several optical set-ups that allow single molecule detection capabilities such as TIRF (total internal reflection fluorescence), HILO (highly inclined and laminated optical sheet illumination), and light-sheet microscopy, among others (reviewed in [5]). In all cases, the goal is to decrease the background signal by eliminating the excitation of out-of-focus molecules. The most common method used to study TF dynamics is HILO [11, 14, 15, 19–21, 34–37], most likely due to its easy implementation. In principle, any microscope capable of a TIRF configuration can do HILO illumination. In HILO (Figure 3A–B), the incident laser beam is highly inclined by moving it to the periphery of the objective lens, and a thin optical sheet at the specimen side is generated by passing the laser through an aperture in a location conjugate to the specimen plane [35]. The background is reduced since only a thin layer of the cell interior is illuminated. This approach succeeds in a limited depth range and in the center of the object field [32], which is acceptable to study intranuclear events. However, due to the use of inclined illumination in HILO, the sample is subjected to non-homogenous illumination (Figure 3C–F). Hence, the photobleaching rate at certain regions of the cell can be higher than others (Figure 3G). This, in turn, may have an effect on the estimations of bound populations.
Figure 3. Schematic representation of epifluorescence (EPI) and highly inclined and laminated optical sheet (HILO) illumination.
(A) In EPI illumination the whole cell is illuminated, resulting in the presence of out-of-focus background fluorescence which can be similar in strength to the signal obtained from a single fluorophore. (B) In HILO illumination only a thin (i.e. a few μm) sheet of the cell is illuminated. This reduces the out-of-focus background fluorescence, which enables the visualization of fluorescent single molecules. Dashed line depicts the focal plane, and transparent yellow section indicates the illuminated region from the cell. (C–F) Representative image of non-homogenous HILO illumination of single HaloTag–GR (+JF549) molecules (C–D) or EGFP-NF1 as marker of the nucleus (E–F). Image shown in grayscale (C, E), and in 16-bit color (D, F) with intensity increasing from blue-to-green-to-yellow-to-red-to-white. Scale bar 5 μm. (G) The single-cell in (D, F) was divided in to two sections of equal size with green representing high illumination and orange representing low illumination. Photobleaching decay of high illumination (green line) and low illumination (orange line) sections were plotted by comparing the detected particles as a function of time. Total count of molecules was normalized to 1.
2.2.3 The camera
The choice of imaging detector is probably the most important decision in tracking single molecules. The two obvious options are the electron-multiplying charge-coupled device (EMCCD), and the scientific complementary metal oxide semiconductor (sCMOS) [38]. Although significant improvements have been made to sCMOS cameras in recent years, and single molecule studies have begun to take advantage of the faster readout and larger chip sizes of sCMOS detectors [39], in our hands EMCCD still provides superior performance for SMT experiments.
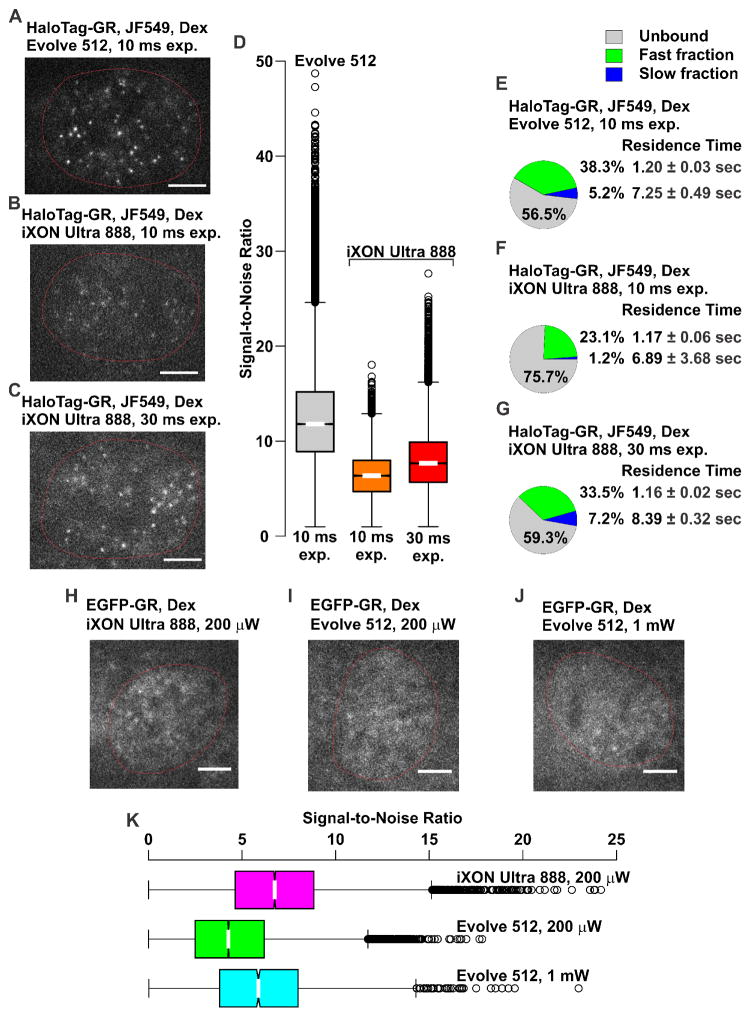
We have also observed slight, but important differences in the performance of EMCCDs from different manufacturers. For example, we have tested both an Evolve 512 (Photometrics), and an iXON Ultra 888 (Andor) on the same system with the same sample. Interestingly, imaging JF549 could be achieved on the Evolve 512 (EM Gain set to 300) with an exposure of 10 ms (Figure 4A), while with the same gain and exposure, barely any signal was visible on the iXON Ultra 888 (Figure 4B), and only after increasing the exposure to 30 ms were we able to achieve results comparable to the Evolve 512 (Figure 4C). Comparison of the SNR for the different cameras and exposures are displayed as Box-plots in Figure 4D. The Evolve 512 with 10 ms exposures gives approximately a two-fold improved SNR compared to the iXON Ultra 888 with 10 ms exposures, median from 12.3 to 6.3. Increasing the exposure to 30 ms on the iXON Ultra 888 improves the SNR to median of 7.9. Furthermore, quantification of a single-molecule dataset of HaloTag–GR (+JF549) tracks showed similar results. The residence time and percentage of bound molecules at the slow long-lived fraction was similar between the data collected using 10 ms exposure on the Evolve 512 camera (Figure 4E) and using 30 ms exposure on the iXON Ultra 888 camera (Figure 4G). Using 10 ms exposure on the iXON Ultra 888 camera showed very low slow long-lived fraction (Figure 4F) and high percentage of unbound molecules. In addition, similar number of tracks were captured from a single cell imaged with the Evolve 512 using 10 ms exposure or iXON Ultra 888 using 30 ms exposure. Conversely, when imaging GFP or other green fluorophores with a 30 ms exposure (EM gain set to 300), the iXON Ultra 888 produced high quality images with a laser power of 200 μW, while with the same exposure and gain conditions the Evolve 512, required 1 mW of power (Figure 4H–I). The iXON Ultra 888 with a laser power of 200 μW has a higher SNR compared to the Evolve 512 with a laser power of 200 μW, median of 6.7 versus 4.3 (Figure 4K). Increasing the laser power to 1 mW on the Evolve 512 improves the SNR to a median of 5.9. These results indicate that the Evolve 512 is capable of imaging with shorter exposures than the iXON Ultra 888, but is likely less sensitive at shorter wavelengths. Therefore, it is crucial to test several cameras with the desired sample prior to committing to a purchase.
