i. Summary/Abstract
Transcriptional enhancers are DNA regulatory elements that are bound by transcription factors and act to positively regulate the expression of nearby or distally-located target genes. Enhancers have many features that have been discovered using genomic analyses. Recent studies have shown that active enhancers recruit RNA polymerase II (Pol II) and are transcribed, producing enhancer RNAs (eRNAs). GRO-seq, a method for identifying the location and orientation of all actively transcribing RNA polymerases across the genome, is a powerful approach for monitoring nascent enhancer transcription. Furthermore, the unique pattern of enhancer transcription can be used to identify enhancers in the absence of any information about the underlying transcription factors. Here we describe the computational approaches required to identify and analyze active enhancers using GRO-seq data, including data pre-processing, alignment, and transcript calling. In addition, we describe protocols and computational pipelines for mining GRO-seq to identify active enhancers, as well as known transcription factor binding sites that are transcribed. Furthermore, we discuss approaches for integrating GRO-seq-based enhancer data with other genomic data, including target gene expression and function. Finally, we describe molecular biology assays that can be used to confirm and explore further the function of enhancers that have been identified using genomic assays. Together, these approaches should allow the user to identify, and explore the features and biological functions of new cell type-specific enhancers.
Keywords: GRO-seq, groHMM, Enhancer, Enhancer RNAs (eRNAs), Enhancer prediction, Gene regulation, Looping, Motif, Motif search, Promoter, Response element, Transcription, Transcription factor, Transcription unit
1. Introduction
1.1. Transcriptional Enhancers Function as Genomic Regulatory Elements
Transcriptional enhancers (‘enhancers’) are DNA regulatory elements that are bound by transcription factors (TFs) and act to positively regulate the expression of nearby or distally-located target genes (1, 2). Enhancers are located throughout the genome, including promoters, gene bodies, and intergenic regions, and they function independent of their orientation and location with respect to their target gene (3–5). They also function in a cell type-specific manner; an enhancer that is active in one cell type might not be in another (1, 6). By controlling unique patterns of gene expression in different cell types, enhancers drive the unique biology of those cells types. Thus, identifying the repertoire of enhancers that are active in a given cell type, the set of target genes regulated by those enhancers, and the molecular mechanisms controlling enhancer function provide important clues for understanding biological outcomes.
1.2. Properties and Features of Active Enhancers
TF binding to a specific locus in the genome does not necessarily lead to the formation of an ‘active’ enhancer (i.e., an enhancer that can drive the transcription of a target gene by RNA polymerase II, Pol II). In fact, TF binding events that fail to promote the formation of an active enhancer have been observed for a variety of transcription factors (7–9). Active enhancers exhibit unique properties and features, many of which have been defined using deep sequencing-based genomic assays. These assays include:
-
(1)
chromatin immunoprecipitation-sequencing (ChIP-seq), which determines the enrichment of TFs, chromatin- and transcription-related factors, and posttranslational modifications of histones across the genome (10).
-
(2)
deoxyribonuclease digestion-sequencing (DNase-seq) and assay for transposase-accessible chromatin-sequencing (ATAC-seq), which determine the ‘openess’ or accessibility of chromatin at specific loci across the genome (11–13).
-
(3)
deep sequencing-based chromosome conformation capture (3C)-related assays (e.g., Hi-C), which monitor the formation of chromatin loops across the genome (14–16).
-
(4)
global run-on-sequencing (GRO-seq) and related assays, which detect the location of active RNA polymerases and the production of nascent transcripts across the genome (17, 18). These assays have been used to identify common features shared by active enhancers (Fig. 1).
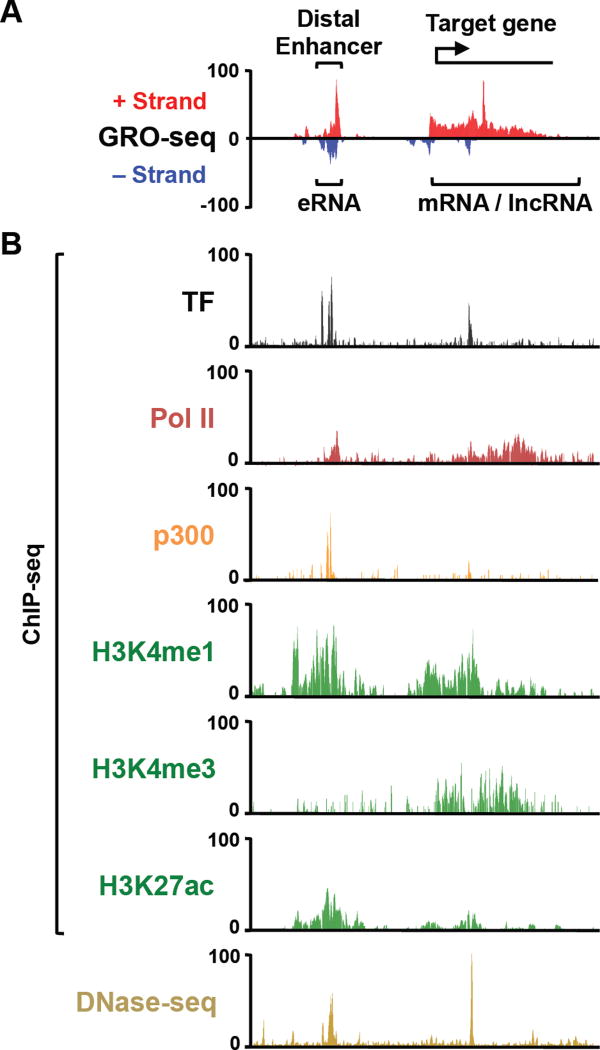
Figure 1. Genomic Features of Active Enhancers and Promoters.
Genome browser tracks showing (A) GRO-seq and (B) ChIP-seq and DNase-seq data at a representative locus of the human genome. Bidirectional transcription at the enhancer is evident, as is TF and p300 binding, recruitment of Pol II, and enrichment of histone modifications.
