Abstract
MicroRNAs play important roles in the process of cancer, which microRNA-520b (miR-520b) has been reported to play critical roles in tumor progression in many types of cancers. However, its role in glioma remains unknown. In this study, we found that miR-520b could inhibit growth and progression in glioma by targeting methyl-CpG-binding domain 2 (MBD2). First, we analyzed the expression of miR-520b in different glioma grades and different cell lines (U87, U251 and astrocyte). Then we assessed the effect of miR-520b on glucose metabolism, invasion, angiogenesis and chemosensitivity in U87 and U251 cells. By using an online database, miR-520b was found to directly bind to the 3’-untranslated regions (3’-UTR) of MBD2 and reduce its expression at the protein level, which further inhibits the development of glioma. MBD2 was also found to be over-expressed in human glioma tissues and in U87 and U251 cells and its level was inversely correlated with that of miR-520b. Furthermore, restoration of MBD2 partially rescued the miR-520b-induced inhibitory effect on glucose metabolism, invasion, angiogenesis and chemosensitivity in glioma cells. In summary, to date, this is the first study to demonstrate that miR-520b functions as a tumor suppressor in glioma by directly targeting MBD2, suggesting that MBD2 may be a potential therapeutic target for glioma.
Keywords: miR-520b, MBD2, glucose metabolism, invasion, angiogenesis, chemosensitivity, glioma
Introduction
Glioma is the most common primary tumor of the brain and is divided into four clinical grades on the basis of histology and prognosis [1,2]. Several gene expression changes and chromosomal abnormalities are regularity found in gliomas and great progress in standard therapies, including surgical resection, chemotherapy and radiotherapy have been made [3]. However, the prognosis of glioma remains poor. Thus, there is an urgent need to develop new treatment strategies for this disease.
MicroRNAs are a family of endogenous non-coding RNA molecules approximately 20 nucleotides [2]. They regulate gene expression at the transcriptional and posttranscriptional level by completely or incompletely binding to the 3’-UTR of their target messenger RNA (mRNA), and then repress translation or promote degradation of the mRNA to exert biological functions [4]. By modulating different target genes, miRNAs have been found to play vital roles in a variety of cellular behaviors in cancer, such as apoptosis, proliferation, invasion, migration and glucose uptake [5-7]. Moreover, miRNAs have been shown to be useful as diagnostic and prognostic indicators of disease type and severity [1]. Aberrant expression of miR-520b has been reported in several types of cancer. For example, miR-520b acts as a tumor suppressor by regulating epidermal growth factor receptor in gastric cancer and provides a novel marker and insights into potential therapy for gastric cancer [8]. Zhang reported that Hepatitis B virus X protein accelerates hepatocarcinogenesis by partnering with survivin through the modulation of tumor suppressor miR-520b and hepatitis B X-interacting protein [9]. Another study has shown that miR-520b is involved in regulating the migration of breast cancer cells by targeting HBXIP to stimulate NF-κB-mediated IL-8 expression [10]. However, the underlying role and mechanism of miR-520b in development of glioma remain unclear.
The methylation status of DNA influences many biological processes during development, and methylation of cytosine residues of promoter-proximal regulatory sequences in human is usually considered to silence gene expression [11]. One of the most commonly studied families of effectors of cytosine methylation the methyl-CpG-binding domain family of proteins, most of which specifically bind methylated CpG dinucleotides and play crucial roles in determining the transcriptional state of the human epigenome [12]. The MBD has the ability to bind single symmetrically methylated CpG dinucleotides, and the functions of MBD1-4 family members are well-studied [13,14]. Apart from MBD3, these proteins preferentially bind 5-methylcytosine over unmethylated cytosine [15]. MBD2 is a subunit of the Mi2-NuRD complex and has been shown to mediate gene silencing via recruitment of the complex to methylated promoters [16]. In particular, there is emerging evidence linking the function of MBD2 and the abnormal hypermethylation of CpG islands and shores in cancer.