Figure 4. Optimization of exposure conditions for different EMCCD cameras.
Example of HaloTag–GR (+JF549) images illustrating the difference obtained with the same cell-line on (A) a Photometrics Evolve 512 with 10 ms exposure where single molecules are clearly visible, and an Andor iXON Ultra 888 with 10 ms exposure (B) where molecules are barely detectable, and (C) 30 ms exposure where single molecules are visible. EM gain was set to 300, and the laser power density was 140 W/cm2 for all images. Nuclear boundaries are shown as red, non-continuous lines. Scale bar 5 μm. (D) SNR comparison of JF549 labeled single-molecule data collected with the Evolve 512 (10 ms exposure), or the iXON Ultra 888 (10 ms and 30 ms exposure) camera. Distribution of SNRs is shown as a Box-plot. (E–G) Pie-charts represent the percentage of molecules unbound (grey), bound at the fast short-lived fraction (green), and bound at the slow long-lived fraction (blue) of HaloTag–GR (+JF549) imaged with the Evolve 512 camera using a 10 ms exposure (E), with the iXON Ultra 888 camera using 10 ms (F), or 30 ms (G) exposure. Interval time of 200 ms was used in each condition. The average residence time of fast short-lived and slow long-lived fraction is presented next to their representative fractions. For each condition, the data has been corrected for photobleaching. Data in each pie-chart represent at least 15 cells and 1900 tracks. The average number of tracks captured per cell are: Evolve 512 [10 ms exp.], 160; iXON Ultra 888 [10 ms exp.], 100; iXON Ultra 888 [30 ms exp.], 164. Exp., exposure. Images of single EGFP-GR molecules were imaged using (H) the iXON Ultra 888 with a laser power of 200 μW, where single molecules can be seen, and the Evolve 512 with a laser power of (I) 200 μW, with molecules barely visible above background, and (J) 1 mW, with the molecules visible. Exposure of 30 ms was used in each condition. Nuclear boundaries are shown as red, non-continuous lines. Scale bar 5 μm. (K) The comparison of the SNR for the three conditions is shown in (H–J), displayed as Box-plots.
2.2.4 Acquisition parameters
There are two important temporal variables to be considered when acquiring single-molecule data: the exposure time at each frame, and the interval time between frames. Ideally, to obtain the maximum amount of dynamic information, one should acquire images as fast as technically possible. However, very short exposure times will lead to a poor SNR, and short or no interval times will lead to the bleaching of individual fluorophores and consequently to an underestimation of the residence time. Hence, a careful balance between temporal resolution and dynamic range must be obtained. Furthermore, it is also necessary to use the lowest possible laser intensity that provides adequate SNR to reduce the amount of photobleaching.
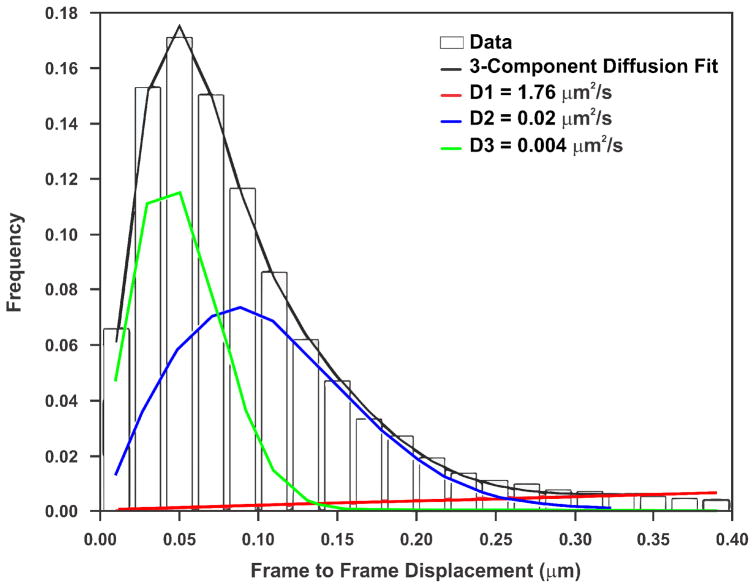
The exposure time is ultimately limited by the brightness of the fluorophore and the sensitivity/speed of the camera. With the optical setup described in section 3.2, 10 ms is the minimum usable exposure time. Since a Gaussian-fitting algorithm is used for identifying and tracking single-molecules (see section 3.3), unless the spot resembles a 2D Gaussian, the algorithm will miss it. It is therefore very important that the fastest component of the studied TF will still resemble a 2D Gaussian during the exposure time. For 2D diffusion, one can calculate the average distance d that a molecule with a diffusion coefficient D will travel according to 〈d2〉 = 4Dt, where t is time. As an example, the JF549-HaloTag-GR’s fastest diffusion coefficient is 1.76 um2/s (Figure 5). Applying the above equation, the average distance that a diffusing GR will travel in 10 ms is ~265 nm, which is very close to the PSF width. As this is the average, some blurring will occur, which will lead to an underestimation in the bound fraction. Recently developed particle identification schemes that take into account this motion hold the promise of alleviating this problem [14].