Properties and features of active enhancers include (1) binding of one or more TFs to DNA sequence motifs specific for those TFs, (2) enhanced chromatin accessibility, (3) enrichment of specific histone modifications, including histone H3 lysine 4 mono/dimethylation (H3K4me1/me2) and H3 lysine 27 acetylation (H3K27ac), (4) binding of transcriptional coactivators, histone-modifying enzymes, and chromatin-modulating enzymes (e.g., the protein acetyltransferases p300 and CBP; the multipolypeptide Mediator complex), (5) recruitment of Pol II and active transcription of nascent enhancer RNAs (eRNAs) (19, 20), (6) and looping to target gene promoters (14, 21) (Fig. 1). While some of the features noted above are also shared with promoters, such as enrichment of coregulators and Pol II, others are more enriched at enhancers than promoters (e.g., H3K4me1/me2) (1, 3, 4, 22). Although these enhancer features have been known for some time, how they contribute to the regulation and function of enhancers remains to be determined.
1.3. Identifying and Characterizing Enhancer Transcripts
Active transcription at enhancers was first observed over a decade ago in locus-specific molecular biology experiments (23–25). These observations were extended by the initial observation using ChIP-seq that Pol II is recruited to enhancers across the genome (22). Subsequent studies using total RNA-seq in neurons and macrophages demonstrated that the Pol II bound at enhancers is indeed engaged in active transcription, producing short, bidirectional, non-coding transcripts called enhancer RNAs (eRNAs) (19, 20). These studies also showed that the production of eRNAs correlates with the recruitment of transcription factors in response to neuron and macrophage activation (19, 20). The genome-wide identification of transcription start sites in intergenic regions using TSS-seq and CAGE technology added further support for enhancer transcription (20, 26). Taken together, these studies provide strong evidence for enhancer transcription as a general biological event.
Additional studies aimed at understanding signal-dependent transcriptional responses have used GRO-seq, a method for identifying the location and orientation of actively transcribing Pol II (and Pol II and Pol III) across the genome, to characterize signal-dependent transcription at enhancers (7, 8, 18, 27–29). GRO-seq has been used to distinguish between TF binding sites (e.g., for estrogen receptor alpha, ERα, and NF-κB) that produce transcripts and those that do not (7, 8). Only the former (i.e., TF binding sites that are transcribed) are enriched for genomic features associated with active enhancers (e.g., H3K4me1, DNaseI accessibility, p300/CBP binding) (7, 8). In more recent studies, derivatives of GRO-seq (i.e., GRO-cap or 5’ GRO-seq), which enrich for 5’-capped nascent transcripts, have been used to study enhancer transcription (27, 28). Collectively, these studies have shown that GRO-seq is an effective means to identify, characterize, and understand the regulation of enhancer transcription. Furthermore, these studies have shown that enhancer transcription is an early event in enhancer activation after TFs binding (which, of course, may require the prior binding of pioneer factors and chromatin remodeling). As such, enhancer transcription, as detected by GRO-seq, is a highly reliable mark of active enhancers, which can be exploited to identify and study these enhancers. In fact, it may be the most robust indicator of enhancer activity, even more so than the histone modifications typically enriched at enhancers (7, 19).
1.4. Using GRO-seq and Related Approaches to Identify and Study Active Enhancers
GRO-seq and related approaches, such as PRO-seq (30), GRO-cap (27), and 5’ GRO-seq (28), are powerful techniques to identify actively transcribed regions of the genome, whether or not those regions have been annotated previously. As we describe below, GRO-seq data can be mined to identify active enhancers in an unbiased way in the absence of any prior information about the initiating TF. In addition, once enhancers are identified, they can be mined using bioinformatic approaches to identify putative underlying TF motifs. In addition, the GRO-seq data can be integrated with other types of genomic data relating to enhancer function (e.g., ChIP-seq for TFs and histone modifications, DNase-seq, looping data; see for example (7, 31).
Recently, software has been developed to analyze GRO-seq (and related) data to search for enhancers and other regulatory elements. For example, groHMM, a software package in the R programing language that is available in Bioconductor (32), uses a two state Hidden Markov Model to define the boundaries of transcription units. Using groHMM, one can identify actively transcribed regions of the genome from GRO-seq data. Furthermore, dREG (discriminative regulatory-element detection from GRO-seq), a computer program that uses read counts to employ support vector regression, can be used to identify active transcriptional regulatory elements from GRO-seq or PRO-seq data (33).
2. Materials: Computer, Data, and Software
Herein, we describe the use of computational tools, approaches, and pipelines to identify and characterize cell type-specific enhancers using GRO-seq and other genomic data. For executing these analyses, you will need a source of GRO-seq data, a suitable computer, and a variety of software.
A high capacity computer suitable for analyzing high content, high complexity data sets.
GRO-seq data from a cell or tissue type of interest.
Additional genomic data for integration and comparison, as desired.
R, a programming language and software environment for statistical computing and graphics (www.r-project.org/).
Perl, a high-level, general-purpose, interpreted, dynamic programming language (https://www.perl.org).
Cutadapt, a python module to remove adapter sequences from high-throughput sequencing data (http://cutadapt.readthedocs.org/en/stable/index.html) (34), used here to trim the polyA tail and adapter sequences from GRO-seq reads.
Burrows-Wheeler aligner (BWA), a software package for mapping low-divergent sequences against a large reference genome (http://bio-bwa.sourceforge.net)(35).
groHMM, an R package from Bioconductor for analyzing GRO-seq data (http://www.bioconductor.org/packages/release/bioc/html/groHMM.html) (32).
Bedtools, a suite of computational tools for a wide-range of genomic analysis tasks (http://bedtools.readthedocs.org/en/latest/) (36).
Python, a general-purpose, high-level programming language (https://www.python.org/)
SAMtools, is a set of utilities that manipulate alignments in the BAM format. (http://samtools.sourceforge.net/)(37)
3. Methods
3.1. Preparation of GRO-seq Libraries
Detailed protocols for the preparation of GRO-seq libraries can be found in the published literature (17, 18, 29, 30); here we outline the key steps. Intact and transcriptionally competent nuclei are isolated from the cells of interest (8, 17, 29, 30, 38). The nuclei are subjected to transcriptional run-on in the presence of bromo-UTP (Br-UTP). The labeled nascent transcripts are isolated from nuclei and enriched by multiple rounds of bead binding using anti-Br-UTP antibody-conjugated agarose beads.