In our study, we discovered that miR-520b is down-regulated in glioma tissues, and that its expression serves as a protective factor for the prognosis of glioma patients. We suggested that the exogenous expression of miR-520b in glioma cells inhibits proliferation, decreases invasion, and promotes apoptosis. In addition, we demonstrated that miR-520b directly targets the 3’-UTR of MBD2 to regulate its expression and downstream signaling proteins. Our study provided the first evidence of the role for miR-520b as a tumor suppressor in glioma via MBD2. According to these findings, miR-520b could be a new therapeutic target and predictor of survival in glioma patients.
Materials and methods
Human tissue samples
Human normal brain tissues and glioma samples were obtained from the Department of Neurosurgery at Nanjing First Hospital, Nanjing Medical University. This study was approved by the hospital’s Institutional Review Board, and written informed consent was obtained from each patient. Tissue samples were collected during surgery and immediately frozen in liquid nitrogen and stored until total RNAs or proteins were extracted.
Cell culture
The human glioma cell lines U87 and U251 were purchased from the Chinese Academy of Sciences Cell Bank. All of the cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) with high glucose supplemented with 10% fetal bovine serum (ScienCell, USA). Human umbilical vein endothelial cells (HUVECs; purchased from GeneChem, China) were cultured in endothelial cell basal medium (Medium 200, Gibco, USA) supplemented with Low Serum Growth Supplement (Gibco, USA).
Plasmid constructs, oligonucleotides and cell transfection
Oligonucleotides were purchased from GenePharma (Shanghai, China). The sequences were as follows: MBD2-small interfering RNA (si-MBD2): 5’-GAT GAT GCC TAG TAA ATT A-3’ (865-894 bp) and scrambled control: (5’-GTG AAT ACA GGC TTT AAA TAG-3’). The hsa-miR-520b mimic and hsa-miR-520b inhibitor were also purchased from GenePharma.
Real-time quantitative PCR
RNA was isolated from harvested cells and tissues with TRIzol (Invitrogen, USA) following the manufacturer’s protocols. To detect the expression of MBD2. RT-qPCR was performed using Fermentas reverse transcription reagents and SYBR Green PCR Master Mix (Applied Biosystems, USA) according to the manufacturer’s protocols. To detect the expression of miR-520b, RT-qPCR was performed using TaqMan miRNA assays (Applied Biosystems, USA). U6 was used for normalization. In addition, analyses were performed using the 2-ΔCt or 2-ΔΔCt method. Each experiment was performed in triplicate.
Western blot analysis
Total proteins were isolated from the indicated cells using RIPA lysis buffer (KeyGEN, Nanjing, Jiangsu, China) and quantified using a BCA Protein Assay Kit (Beyotime, Nanjing, Jiangsu, China). The obtained proteins were then separated by SDS-PAGE and transferred onto a 0.45-μm cellulose acetate membrane (Immobilon, USA). After blocking with 5% nonfat milk (BrightDairy, Shanghai, China), the membrane was incubated with primary antibodies recognizing MBD2 (1:1000, Sigma, M7318) or GAPDH (1:1,000, YIFEIXUE BIO TECH. Jiangsu, China), followed by an incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody (YIFEIXUE BIO TECH Jiangsu, China). Finally, the bands were visualized with Image Quant LAS 4000 mini imager (GE, USA).
Glucose uptake assay
Glucose uptake was measured using a 2-Deoxyglucose Glucose Uptake Assay Kit (Fluorometric, Abcam, USA) based on the protocol provided by the manufacturer. U87 and U251 cells were seeded in 96-well plates (1000 cells/well) and incubated overnight. After 24 h, the cells were incubated in the dark with 2-deoxyglucose (10 mM) for 20 min at 37°C in CO2 containing humidified atmosphere and the uptake of 2-deoxyglucose was measured using a fluorescence microplate reader at Ex/Em=535/587 nm.