Figure 5. GR is best fit with a 3-component diffusion model.
Frame to frame displacements for JF549-HaloTag-GR imaged at 5 fps (bars), can be modeled with three populations (F-test comparing to two-component model has a p-value < 0.0001) that are slow diffusion (D = 0.004 μm2/s, green), moderate diffusion (D = 0.02 μm2/s, blue), and fast diffusion (D = 1.76 μm2/s, red). During the exposure time used (10 ms), even the fast population will move, on average, roughly the same distance as the diffraction-limited spot.
The interval time will define which population of molecules is going to be sampled. If short intervals are used, diffusing and short-bound molecules are easily tracked but bleaching becomes the predominant factor in SMT. Hence, the most stable binding events are no longer recorded and residence time is underestimated. On the contrary, if longer imaging intervals are used, faster transiently bound molecules cannot be accurately tracked; hence, there might be an overestimation of the residence times. Therefore, the dwell time for a given factor should be evaluated over a range of conditions to develop a rigorous understanding of its dynamic behavior. This sampling problem has been started to be recognized as a limitation of the methodology to provide an absolute residence time, particularly where the protein is expected to stably bind chromatin [29]. However, other investigators have measured the same TF with different interval times and integrated all data to extract a single residence time [10, 18].
2.3. Data Analysis
2.3.1. Software for extracting particle dynamics from images
The generation of binding data from time-lapse imaging is a three-step process that involves i) locating particles in individual movie frames, ii) linking the particle locations between frames to produce tracks, and iii) analyzing individual tracks to identify time periods where the particle remained stationary. Clearly, the reliability of the binding estimates depends strongly on the performance of the two tracking steps (particle identification, and linking).
There currently exists a wide-range of freely available software options that can be used to extract trajectories of single molecules from images; examples include the ImageJ plugin SOS [40], uTrack [41], and TrackRecord [19, 42]. For a more comprehensive review of available tracking algorithms, including comparisons in various contexts see [43, 44]. We prefer to use TrackRecord due to its integration of binding analysis, its use of parameters that are more readily interpretable by biologists, and its low computational demands (see section 3.3 for a detailed description of the software). The tracking algorithms analyzed in [43] use similar approaches as TrackRecord for locating particles; filtering used to reduce noise, prominent particles identified, and center of the particles are estimated using e.g. Gaussian fitting. However, those algorithms also rely on more sophisticated methods such as wavelet filtering and adaptive thresholding. Further, for tracking purposes the algorithms use such methods as motion models, and multiple target tracking.
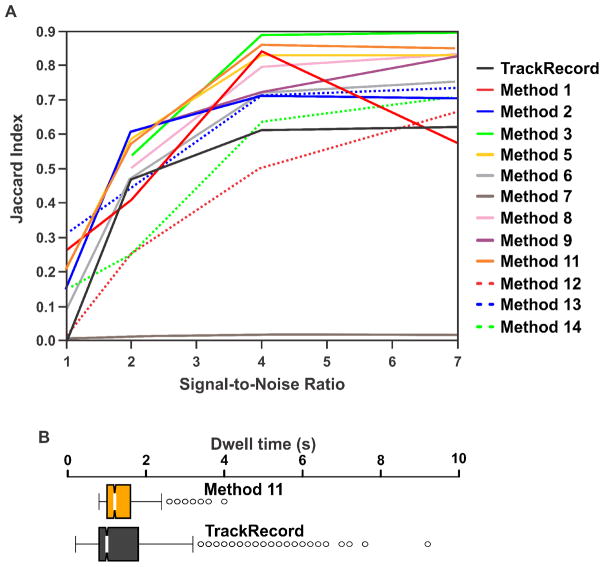
It is important to verify that TrackRecord produces adequate tracking results compared to other software. To this end, we used the simulated data and performance metrics described in [43] to compare TrackRecord to the reviewed algorithms based on the Jaccard similarity coefficient, JSC = TP/(TP + FN + FP), where TP is the number of correct tracks, FN is the number of tracks missed, and FP is the number of spurious tracks. Figure 6A shows the JSC value for several SNRs obtained from TrackRecord (black line) in comparison to the reviewed methods (colored lines) when analyzing data simulating a medium density of receptors. Although some of the more powerful algorithms outperform TrackRecord at high SNR, at low and moderate SNRs, TrackRecord compares favorably with the other methods. Furthermore, the binding rates extracted from TrackRecord tracking results were similar to the more robust tracking methods (Figure 6B). We attribute this to the fact that the improved performance of the powerful methods is due to their ability to track diffusing molecules that TrackRecord cannot follow, but these highly mobile molecules do not contribute significantly to the analysis of binding events. However, it should be noted that for studies of diffusion characteristics, the use of tracking methods based on more sophisticated algorithms, such as those described in [39], or Bayesian treatments [45], would likely produce more reliable results than can be obtained from TrackRecord.
Figure 6. Comparison of TrackRecord with various other tracking software.