The nascent transcripts are then converted to high-throughput sequencing libraries through a series of molecular biology manipulations for annealing/reverse transcription-based addition of sequencing adapters (8, 17, 29). The steps include :
-
(1)
polyA tailing of nascent RNA using polyA polymerase, which adds a polyA tail to allow annealing of the sequencing adapters (to circumvent an inefficient RNA ligation step in the original protocol).
-
(2)
annealing of an DNA oligonucleotide containing an oligo dT sequence followed by the 3’ and 5’ sequencing adapters separated by an abasic site, which is used for later cleavage of the sequencing adapters.
-
(3)
reverse transcription of the polyadenylated nascent RNA using the annealed oligonucleotide primer.
-
(4)
digestion of the excess oligonucleotide primer using exonuclease I and degradation of the nascent RNA using base hydrolysis leaving single-standed cDNA with the adapter sequences incorporated.
-
(5)
circular ligation of the single-stranded cDNA using CircLigase (a single-stranded DNA ligase).
-
(6)
cleavage at the abasic site between the 3’ and 5’ sequencing adapters using an abasic lysase,.
-
(7)
PCR amplification with primers that add unique sequencing barcodes to each sample to allow for sample multiplexing.
After purification, quantification, and quality control of the final libraries, they are subjected to deep sequencing (we typically use the Illumina HiSeq platform). The resulting raw data are analyzed as described below.
3.2. Processing and Aligning GRO-seq Data
The following are a standard set of computational approaches that can be used to process GRO-seq data. The analytical steps involved include: (1) quality control analysis of the GRO-seq data, (2) pre-processing of the GRO-seq data depending on the information from the quality control analysis to improve the usability of the dataset, and (3) aligning the processed GRO-seq reads to a reference genome (‘mapping’) to associate the signals with specific genomic locations. These steps are performed using a variety of open source software, some of which have user-friendly graphical user interfaces, while others require the use of command lines. Below, we have provided commands that can be cut and pasted into the command line versions of the software noted.
1. Quality control and trimming the adapter and polyA sequences from the GRO-seq reads
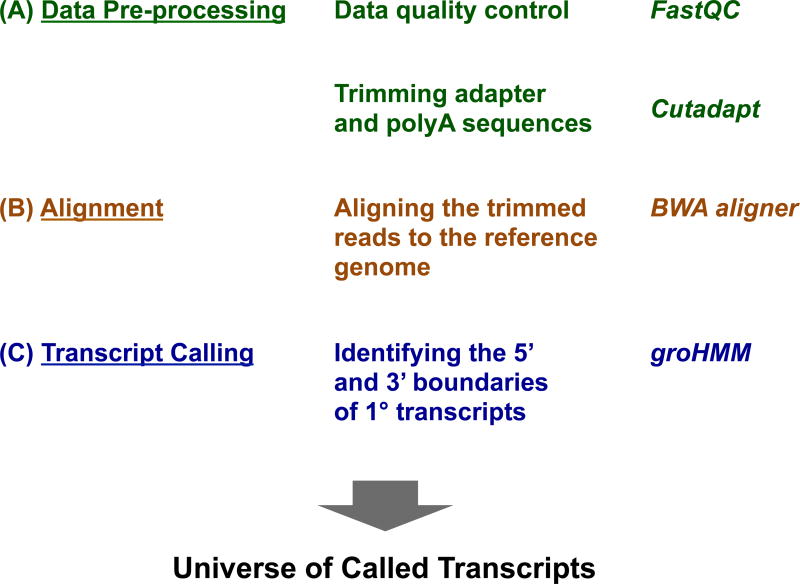
Quality control is an important first step in processing high throughput sequencing data, including GRO-seq. The GRO-seq data should be checked for contamination from the sequencing adapters or the polyA addition (“pre-processing”). Quality control analysis can be performed using tools like FastQC, a quality control tool for raw high throughput sequencing data (39) (Fig. 2). In order to improve the alignment of reads to the reference genome for the species in which you are working, adapter and polyA trimming should be performed (Fig. 2). The adapter and polyA sequences should be trimmed from the GRO-seq reads to increase the fraction of reads that can be aligned to the reference genome. This can be done using various publicly available trimming tools, such as Cutadapt and Trimmomatic (40).
Figure 2. Pre-processing, Alignment, and Transcript Calling for GRO-seq Data.
Overview of GRO-seq data analysis, as well as software that can be used for the key steps in the analysis.
Here we show how adapter and polyA sequences can be trimmed using Cutadapt. Only reads which are > 32 bp in length (--minimum-length) after adapter trimming are retained for further analysis. A default maximum error rate (−e) of 0.1 is used. In order to comply with the input format necessary for futher steps, all negative quality values are changed to zero (−z). The statistics regarding the reads that are trimmed in this step are redirected (2>&1) to an output statistics file.
The following example can be executed in the command line version of Cutadapt to trim adapter and polyA sequence contamination resulting from the GRO-seq protocol. An implementation of the commands in Bash scripts are available through the GitHub repository (see below). Trimming of the adapter sequence (1, below) should be sequentially followed by the execution of trimming polyA tail (2, below).
- # (1) Trimming adapter sequence: GRO-seq data in the fastq format is provided as input for this step.
$ cutadapt -a <adapter sequence> -z -e 0.10 --minimum-length=32 -- output=filename.noAdapt.fastq.gz inputfile.fastq.gz 2>&1 >> RunCutadapt.out
- # (2) Trimming polyA tail: After trimming the adapter sequence, the output file from the above step (reads trimmed for adapter sequence) is now processed in this step to trim the polyA contamination.
$ cutadapt -a AAAAAAAAAAAAAAAAAAAA -z -e 0.10 --minimum-length=32 -- output= filename.noPolyA.noAdapt.fastq.gz filename.noAdapt.fastq.gz 2>&1 >> RunCutadapt.out
2. Aligning the trimmed GRO-seq reads to the reference genome
After trimming the sequencing reads, the data should be aligned to the appropriate reference genome to provide the map of the sites of active transcription across the genome. The alignment can be accomplished using publicly available software, such as BWA (35) and SOAP (41) (Fig. 2).
Here we show the trimmed reads can be aligned using the BWA aligner. We find that it works better for handling the unequal read lengths that are produced after the pre-processing step. A maximum of two mismatches (−n) and a subsequence seed length of 32 bp (−l) are used as parameters for alignment in this step. The ‘samse’ command will produce an output with a maximum of one alignment per read (−n). After alignment the files containing the aligned reads will have to be in a specific format (i.e., bam, -b) to perform subsequent transcript calling and tuning using the groHMM package.