Transwell assay
To measure cell invasion, 8 mm pore 24-well Matrigel invasion chambers (Corning, NY, USA) were used according to the protocol provided by manufacturer. 20000 cells were seeded into each well. Two hundred microliters of DMEM supplemented with 0.1% was added to upper chambers, while 400 µl DMEM supplemented with 10% FBS was added to the lower chambers. After incubation at 37°C in 5% CO2 for 48 hours, the non-invading cells were removed from the upper chamber. Then, the migrating cells were stained with 0.1% crystal violet (YIFEIXUE BIO TECH, Jiangsu, China). The migrated cells were counting the number of cells in 3 independent visual fields under a microscope.
Angiogenesis assay
Matrigel (BD, USA) was thawed at 4°C overnight, and each well of pre-chilled μ-Slide Angiogenesis plate (Ibidi, Germany) was coated with 10 µl of Matrigel. The plates were then incubated at 37°C for 2 hours to form a layer of Matrigel. Cells were cultured to 90-100% confluence. The old medium was discarded and replaced with serum-reduced medium (1% FBS) for 24 h. The conditioned medium was collected and stored at -80°C. HUVECs were switched to basic medium containing 0.2% FBS. After 24 h, the starved HUVECs were trypsinized, collected, counted, and resuspended in endothelial cell growth medium (Gibco, USA) supplemented with Gibco LSGS (Low Serum Growth Supplement, Gibco, USA). Then, the cells were mixed with equal volumes of the conditioned medium and seeded in the Matrigel-coatedμ-Slide Angiogenesis plates at 3×104 cells/well. After 12 h, tube formation was examined under a light microscope. The length of the tubes was measured using the Soft Imaging System (Soft Imaging System GmbH, Germany).
In vitro chemosensitivity assay
Cancer cells were seeded at a density of 4000 cells per well in a 96-well plate and incubated overnight. Freshly prepared temozolomide (TMZ) was added at final concentrations ranging from 25 to 400 mM. Forty-eight hours later, cell viability was assayed using a CCK8 kit.
Xenograft tumor assay
Ten immunodeficient female nude mice (Beijing Laboratory Animal Center, Beijing, China) were used to test the effect of miR-520b on glioma in vivo. These nude mice were randomly divided into two groups (5 mice per group). Approximately 2×106 logarithmically growing U87 cells stably expressing negative control or miR-520b mimics-transfected cells were subcutaneously injected into the nude mice. After 21 days, the subcutaneous tumors were dissected and weighed. Tumor volume was calculated according to the formula: V (mm3)=0.5 * a * b2 (a represents the longest axis, and b represent the shortest axis).
Statistical analysis
All experiments except the animal experiments were conducted at least three times. All values in this study are shown as the means ± SD. The difference between the groups was considered significant and very significant at P < 0.05 (* or #) and P < 0.01 (** or ##), respectively.
Results
MiR-520b is down-regulated in glioma tissues and cell lines
To reveal the expression of miR-520b in glioma, we examined the expression patterns of miR-520b in the Chinese Glioma Genome Atlas (CGGA) database and found that miR-520b expression was significantly lower in high-grade glioma tissues than in low-grade glioma tissues, indicating that miR-520b expression correlated with malignancy (Figure 1A). Next, RT-qPCR was performed to examine the expression levels of miR-520b in 5 normal brain tissues and tumor samples of 20 patients with glioma (Figure 1B). As shown in Figure 1B, miR-520b was down-regulated in glioma tissues compared with the non-tumor tissues. The expression levels of miR-520b in glioma cell lines (U87, U251) were also investigated and the human astrocytic cell line was used as the control (Figure 1C). The results revealed that miR-520b was significantly down-regulated in glioma cell lines compared with the astrocytic cells. These findings demonstrated that low expression of miR-520b might be a prognostic marker for glioma patients.
Figure 1.