(A) Performance based on the Jaccard Index of TrackRecord compared to the various methods described in, and using the simulated, medium density receptor data from Chenouard, et al [43] at several signal-to-noise values. (B) Extracted residence times from the same data as shown in (A) with a signal-to-noise value of 4 for Method 11 from Chenouard, et al [43], and TrackRecord. Although Method 11 performs better at tracking than TrackRecord based on the Jaccard Index in this scenario, TrackRecord is able to capture a wider range of residence time distributions than Method 11.
2.3.2. Obtaining dissociation rates
TFs bind to DNA randomly while searching for their target sequences. In the current paradigm, this results in at least two populations; one consisting of short-lived non-specific events, and a second population of longer lived events when bound to a target sequence. As both of these processes can be modeled with exponential decays, the entire distribution of residence times can be modeled with a multi-exponential decay [9]. Although there are well-known issues when attempting to extract the number of existing exponential components and their parameters, these can be averted by assuming a fixed number of components [46]. We therefore fit with least-squares to mono-, and bi-exponential decay models, and use an F-test to determine if the bi-exponential provides a significantly improved fit. It should be noted that even if the experimental decay can be adequately fit with one or two components, it is possible that there exist more components to the underlying distribution that cannot be obtained due to experimental (i.e. low frame rate, short experiment) or biological (i.e. small population) factors. In fact, non-specific binding may be better modeled with power law statistics [18], however this greatly complicates the analysis of residence times.
If residence times are to be determined from fitting to exponential decays, the question arises whether the residence times should be described by the probability density function (PDF), or the cumulative distribution function (CDF). The PDF is the conventional histogram where the frequency of bound events is plotted for their respective bound-times and requires some arbitrary binning. On the other hand, the CDF, also known as the survival distribution, corresponds to the probability of having molecules bound longer than the plotted time. It requires no binning and therefore allows a more direct comparison of different experiments [47].
The PDF and CDF are both exponential distributions, and therefore, in theory, either should be adequate for extracting the mean residence times. However, those that perform fitting on the PDF generally report binding events that contain a single component [10, 18], whereas those fitting the CDF always obtain two distinct populations that consist of specific and nonspecific binding [9, 11, 19–21]. We therefore used simulated decay curves of bi-exponentials to objectively compare results obtained from fitting the two types of distributions.
Two-component exponentials were simulated by taking random numbers from two independent exponential distributions, such that the probability of finding a residence time, P(t), was given by
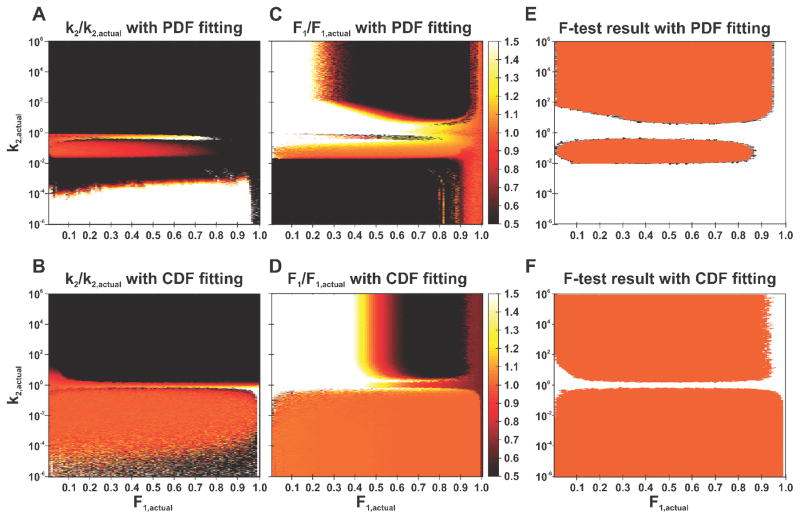
Where k1 and k2 are the rate constants and F1 is the fraction of events originating from the exponential with rate constant k1. For all simulations, k1 was fixed at 1 s−1, while k2 was varied between 10−6–106s−1, and F1 was varied between 0–1. For each parameter set, 104 residence times were generated to obtain a smooth exponential decay with a time resolution of 100 ms, and a maximum time of 120 s, to replicate the conditions of experimental data. Both the PDF and CDF were generated and both were fit with non-linear least-squares to both a single- and double-component exponential. An F-test was used to determine if the double exponential provided a statistically significant improvement in fitting compared to the single exponential. Figure 7A–D shows the results of this analysis in the form of heat maps of the ratio of the fitted value to the actual value in the k2 – F1 parameter space. For clarity all fitted values representing ≤ 50% of the actual value are shown as black, while those representing ≥ 150% of the actual value are shown as white. The results of the F-test are also shown in Figure 7E–F, with white indicating that the single exponential fit is sufficient, and orange indicating that the double exponential fit is required to describe the data. A successful two-component fit would therefore consist of an orange colored pixel in the heat maps and an orange pixel in the F-test map.
Figure 7. Fitting survival times can explore a larger portion of parameter space than fitting residence times.
(A–D) Heat maps showing the ratio of extracted to expected values of (A, B) k2 and (C, D) F1 from simulations with a fixed value of k1 = 1 s−1, while varying k2 and F1 for fitting of the (A, C) residence times, and (B, D) survival times. The color scale saturates at ±50% difference to highlight the regions of parameter space where the fit values are approximately equal to the input values. Orange color thus indicates that the extracted and expected values are the same. White and black colors indicate that the difference between extracted and expected values are over 50%. (E–F) Results of an F-test with the null hypothesis that the double exponential does not provide a better fit than the single exponential for (E) residence time, and (F) survival time distributions. Regions with a p-value >0.05 (and therefore the null hypothesis cannot be rejected) are colored white, and p-values ≤0.05 are colored orange. Fitting of the survival times allows one to extract populations and decay rates at least one order of magnitude lower than can be obtained from fitting the residence times.