The following examples can be executed in the command line version of the BWA aligner, followed by conversion to the bam format using ‘samtools’. An implementation of the commands in a single Bash script is available from the GitHub repository (see below).
- # Aligning to the reference genome index: The output from Cutadapt after adapter and polyA trimming (‘filename.noPolyA.noAdapt.fastq.gz’) is provided as input to the BWA aligner. The final reads passing these criteria are aligned to the reference genome and are written to the ‘alignedFile.sam’ file.
$ bwa aln -n 2 -l 32 -t 8 Genome_INDEX.fa filename.noPolyA.noAdapt.fastq.gz > alignedFile.sai $ bwa samse Genome_INDEX.fa -n 1 alignedFile.sai inputfile.fastq.gz > alignedFile.sam
- # Converting aligned files from sam to bam format using Samtools.
$ samtools view -bh -S alignedFile.sam > alignedFile.unsorted.bam $ samtools sort alignedFile.unsorted.bam alignedFile.sorted.bam
3.3. Analyzing GRO-seq Data Using groHMM and Other Computational Tools
GroHMM is a software package in R that can be used to define the boundaries of transcription units from a GRO-seq data using a two-state Hidden Markov Model (HMM) (32). It also provides additional tools for visualizing and analyzing GRO-seq data. The groHMM package covers basic steps of GRO-seq data analysis, including the generation of wiggle files using the ‘writeWiggle’ function and the creation of metagene (data average) plots using the ‘runMetaGene’ function, as well as more advanced steps, such as predicting the boundaries of actively transcribed regions (‘transcription units’) across the genome de novo (Fig. 2).
The aligned files from the section 3.2-1 serve as the input to groHMM. Since GRO-seq data is strand-specific, one can visualize the signals from the plus and minus strands separately. The pipelines for calling transcription units (using ‘detectTranscripts’), as well as evaluating (using ‘evaluateHMMInAnnotations’) and tuning the transcript calling, are explained in detail in the tutorial associated with the groHMM package (32). In a systematic comparison of the performance of groHMM versus other transcription unit callers, such as SICER and HOMER (42, 43), groHMM performed better with respect to coverage of genic and intergenic regions, as well as transcription unit accuracy for both short and long transcripts (32).
3.4. Identification of Active Enhancers from GRO-seq Data
Transcription from GRO-seq data can be used as a signature to identify active enhancers (here, by ‘active enhancer’, we mean those that are actively transcribed) (7, 33, 38). This can be accomplished using two approaches: (1) de novo identification of active enhancers using short bidirectional transcript pairs and (2) identification of TF binding sites (from ChIP-seq data) that are actively transcribed. For the de novo identification, bioinformatic approaches can be used to identify motifs for putative transcription factors that drive the formation of those enhancers (7). In the sections below, we describe how active enhancers can be identified using groHMM, open source software, and additional scripts in the R and perl programming languages.
1. De novo identification of enhancers using GRO-seq data
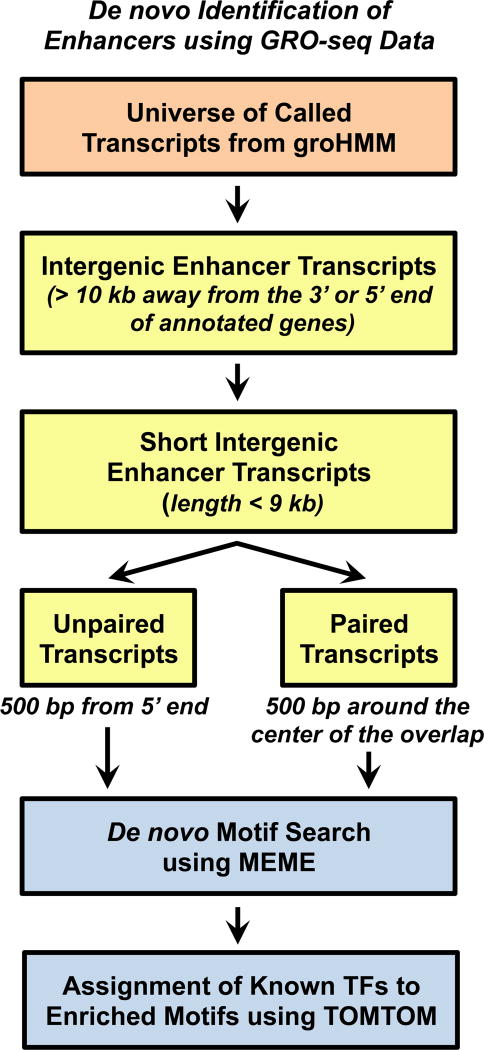
We have shown previously that the production of enhancer transcripts can be used to identify active enhancers de novo in the absence of any other genomic information (7). For these analyses, we have focused on intergenic enhancers to avoid complications in the analysis associated with overlapping gene body transcription. For our purposes, we have searched > 10 kb away from the 5’ or 3’ end of an annotated gene (7), although this can be adjusted to recover a greater number of enhancers or those closer to promoters (8). We have also defined the enhancer transcripts as ‘short’ (i.e., ≤ 9 kb), as well as unidirectional (i.e., transcript produced from one strand of DNA, but not the other) or bidirectional (i.e., transcript produced both strands of DNA) (7) (Figs. 3 and 4).
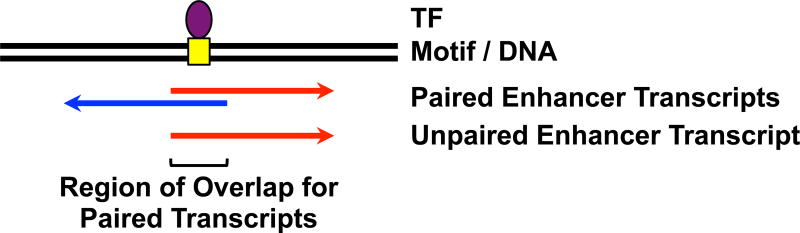
Figure 3. Schematic Representation of an Actively Transcribed Enhancer.
Actively transcribed enhancers that form at TF binding sites may produce paired or unpaired enhancer transcripts.