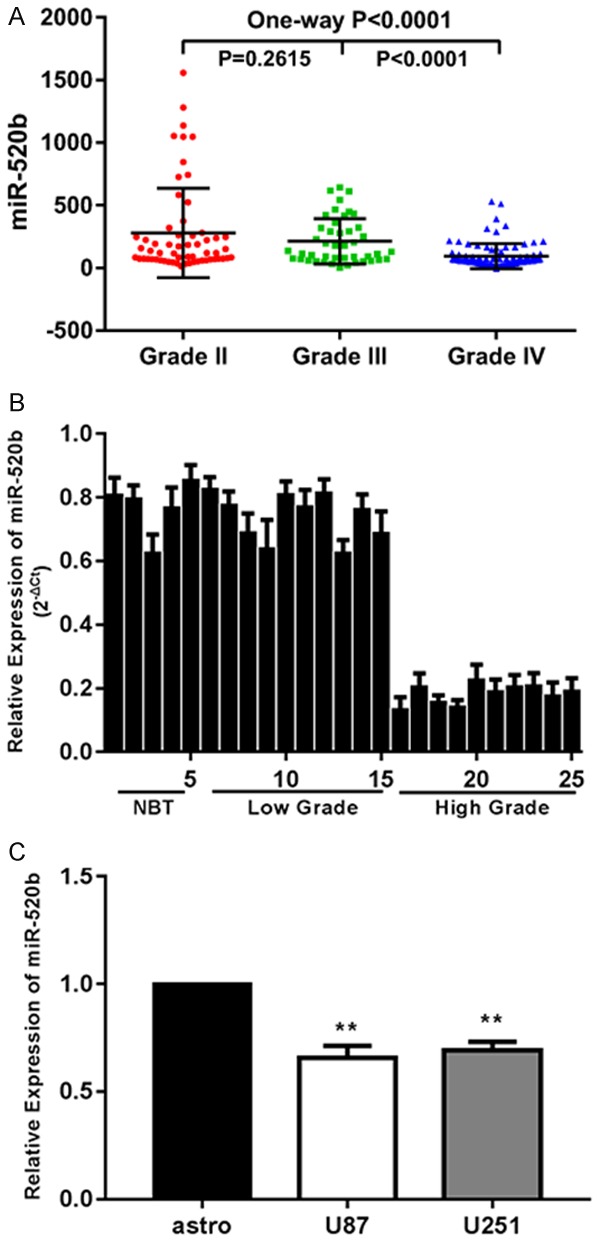
MiR-520b is down-regulated in glioma tissues and cell lines. A. Expressions of miR-520b in CGGA public database. B. Relative expressions of miR-520b in glioma tissues and normal brain tissues. C. Relative expressions of miR-520b in astrocyte and glioma cell lines U87, U251 (*P < 0.05, **P < 0.01).
MiR-520b suppresses glucose metabolism, invasion and angiogenesis and enhances chemosensitivity glioma cells in vitro
The abnormal expression of miR-520b in gliomas and glioma cell lines led us to investigate its biological roles. Chemically synthesized miR-520b mimics were transfected into U87 and U251 cells to alter the expression of miR-520b. In both cells, the transfection efficiency was confirmed by qPCR (Figure 2A). We treated the cell lines with increasing concentrations of glucose (0, 10 or 20 mM) for 12 h and examined the levels of glucose uptake in these cells (Figure 2B). Consistently, up-regulation of miR-520b significantly inhibited glucose uptake by the glioma cell lines. Next transwell assays were adopted to assess the invasion of glioma cells. Significant differences were observed in cell invasion between the cells transfected with miR-520b and the control (Figure 2C). The results of the angiogenesis assay showed that the up-regulation of miR-520b decreased the angiogenic capacity of the HUVECs (Figure 2D). As expected, our results showed that the overexpression of miR-520b in U87 and U251 cells significantly increased their chemosensitivity to treatment with TMZ (Figure 2E). Collectively, these results suggested that miR-520b functions as tumor suppressor in glioma.
Figure 2.