There are several interesting observations that can be made from these analyses. First, it is impossible to obtain a bi-exponential decay if the two rate constants are within a factor of 2 relative to each other (Figure 7F); this is even worse when fitting the PDF (Figure 7E). Second, fast decays (k2 > 2 s−1, corresponding to a mean residence time of 0.5 s), cannot be captured with the time-resolution of 100 ms. Finally, by fitting the CDF, we are able to obtain accurate two-component fits for both very slow rate constants (k2 ≥ 10−4 s−1) and very small fractions ((1-F1) ≥ 1%). In contrast, two components could be extracted when fitting the PDF only if k2 > 10−2 s−1, and (1-F1) > 20%.
3. Materials and methods
3.1. Sample preparation for single-molecule imaging
3.1.1. Plasmid constructs
The pHaloTag–GR has been previously described [11]. Briefly, the construct expresses the rat GR with HaloTag protein (Promega, Madison, WI, USA) fused in the C-terminal domain under the CMVd1 promoter. The SNAP-tag–GR expresses the rat GR with SNAP-tag protein fused in the C-terminal domain under the CMV promoter. This was generated by PCR amplification from HaloTag–GR and sub cloned into the pSNAPf (N9183S, New England Biolabs, Ipswich, MA, USA) backbone with NheI and AgeI sites. The CLIP-tag–GR expresses the rat GR with CLIP-tag fused in the C-terminal domain under the CMV promoter. This was generated by PCR amplification from HaloTag–GR and sub cloned into the pCLIPf (N9215S, New England Biolabs) backbone with NheI and AgeI sites. The mEOS3–GR was generated by purification of the rat GR sequence from a AgeI/XhoI digested fragment of the pEGFP–GR vector [48], and subsequent sub-cloning into a pre-digested AgeI/XhoI pmEOS3 plasmid, kindly provided by the Lippincott-Schwartz lab. Finally, pEGFP-NF1[49] was used as an homogenous nuclear marker in Figure 3.
3.1.2. Cell culture and transfection
The generation of 3617 mouse mammary adenocarcinoma cell line utilized in this study has been previously described [50]. The cell line was routinely cultured in high glucose DMEM supplemented with 10 % fetal bovine serum (FBS) (Life Technologies, Grand Island, NY, USA) and 2 mM L-glutamine (Life Technologies) at 37 °C in a CO2-controlled humidified incubator. The cell line contains stably integration of rat GFP–GR under tetracycline regulation. To prevent expression of GFP–GR the cells were grown in the presence of 5 μg/ml tetracycline (Sigma-Aldrich, St. Louis, MO, USA). The “leaking” of the Tet-system provided enough molecules to performed SMT of the GFP–GR, and to perform the camera comparisons in Figure 4. For the rest of the single-molecule imaging experiments the following protocol was utilized:
100 000 cells were seeded onto each well of 2-well Lab-Tek chamber (1.5 German borosilicate coverglass, Thermo Fisher, Waltham, MA, USA) in high glucose DMEM supplemented with 10 % charcoal-stripped FBS (Life Technologies), 2 mM L-glutamine and 5 μg/ml tetracycline.
Cell were incubated 16 h at 37 °C before transient transfections.
500 ng of plasmid DNA was diluted to 400 μl of jetPRIME® buffer (PolyPlus, New York, NY, USA). To mix DNA with buffer, the samples were gently vortex and centrifuged.
1:3 (w/v) ratio of DNA/jetPRIME® reagent (PolyPlus) was added to each tube. The samples were gently vortex, centrifuged, and incubated at RT for 10 min.
The DNA/ jetPRIME mix were added dropwise to each well on the chamber slide and gently mixed.
After 4 h incubation at 37 °C, the medium was replaced without any additional washing steps.
The transfected cells were incubated for 16 h at 37 °C before fluorescent labeling.
The goal of this transfection protocol is to obtain a wide range of expression levels in the cell population. Individual cells with proper labeling density are easily identified under the microscope (Movie S3). Modifications of the above protocol have been used for other cell lines. For MCF-7 cells [20], 250000 cells were seeded onto wells in high glucose DMEM without phenol red supplemented with 10 % charcoal-stripped FBS, 2 mM L-glutamine, 1 mM Na-Pyruvate (Life Technologies), and 1× Non-essential amino acids (Life Technologies). The rest of the transfection was performed as indicated. For HepG2 cells [51], 300000 cells were seeded onto wells in high glucose DMEM supplemented with 10 % charcoal-stripped FBS, 2 mM L-glutamine, 1 mM Na-Pyruvate. Transfection was done 6 h after seeding of the cells, using 500 ng of plasmid DNA. Instead of using jetPRIME ®, HepG2 cells were transfected using Lipofectamine® 2000 (Thermo Fisher) and Opti-MEM (Life Technologies) according to manufacturer’s instructions. Medium was replaced after 16 h incubation at 37 °C, just before starting fluorescent labeling.
3.1.3. Fluorescent labeling of tagged molecules and hormone treatments
For the fluorescent labeling of HaloTag, SNAP-tag and CLIP-tag containing proteins, the cognate cell-permeable JF ligands were utilized. The JF549-HaloTag ligand has been described previously; the JF549-cpSNAP-tag and JF549-CLIP-tag were prepared from 6-carboxy JF549 in an analogous fashion [25]. Cells were labeled using the following protocol:
-
Transfected cells were treated with JF ligands by adding them to the media.
5 nM JF549-HaloTag ligand for HaloTag-constructs
10 nM JF549-cpSNAP-tag ligand for SNAP-tag-constructs (Label density was not high enough at 5 nM JF549-cpSNAP-tag ligand, data not shown)
10 nM JF549-CLIP-tag ligand for CLIP-tag-constructs (Label density was not high enough at 5 nM JF549-CLIP-tag ligand, data not shown)
JF549 ligand treated cells were incubated 20 min at 37 °C.
-
To remove unbound JF ligands, the cells were washed 3 times with phenol red free DMEM media supplemented with 10% charcoal-stripped FBS and 5 μg/ml tetracycline.