Figure 4. De novo Identification of Enhancers using GRO-seq Data.
Details are provided in the text.
The first step in this analysis is to identify intergenic transcripts from the universe of all transcripts obtained from groHMM (7, 32). As noted above, we use a cutoff of > 10 kb away from either end of annotated genes in order to distinguish enhancer transcription from genic transcription. Here we show how a set of intergenic transcripts can be identified from a transcript universe using the ‘intersect’ function in BEDtools, a suite of different analysis tools that can be used to modify, convert, or compare bed files (36). The following example illustrates the use of ‘intersect’ to isolate transcripts that do not intersect (−v) with genic regions. An implementation of the command in a single Bash script is available from the GitHub repository (see below).
- # Identify intergenic transcripts: The ‘genic_regions_to_avoid.bed’ file contains the genomic coordinates extending 10 kb from the 5’ and 3’ ends of annotated genes. The input files should be sorted before running the bedtools intersect function using the following unix command.
$ sort-k1,1-k2,2n ip.txt ip_sorted.txt $ bedtools intersect -a transcript_universe_from_groHMM.txt -b genic_regions_to_avoid.bed -v > intergenic_transcripts.txt
After filtering for transcripts that are intergenic, we use a length cutoff to define and identify enhancer transcripts (Fig. 4). In a previous study, we observed that the median length of transcripts originating from distal ERα enhancers in MCF-7 breast cancer cells is ∼9 kb (7). Hence, we use 9 kb as the length cutoff to define ‘short’ eRNA transcription units and hypothesize that longer transcripts originating from the enhancers are more likely to be bona fide long non-coding RNAs (lncRNAs) (7, 44). As noted above, enhancer transcription can be unidirectional or bidirectional, depending on the nature of the enhancer. Furthermore, the magnitude of enhancer transcription may correlate directly with the activity of the enhancer (7). A comparison of active enhancers (with robust uni- or bidirectional transcription) with ‘inactive’ enhancers, as well as their associated genomic features, suggests that it is informative to distinguish these different categories of enhancers (7).
The provided Perl script can be used to identify short intergenic transcripts (i.e., putative enhancer transcripts) and then divide them into short paired (bidirectional) enhancer transcripts. The transcripts remaining in the universe of short intergenic transcripts are considered to be “short unpaired transcripts” (7). The Perl code is available for download from the GitHub repository (see below; https://github.com/Kraus-Lab/active-enhancers/blob/master/scripts/Define_enhancer_transcripts.pl). It will produce an output of short paired intergenic transcripts together with information about the overlap of the transcript pair.
- # Identify short intergenic transcripts: The output from bedtools intersect after identifiying intergenic transcripts (‘intergenic_transcripts.txt’) is provided as input. The final transcripts passing these criteria are written to the ‘paired_transcripts.txt’ file, along with length of overlap ‘paired_transcripts_overlap.txt’ and coordinates of a 1kb window around the center of the overlap ‘paired_transcripts_1kb_window_overlap’.
$ ./Define_enhancer_transcripts.pl -i intergenic_transcripts.txt -a short_paired_transcripts.txt -b short_paired_transcripts_overlap.txt -c short_paired_transcripts_1kb_window_overlap.txt
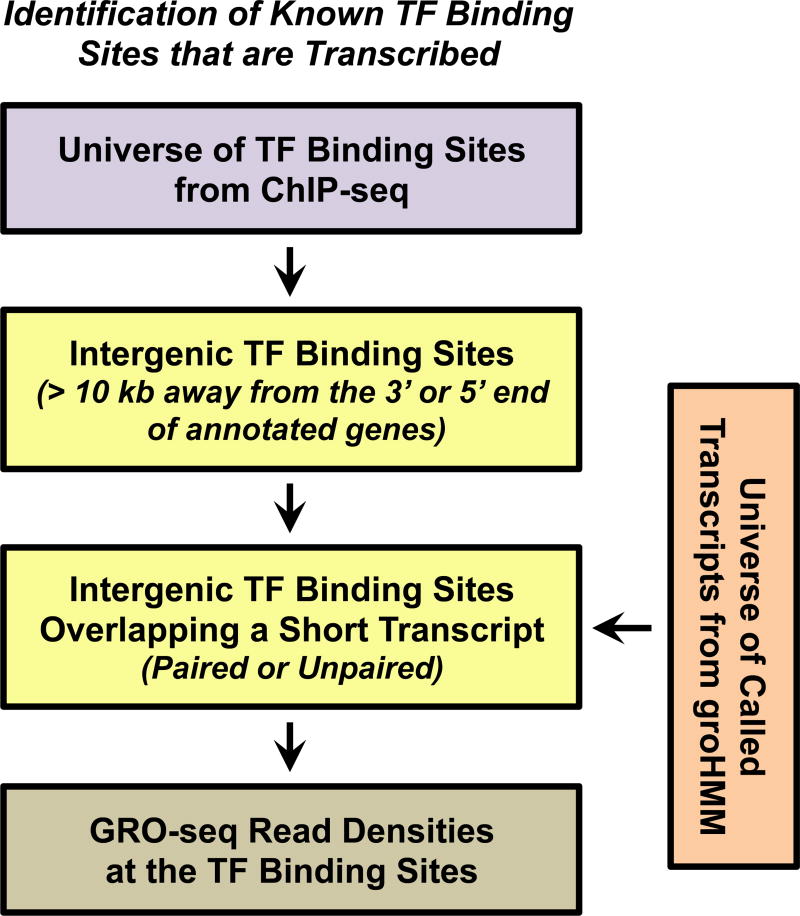
2. Identification of known TF binding sites that are actively transcribed using GRO-seq data
GRO-seq data can be used to identify known TF binding sites (from ChIP-seq data) that are actively transcribed. This can be accomplished in two ways: 1) by comparing the overlap of transcripts in the universe of transcripts from groHMM with known TF binding sites of interest or 2) by collecting and quantifying the GRO-seq reads that fall within in a specified window around known TF binding sites of interest (Fig. 5). With respect to the former, criteria for the location of the TF binding site relative to the cognate enhancer transcript(s) (or vice versa) can be specified. For example, if the focus is on paired/bidirectional enhancer transcripts, one might specify that the TF binding site must be located within the region of overlap of the + strand and – strand transcripts (7).