Washed cells were incubated 15 min at 37 °C.
The washing step and 15 min incubation at 37 °C was repeated two more times.
Medium was replaced to phenol red free DMEM media supplemented with 10 % charcoal-stripped FBS and 5 μg/ml tetracycline before hormone treatments.
The cells were treated with 100 nM of Dex (Sigma-Aldrich).
Hormone treated cells were incubated 20 min at 37 °C before imaging.
3.2 Image acquisition for single-molecule tracking
An Olympus IX81 inverted microscope (Olympus Scientific Solutions, Waltham, MA) with a 150x 1.45 NA oil immersion objective lens was adapted to perform HILO illumination for single-channel, single-molecule tracking by adding the following elements:
-
Four lasers were added to image a wide-range of fluorophores:
405 nm (iFLEX-2000, Excelitas Technologies Corp., Waltham, MA, not used in this study).
473 nm (FB-473-500, RGBLase, LLC, Fremont, CA)
561 nm (iFLEX-Mustang, Excelitas Technologies Corp., Waltham, MA)
647 nm (OBIS 647 LX, Coherent, Inc., Santa Clara, CA, not used in this study).
-
The lasers are combined with a series of 3 dichroic mirrors:
573 nm longpass beamsplitter (Di02-R561-25x36, Semrock, Rochester, NY)
480 nm longpass beamsplitter (LM01-480-25, Semrock, Rochester, NY)
414 nm longpass beamsplitter (Di02-R405-25x36, Semrock, Rochester, NY)
The combined lasers then pass through an acousto-optic tunable filter (AOTFnC-400.650, AA Optoelectronic, Orsay, France) is used to select the laser line and control laser power. Computer control is facilitated with the use of an AOTF controller (ESio AOTF Controller, ES Imaging, Kent, UK).
Downstream of the AOTF, the lasers pass through a 3X beam expander, consisting of a 50 mm focal length lens (AC254-050-A, Thorlabs, Inc., Newton, NJ), followed by a 150 mm focal length lens (AC254-050-A, Thorlabs, Inc., Newton, NJ).
The lasers are then directed to the illumination port of the IX81 by way of a mirror mounted on a translation stage. Moving the mirror adjusts the position that the lasers hit the objective lens, and therefore the angle that the beam is incident upon the sample. Each laser is independently aligned to ensure that the illumination angle is consistent for the various colors.
A quad-band dichroic (ZT405/473/561/640rpc-UF2, Chroma Technology Corp., Bellows Falls, VT) is mounted in the filter turret beneath the objective lens.
Imaging is performed with an Evolve 512 EMCCD camera (Photometrics, Tucson, AZ). In addition, the iXON Ultra 888 EMCCD camera (Andor, Belfast, UK) was also utilized in Figure 4 comparisons.
3.3 Data analysis
Tracking was performed with the custom software TrackRecord [42] in MATLAB (The MathWorks, Inc., Natick, MA). The latest version of this software is freely available for download at: www.sourceforge.net/projects/single-molecule-tracking. Briefly, the software performs the following steps, with typical computation times for a 600 frame movie containing ~7000 particles performed on a Windows 64-bit PC with an Intel Core i7-4770 CPU and 16 GB of RAM shown in parentheses:
Each image in the time-series is filtered using top-hat, Wiener, and Gaussian filters, to smooth out uneven illumination, remove speckle noise, and accentuate objects of a particular size, respectively (~16 s).
One or more Region of Interest (ROI) is selected by the user to limit tracking inside of the nucleus.
Particles are found in each frame by first looking for intensity maxima in the filtered image, and then fitting the area around the peaks in the raw image to a two-dimensional Gaussian function (~180 s).
Particle positions are linked between frames using a nearest neighbor algorithm (~1 s).
Automatically generated tracks are manually inspected to correct for gaps within tracks and misidentified assignments.
After tracking has been performed on at least 20 movies, the data from these is merged to extract the mean residence times.
Survival time histograms are built up by finding non-moving portions of each track. These motionless, or bound, track segments are defined by frame-to-frame displacements that are less than 220 nm. To reduce the possibility that diffusing molecules moving less than this amount are included in the analysis, bound molecules must not move more than 270 nm for a specific number (Nmin) of consecutive time-points, which depends on the temporal resolution of the movie. For 200 ms resolution, for example, Nmin = 2 frames. The bars in the survival histogram indicate the fraction of track segments that survive for the indicated time or longer.
The survival time histogram is normalized to the bound fraction, which is defined as the ratio of the bound track points to the total number of detected particles.
The survival time histogram is corrected for photobleaching by dividing it by the decay in the number of particles detected over time [9, 42, 52].
The photobleaching-corrected survival histogram is then fit with a single and double exponential function, and the appropriate fitting function is chosen by use of an F-test with the null hypothesis that the double exponential does not provide a significant improvement in fit over the single exponential.
4. Conclusion and perspectives
TFs are key players to initiate the transcriptional program and their interaction with chromatin has been studied for many years. From cell-average population assays such as chromatin immunoprecipitation to single-cell studies (FRAP, FCS), it becomes clear that most of the TFs interact with DNA in a very dynamic fashion [2]. Now that is technically possible to investigate TFs dynamics at the single-molecule level, new answers and most likely new questions will emerge in the field.
For example, the three population of molecules detected by SMT (Figure 1D, unbound, fast bound and slow bound) has been interpreted as the TF diffusing in the nucleus or binding to non-specific sites in chromatin (fast component), or to specific response elements (slow component) [13, 53]. However, it remains to be answered what proportion of the slow component is responsible for productive transcription, that means, responsible for the successful recruitment and initiation or RNA polymerase II [22]. Answering this question will likely require a trackable, single-copy targeted promoter and multi-color SMT imaging, as well as the possibility of tracking in 3D space.