Figure 5. Identification of Known TF Binding Sites that are Transcribed.
Details are provided in the text.
Pipelines for the global identification of enhancer transcripts associated with known TF binding sites using ERα as an example has been described previously (7). The analysis is similar to the one described in 3.4-1. However, in this case, the starting point is a set of known TF binding sites, rather than a set of known enhancer transcripts. As described above, the first step is to define intergenic TF binding sites and then search for those that overlap with an enhancer transcript to identify active intergenic enhancers.
3.5. Associating Newly Identified Enhancers with TF Motifs
After completing the pipeline for de novo identification of active enhancers using GRO-seq data, as in 3.4-1 above, one can search in the transcribed region for an enrichment of motifs that suggest putative TFs that may drive the formation of those enhancers (7). In our analyses, we have focused on (1) a region (e.g., 500 bp) surrounding the center of the overlap between the enhancer transcript pairs for bidirectional/paired enhancer transcripts or (2) a window (e.g., 500 bp) at the 5’ end of unidirectional/unpaired enhancer transcripts (Fig. 3 and 4). The sequences of the genomic regions specified above are extracted from the UCSC genome browser.
Within the regions specified above, motifs for putative TFs can be identified in two ways: 1) a directed approach using software, such as FIMO (45) or MotifScanner (46), which searches for enrichment of known, user-provided TF motifs in the region of interest and (2) a de novo approach using software, such as MEME (47), which searches for the enrichment of specific DNA sequences that can then be matched to known TF motifs using software, such as STAMP (36) or TOMTOM (48). Motif searches in genomic regions where enhancer transcripts originate, such as those described here, can help in uncovering the TFs that mediated the formation and activity of the enhancers of interest.
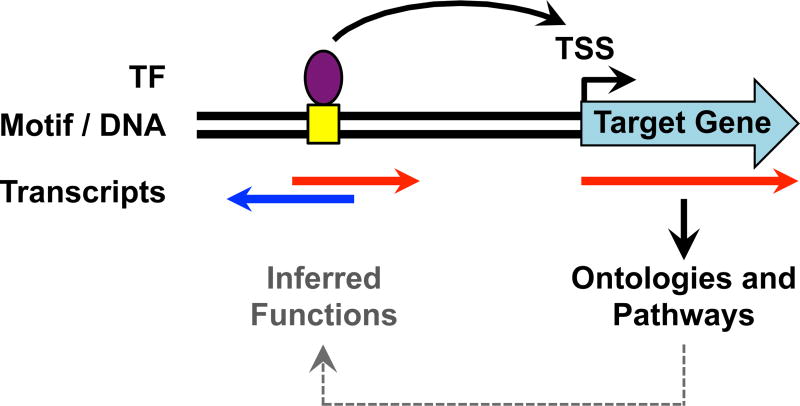
3.6. Associating Newly Identified Enhancers with Putative Target Genes
How an enhancer targets and promotes the transcription of its target genes is a fundamental question in gene regulation biology. Such analyses can be readily performed by using a ‘nearest-neighboring gene’ approach. In this approach, the actively transcribed gene (e.g., mRNA gene or lncRNA gene) nearest to an enhancer is assumed to be a target of the enhancer (Fig. 6). While not perfect, this assumption holds well enough to be informative with respect to enhancer function and target gene activation (7, 31). Alternatively, if genome-wide looping data are available for a particular TF (e.g., from ChIA-PET analyses; (15, 49, 50), then direct associations between enhancers and target genes can be discerned. In either case, the relationship between enhancer transcription and target gene transcription can be determined from GRO-seq data. Furthermore, potential biological functions of a set of enhancers identified using GRO-seq data can be explored by gene ontology (GO) or pathways analyses of the target gene set (31). Such analyses can reveal the likely biological functions of the target genes and, by extension, the likely biological functions of the enhancers as well (Fig. 6).
Figure 6. Analysis of Target Gene Activation and Functions.
Active enhancers may promote the transcription of nearby genes through looping mechanisms that bring the enhancers and target gene promoters in proximity. Knowledge of the functions of the target genes from ontology analyses can provide clues about the biological functions of the enhancers.
3.7. Identifying Cell Type-Specific Enhancers Using GRO-Seq Data
The profiles of enhancer transcripts are highly cell type-specific (32), more so than the profiles of other genomic enhancer data. This cell-type specificity can be used to discern important biological insights. The groHMM-based enhancer identification pipelines described above can be used to identifying cell type-specific enhancers by comparing GRO-seq data derived from different cell types. Using an approach similar to the one described in section 3.4 above, one can identify the universe of enhancer transcripts expressed in a particular cell type and then compare that universe to the universes of enhancer transcripts expressed in other cell types. These comparisons allow for the identification of enhancer transcripts that (1) are common across various cell types or (2) are unique to a particular cell type. Motif analysis, as described in section 3.5 above, can be performed for the enhancers producing common or unique transcripts to identify putative TFs that might drive the formation of those enhancers.
3.8. Integration with Other Genomic Data and Other Bioinformatic Analyses
After identifying the set of active enhancers in a particular cell type, the enhancer information from the GRO-seq data, which includes the genomic location and the magnitude of transcription, can be integrated with data from other genomic approaches. For example, the enrichment of enhancer-related histone modifications (e.g., H3K4me1, H3K27ac) and TF binding from ChIP-seq data or the chromatin state from DNase-seq can be assessed at the GRO-seq-called enhancers (Fig. 1).
As noted above, nearest neighboring gene analyses can be used to identify putative target genes of the predicted enhancers with subsequent GO and pathway analyses on the potential target genes. The GO and pathway analyses can be performed using tools such as WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) (51) and DAVID (52). Such analyses can provide insights about the biological functions of GRO-seq-identified enhancers. These ‘functional’ analyses can be facilitated by using GREAT (Genomic Regions Enrichment of Annotations Tool), which assigns biological meaning to a set of non-coding genomic regions by analyzing the annotations of the nearby genes (53). Users can provide GRO-seq-defined enhancer locations as input in the GREAT web interface and select the “Single nearest gene” option in the association rule settings.