Several technical challenges still remain in the SMT field. Improvement in fluorophore stability will expand the currently narrow temporal range of SMT measurements as well as decreasing the sample bias due to acquisition conditions. In this sense, great advancements have already been achieved with organic dyes [25, 34], but even the same bright fluorophore (e.g. JF549) can behave very different when bound to different tags (Figure 2), limiting the scope of two-color imaging. Further protein- and dye-engineering will be required to create better pairs for SMT where both the protein microenvironment and the dye can be tuned for highest performance. Finally, another key aspect is the tracking method per se used to extract the binding characteristics. While several algorithms and software packages exist (section 2.3.2), there is still no clear consensus of its uses among different research groups. For example, using the PDF or CDF of the same data will give you different outcomes (Figure 7).
The application of single-molecule approaches to the study TF binding is just getting started. Here, we present in detail one of the many ways to label, acquire and analyze SMT data. Further technological improvements will expand the capabilities of the assay and deepen our understanding of transcriptional regulation.
Supplementary Material
Single-molecule tracking acquires binding properties of transcription factors
A detailed set-up to perform single molecule tracking is presented
Challenges of the methodology are discussed
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), the Center for Cancer Research (CCR) at the National Cancer Institute (NCI), and the Howard Hughes Medical Institute. V.P. was supported, in part, by the Sigrid Jusélius Foundation.
Abbreviations
- CDF
cumulative distribution function
- EMCCD
electron-multiplying charge-coupled device
- FCS
fluorescence correlation spectroscopy
- FRAP
fluorescence recovery after photobleaching
- Dex
dexamethasone
- GR
glucocorticoid receptor
- HILO
highly inclined and laminated optical sheet
- JF
Janelia Fluor
- sCMOS
scientific complementary metal oxide semiconductor
probability density function
- PSF
point spread function
- ROI
region-of-interest
- SMT
single-molecule tracking
- SNR
signal-to-noise ratio
- TF
transcription factor
- TIRF
total internal reflection fluorescence
- TMR
tetramethylrhodamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14(8):572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35(6):741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stasevich TJ, McNally JG. Assembly of the transcription machinery: ordered and stable, random and dynamic, or both? Chromosoma. 2011;120(6):533–545. doi: 10.1007/s00412-011-0340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, English BP, Grimm JB, Kazane SA, Hu W, Tsai A, Inouye C, You C, Piehler J, Schultz PG, Lavis LD, Revyakin A, Tjian R. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 2016;30(18):2106–2118. doi: 10.1101/gad.285395.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z, Lavis LD, Betzig E. Imaging Live-Cell Dynamics and Structure at the Single-Molecule Level. Mol Cell. 2015;58(4):644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 6.Coleman RA, Liu Z, Darzacq X, Tjian R, Singer RH, Lionnet T. Imaging Transcription: Past, Present, and Future. Cold Spring Harbor symposia on quantitative biology. 2016 doi: 10.1101/sqb.2015.80.027201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316(5828):1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336(6088):1595–8. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 9.Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012;40(15):e119. doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10(5):421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morisaki T, Muller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun. 2014;5:4456. doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife. 2014;3:e04236. doi: 10.7554/eLife.04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, Tjian R, Liu Z. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156(6):1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izeddin I, Recamier V, Bosanac L, Cisse II, Boudarene L, Dugast-Darzacq C, Proux F, Benichou O, Voituriez R, Bensaude O, Dahan M, Darzacq X. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife. 2014;3:e02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugo N, Morimatsu M, Arai Y, Kousoku Y, Ohkuni A, Nomura T, Yanagida T, Yamamoto N. Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence. Sci Rep. 2015;5:10662. doi: 10.1038/srep10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speil J, Baumgart E, Siebrasse JP, Veith R, Vinkemeier U, Kubitscheck U. Activated STAT1 transcription factors conduct distinct saltatory movements in the cell nucleus. Biophys J. 2011;101(11):2592–600. doi: 10.1016/j.bpj.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Royen ME, van Cappellen WA, Geverts B, Schmidt T, Houtsmuller AB, Schaaf MJ. Androgen receptor complexes probe DNA for recognition sequences by short random interactions. J Cell Sci. 2014;127(Pt 7):1406–1416. doi: 10.1242/jcs.135228. [DOI] [PubMed] [Google Scholar]
- 18.Normanno D, Boudarene L, Dugast-Darzacq C, Chen J, Richter C, Proux F, Benichou O, Voituriez R, Darzacq X, Dahan M. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat Commun. 2015;6:7357. doi: 10.1038/ncomms8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball DA, Mehta GD, Salomon-Kent R, Mazza D, Morisaki T, Mueller F, McNally JG, Karpova TS. Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swinstead Erin E, Miranda Tina B, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova Tatiana S, Ball D, Mazza D, Lavis Luke D, Grimm Jonathan B, Morisaki T, Grøntved L, Presman Diego M, Hager Gordon L. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell. 2016;165(3):593–605. doi: 10.1016/j.cell.2016.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhen CY, Tatavosian R, Huynh TN, Duc HN, Das R, Kokotovic M, Grimm JB, Lavis LD, Lee J, Mejia FJ, Li Y, Yao T, Ren X. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife 5. 2016 doi: 10.7554/eLife.17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Larson DR. What have single-molecule studies taught us about gene expression? Genes Dev. 2016;30(16):1796–810. doi: 10.1101/gad.281725.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19(11):555–65. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groeneweg FL, van Royen ME, Fenz S, Keizer VI, Geverts B, Prins J, de Kloet ER, Houtsmuller AB, Schmidt TS, Schaaf MJ. Quantitation of glucocorticoid receptor DNA-binding dynamics by single-molecule microscopy and FRAP. PLoS One. 2014;9(3):e90532. doi: 10.1371/journal.pone.0090532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, Lavis LD. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015 doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein T, Proppert S, Sauer M. Eight years of single-molecule localization microscopy. Histochem Cell Biol. 2014;141(6):561–75. doi: 10.1007/s00418-014-1184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3(6):373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 28.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15(2):128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, El Beheiry M, Masson JB, Dahan M, Liu Z, Doudna JA, Tjian R. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350(6262):823–6. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 30.Grbesa I, Hakim O. Genomic effects of glucocorticoids. Protoplasma. 2016 doi: 10.1007/s00709-016-1063-y. [DOI] [PubMed] [Google Scholar]
- 31.Crivat G, Taraska JW. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol. 2012;30(1):8–16. doi: 10.1016/j.tibtech.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manzo C, Garcia-Parajo MF. A review of progress in single particle tracking: from methods to biophysical insights. Reports on progress in physics. Physical Society (Great Britain) 2015;78(12):124601. doi: 10.1088/0034-4885/78/12/124601. [DOI] [PubMed] [Google Scholar]
- 33.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimm JB, English BP, Choi H, Muthusamy AK, Mehl BP, Dong P, Brown TA, Lippincott-Schwartz J, Liu Z, Lionnet T, Lavis LD. Bright photoactivatable fluorophores for single-molecule imaging. Nat Methods. 2016 doi: 10.1038/nmeth.4034. [DOI] [PubMed] [Google Scholar]
- 35.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5(2):159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 36.Basu S, Tan YL, Taylor EJ, Laue ED, Lee SF. Studying the Dynamics of Chromatin-Binding Proteins in Mammalian Cells Using Single-Molecule Localisation Microscopy. Methods Mol Biol. 2016;1431:235–63. doi: 10.1007/978-1-4939-3631-1_17. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt JC, Zaug AJ, Cech TR. Live Cell Imaging Reveals the Dynamics of Telomerase Recruitment to Telomeres. Cell. 2016 doi: 10.1016/j.cell.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quan T, Zeng S, Huang ZL. Localization capability and limitation of electron-multiplying charge-coupled, scientific complementary metal-oxide semiconductor, and charge-coupled devices for superresolution imaging. Journal of biomedical optics. 2010;15(6):066005. doi: 10.1117/1.3505017. [DOI] [PubMed] [Google Scholar]
- 39.Juette MF, Terry DS, Wasserman MR, Altman RB, Zhou Z, Zhao H, Blanchard SC. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods. 2016;13(4):341–4. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuter M, Zelensky A, Smal I, Meijering E, van Cappellen WA, de Gruiter HM, van Belle GJ, van Royen ME, Houtsmuller AB, Essers J, Kanaar R, Wyman C. BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J Cell Biol. 2014;207(5):599–613. doi: 10.1083/jcb.201405014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5(8):695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazza D, Ganguly S, McNally JG. Monitoring dynamic binding of chromatin proteins in vivo by single-molecule tracking. Methods Mol Biol. 2013;1042:117–137. doi: 10.1007/978-1-62703-526-2_9. [DOI] [PubMed] [Google Scholar]
- 43.Chenouard N, Smal I, de CF, Maska M, Sbalzarini IF, Gong Y, Cardinale J, Carthel C, Coraluppi S, Winter M, Cohen AR, Godinez WJ, Rohr K, Kalaidzidis Y, Liang L, Duncan J, Shen H, Xu Y, Magnusson KE, Jalden J, Blau HM, Paul-Gilloteaux P, Roudot P, Kervrann C, Waharte F, Tinevez JY, Shorte SL, Willemse J, Celler K, van Wezel GP, Dan HW, Tsai YS, Ortiz de SC, Olivo-Marin JC, Meijering E. Objective comparison of particle tracking methods. Nat Methods. 2014;11(3):281–289. doi: 10.1038/nmeth.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smal I, Meijering E. Quantitative comparison of multiframe data association techniques for particle tracking in time-lapse fluorescence microscopy. Medical image analysis. 2015;24(1):163–89. doi: 10.1016/j.media.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Persson F, Linden M, Unoson C, Elf J. Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat Methods. 2013;10(3):265–9. doi: 10.1038/nmeth.2367. [DOI] [PubMed] [Google Scholar]
- 46.Istratov AA, Vyvenko OF. Exponential analysis in physical phenomena. Rev Sci Instrum. 1999;70(2):1233–1257. [Google Scholar]
- 47.Mazza D, Stasevich TJ, Karpova TS, McNally JG. Monitoring dynamic binding of chromatin proteins in vivo by fluorescence correlation spectroscopy and temporal image correlation spectroscopy. Methods Mol Biol. 2012;833:177–200. doi: 10.1007/978-1-61779-477-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Presman DM, Ogara MF, Stortz M, Alvarez LD, Pooley JR, Schiltz RL, Grontved L, Johnson TA, Mittelstadt PR, Ashwell JD, Burton G, Ganesan S, Burton G, Levi V, Hager GL, Pecci A. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the Glucocorticoid Receptor. PLoS Biol. 2014;12(3):e1001813. doi: 10.1371/journal.pbio.1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muratcioglu S, Presman DM, Pooley JR, Grontved L, Hager GL, Nussinov R, Keskin O, Gursoy A. Structural Modeling of GR Interactions with the SWI/SNF Chromatin Remodeling Complex and C/EBP. Biophys J. 2015;109(6):1227–39. doi: 10.1016/j.bpj.2015.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287(5456):1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 51.Goldstein I, Baek S, Presman DM, Pakkinaho V, Swinstead EE, Hager GL. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 2016 doi: 10.1101/gr.212175.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki KG, Kasai RS, Fujiwara TK, Kusumi A. Single-molecule imaging of receptor-receptor interactions. Methods Cell Biol. 2013;117:373–90. doi: 10.1016/B978-0-12-408143-7.00020-7. [DOI] [PubMed] [Google Scholar]
- 53.Mueller F, Stasevich TJ, Mazza D, McNally JG. Quantifying transcription factor kinetics: at work or at play? Crit Rev Biochem Mol Biol. 2013;48(5):492–514. doi: 10.3109/10409238.2013.833891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.