Custom multi-dimensional analyses can be used to explore the relationships among multiple enhancer-related parameters. For example, we have recently demonstrated how enhancer transcription (from GRO-seq), target gene transcription (from GRO-seq), and TF binding at the predicted enhancer (from ChIP-seq) increase simultaneously in response to an external signal, an observation that can be visualized in a three-dimensional box plot (31). Of course, the additional analyses described here represent a few of the many ways in which GRO-seq and other genomic data can be mined to explore enhancer functions.
3.9. Validation of Genomic Results Using Enhancer-Specific Molecular Biology Techniques
All of the specific conclusions regarding enhancer formation and function derived from the genomic analyses described here should be validated for individual enhancers using molecular biology approaches. Enhancer features can be tested in locus-specific assays that assess (1) enhancer transcription (e.g., by reverse transcription-qPCR), (2) binding of TFs and enrichment of histone modifications (e.g., by ChIP-qPCR), (3) chromatin accessibility (e.g., by DNase-qPCR), and (4) looping (e.g., by 3C–qPCR) (7). The function of the enhancers identified by GRO-seq can be tested in reporter gene assays, where the DNA sequence from an identified enhancer is inserted into a reporter construct. Upon introduction of the enhancer-reporter construct into cells expressing the cognate TF, the presence of the enhancer DNA element should increase reporter activity if it is a functional enhancer (54).
In addition, the function of putative TFs driving the formation of enhancers identified using GRO-seq can be tested in functional assays. For example, the TF should bind to the enhancer (as determined by ChIP-qPCR) and RNA-mediated knockdown of the TF should abolish enhancer formation and function (e.g., loss of enhancer transcription and a reduction of enhancer-associated histone modifications). Furthermore, the functions of GRO-seq-identified enhancers can be tested using enhancer deletion assays in cells, in which the enhancer DNA is deleted (or mutated) using CRISPR/Cas9 and the impairment of enhancer function and target gene transcription is assessed using the qPCR-based locus-specific assays described above. Ultimately, the function of each enhancer identified and examined in detail should be tested using genetic models in vivo (55).
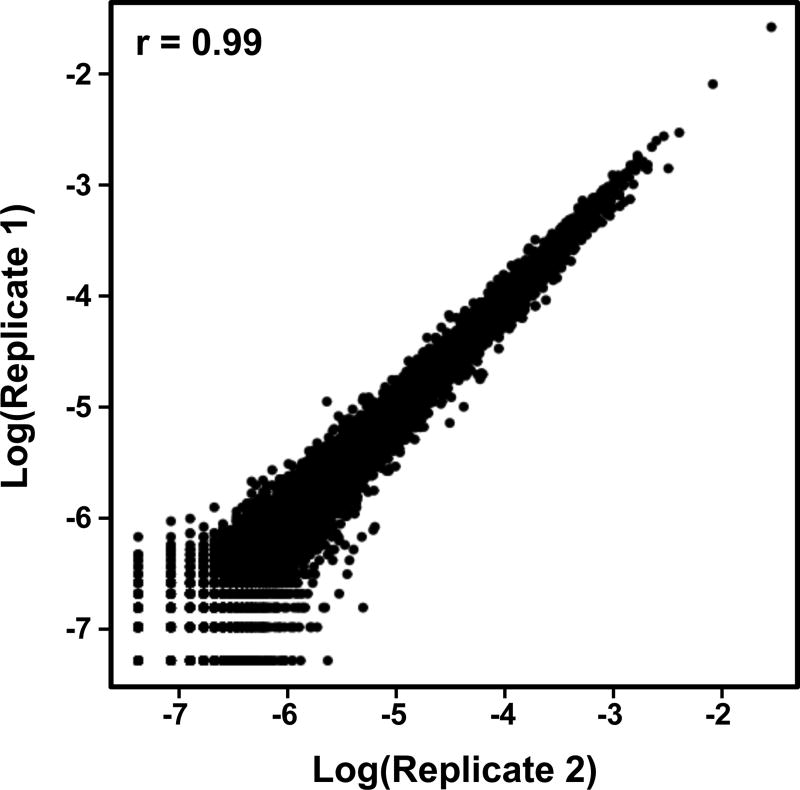
Figure 7. Correlation Plot of Two Biological Replicates of GRO-seq Data.
A typical GRO-seq experiment has two or more replicates for each experimental condition. Hence, it is important to test that the replicates are highly correlated. Shown here is a Pearson’s correlation plot.
Acknowledgments
The authors thank Minho Chae and Hector L. Franco for helpful comments and suggestions about enhancer identification using GRO-seq, as well as this manuscript. The enhancer-related work in the Kraus lab is supported by grants from the NIH/NIDDK and the Cancer Prevention and Research Institute of Texas (CPRIT).
Appendix 1. Perl script for the identification of short intergenic transcripts, with subsequent separation into short paired (bidirectional) enhancer transcripts
The code provided here is for reference only. An executable version is available for download at https://github.com/Kraus-Lab/active-enhancers/blob/master/scripts/Define_enhancer_transcripts.pl. It will produce an output of short paired intergenic transcripts together with information about the overlap of the transcript pair. Save the following code in a separate file named 'Define_enhancer_transcripts.pl' and execute in the command line as described above.
- #!/usr/bin/perl
use strict; use Getopt::Std; my $infile; my $outfile1; my $outfile2; my $outfile3; my $help; my %Options; my $optset=getopts('i:a:b:c:h:',\%Options); my $size = $#ARGV+1; if($size != 0 || !$optset || $Options{h}) { print "Usage: Define_enhancer_transcripts.pl -i <Infile> -a <OutputFile1> -b <OutputFile2> -c <OutputFile3> \n"; print "Options:\n"; print " -i <InputFile>\n"; print " -a <OutputFile1>\n"; print " -b <OutputFile2>\n"; print " -c <OutputFile3>\n"; die("Get ready with the files… \n"); }
- # Declaring the variables
my $infile=$Options{i}; my $outfile1=$Options{a}; my $outfile2=$Options{b}; my $outfile3=$Options{c}; my $length=0; my @coord=();
- # Read the input file with intergenic transcripts
open (FILE1, "<$infile") || die "can't: $!";
- # Output file1 with short intergenic transcripts
open(OUTPUT1, ">$outfile1") or die("Unable to open file"); while(<FILE1>) { my $line1 = $_; chomp($line1); @coord=split(/\t/,"$line1");
- # Calculating transcript lengths
$length = $coord[2]-$coord[1];
- # Selecting the transcripts shorter than 9 kb
if($length < 9000) { print OUTPUT1 "$line1\n"; } }
- # Sorting the short intergenic transcripts based on the chromosome, start position
$a=`sort -k1,1 -k2n,2 $outfile1 >sort_$outfile1`;
- # Identifying intergenic short paired transcripts
open (FILE, "<sort_$outfile1") || die "can't: $!";
- # Output file2 with a list of short paired transcripts, length of the overlap
open(OUTPUT2, ">$outfile2") or die("Unable to open file");
- # Output file3 with the coordinates of a 1kb window around the center of the overlap of intergenic short paired transcripts
open(OUTPUT3, ">$outfile3") or die("Unable to open file");
- # Declaring the variables to be used
my @line=<FILE>; my @firstline=(); my @secondline=(); my $j; my $overlap; my $overlap_center; my $window_start; my $window_end;
-
# Read the lines of the file and store in an array; accessing each line through for loop
for(my $i=1;$i<=$#line;$i++) {- # Comparing two consecutive lines to check overlap
$j=$i+1; @firstline=split(/\t/,"$line[$i]"); @secondline=split(/\t/,"$line[$j]");
-
# Matching the chromosome number of the two consecutive lines and checking if overlap exists to define the universe of paired transcripts
if($firstline[0] eq $secondline[0] && $firstline[1] <= $secondline[2] && $firstline[2] >= $secondline[1]) {- # Calculating overlap, overlap center
$overlap=$firstline[2]-$secondline[1]; $overlap_center = int(($firstline[2]+$secondline[1])/2);
- # Calculating a 500 bp window on either side of the overlap center
$window_start = $overlap_center-500; $window_end = $overlap_center+500;
- # Writing a list of short paired transcripts, length of the overlap to output1
print OUTPUT2 "$line[$i]$line[$j]\t\t\t\t\t\t\t\t\t\t$overlap\n";
- # Writing the coordinates of a 1 kb window around the length of the overlap to output2
print OUTPUT3 "$firstline[0]\t$window_start\t$window_end\t$overlap_ce nter\n"; } }
Footnotes
The various cutoffs described herein may have to be tuned for the particular biological system or the particular data set being analyzed.
A typical GRO-seq experiment has two or more replicates for each experimental condition. Hence, it is important to test that the replicates are highly correlated (Fig. 7).
For the analysis described in section 3.6, which involves the comparison of multiple GRO-seq datasets to identify cell type specific enhancers, the library sizes of all the samples should be compared. Appropriate normalization steps should be used to avoid bias due to differences in sequencing depth.
Script Availability
The enhancer identification pipelines described herein are implemented in Bash, Perl and R. The most up to date version, with full documentation and examples, is available free-of-charge under an open-source MIT license via GitHub at: https://github.com/Kraus-Lab/active-enhancers.
References
- 1.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 2.Wamstad JA, Wang X, Demuren OO, Boyer LA. Distal enhancers: new insights into heart development and disease. Trends Cell Biol. 2014;24:294–302. doi: 10.1016/j.tcb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 6.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X, Chae M, Krishnakumar R, Danko CG, Kraus WL. Dynamic reorganization of the AC16 cardiomyocyte transcriptome in response to TNFalpha signaling revealed by integrated genomic analyses. BMC Genomics. 2014;15:155. doi: 10.1186/1471-2164-15-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savic D, Roberts BS, Carleton JB, Partridge EC, White MA, Cohen BA, Cooper GM, Gertz J, Myers RM. Promoter-distal RNA polymerase II binding discriminates active from inactive CCAAT/enhancer-binding protein beta binding sites. Genome Res. 2015 doi: 10.1101/gr.191593.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores O, Deniz O, Soler-Lopez M, Orozco M. Fuzziness and noise in nucleosomal architecture. Nucleic Acids Res. 2014;42:4934–4946. doi: 10.1093/nar/gku165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 15.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 17.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 22.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 23.Spicuglia S, Kumar S, Yeh JH, Vachez E, Chasson L, Gorbatch S, Cautres J, Ferrier P. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol Cell. 2002;10:1479–1487. doi: 10.1016/s1097-2765(02)00791-8. [DOI] [PubMed] [Google Scholar]
- 24.Vieira KF, Levings PP, Hill MA, Crusselle VJ, Kang SH, Engel JD, Bungert J. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J Biol Chem. 2004;279:50350–50357. doi: 10.1074/jbc.M408883200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J Mol Biol. 2005;350:883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto S, Sugano S, Nakai K, Suzuki Y. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 2011;21:775–789. doi: 10.1101/gr.110254.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franco HL, Nagari A, Kraus WL. TNFalpha signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell. 2015;58:21–34. doi: 10.1016/j.molcel.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chae M, Danko CG, Kraus WL. groHMM: a computational tool for identifying unannotated and cell type-specific transcription units from global run-on sequencing data. BMC Bioinformatics. 2015;16:222. doi: 10.1186/s12859-015-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danko CG, Hyland SL, Core LJ, Martins AL, Waters CT, Lee HW, Cheung VG, Kraus WL, Lis JT, Siepel A. Identification of active transcriptional regulatory elements from GRO-seq data. Nat Methods. 2015;12:433–438. doi: 10.1038/nmeth.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinformatics in Action. 2012;17(1):10–12. Key: citeulike:11851772 17:10–12. [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data 2010 [Google Scholar]
- 40.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 42.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, Annotation, and Functional Analysis of Long Noncoding RNAs Controlling Cell-Cycle Gene Expression and Proliferation in Breast Cancer Cells. Mol Cell. 2015;59:698–711. doi: 10.1016/j.molcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta SSJ, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007 doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heldring N, Isaacs GD, Diehl AG, Sun M, Cheung E, Ranish JA, Kraus WL. Multiple sequence-specific DNA-binding proteins mediate estrogen receptor signaling through a tethering pathway. Mol Endocrinol. 2011;25:564–574. doi: 10.1210/me.2010-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyer MB, Benkusky NA, Onal M, Pike JW. Selective regulation of Mmp13 by 1,25(OH)D, PTH, and Osterix through distal enhancers. J Steroid Biochem Mol Biol. 2015 doi: 10.1016/j.jsbmb.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]