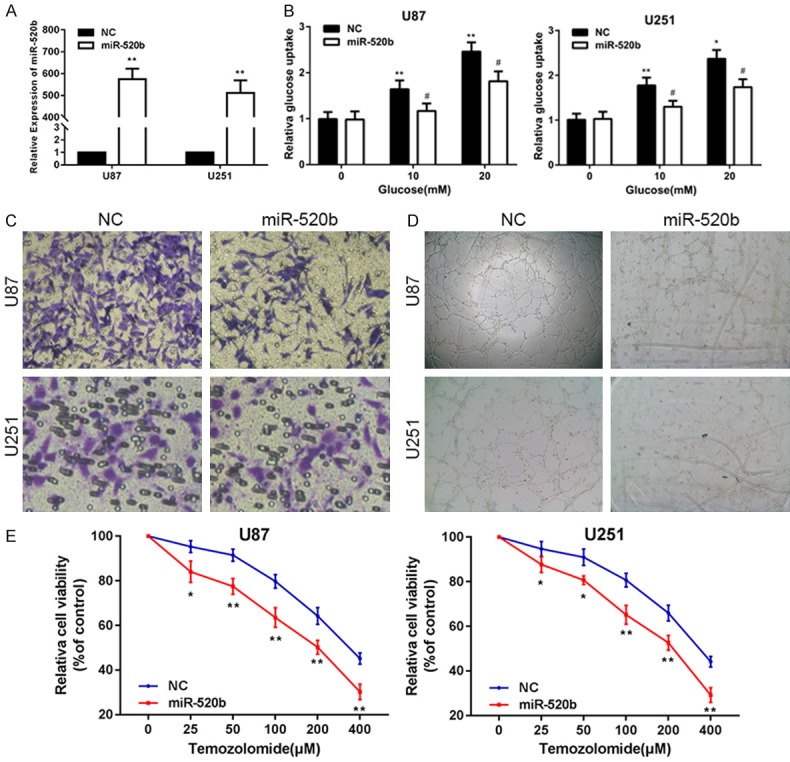
MiR-520b suppresses glucose metabolism, invasion and angiogenesis and enhances chemosensitivity glioma cells in vitro. A. The expression of miR-520b in U87 and U251 cells transfected with NC or miR-520b mimic. B. Relative glucose uptake in U87 and U251 cells transfected with NC or mir-520b mimic and treated with the indicated concentrations of glucose (0, 10, or 20 mM). *P < 0.05, **P < 0.01 compared to cells treated with 0 mM glucose, #P < 0.05 compared to control miR-520b transfected cells treated with the same glucose concentration. C. Transwell assay in U87 and U251 cells transfected with NC or miR-520b mimic. D. Tube formation assay in U87 and U251 cells transfected with NC or miR-520b mimic. E. Cell proliferation was examined in U87 and U251 cells that were expressing NC or miR-520b mimic following TMZ treatments at different doses. CCK8 assay was conducted 48 hours after TMZ treatment (*P < 0.05, **P < 0.01, #P < 0.05, ##P < 0.01).
MBD2 is a direct target of miR-520b in glioma cell lines
It is accepted that miRNAs exert their functions by regulating the expression of downstream target genes. Therefore, we searched for miR-520b targets using TargetScan. According to the prediction, MBD2, which might be a target of miR-520b, was found (Figure 3A). A high level of MBD2 was observed in the CGGA (Figure 3B). Furthermore, the expression of MBD2 was higher in the glioma tissues of high grade than these in low grade and normal tissues (Figure 3C). The expression levels of MBD2 protein in glioma cell lines were remarkably higher than astrocyte cell line by using Western Blot assay (Figure 3D). In addition, Western blot assay showed that expression of MBD2 was increased/decreased in glioma cells with down-regulation/up-regulation of miR-520b (Figure 3E). After over-expressing miR-520b, the luciferase activity of the wild-type MBD2 reporter containing miR-520b binding site was found to be attenuated, which was not observed in the mutant MBD2 reporter or the miR-520b negative control-transfected group of U87 and U251 cells (Figure 3F). As illustrated in Figure 3F, the level of MBD2 mRNA by RT-qPCR was significantly reduced when the cells were transfected with miR-520b mimics. According to the result above, MBD2 was validated as a target of miR-520b via direct binding its 3’-UTR.
Figure 3.
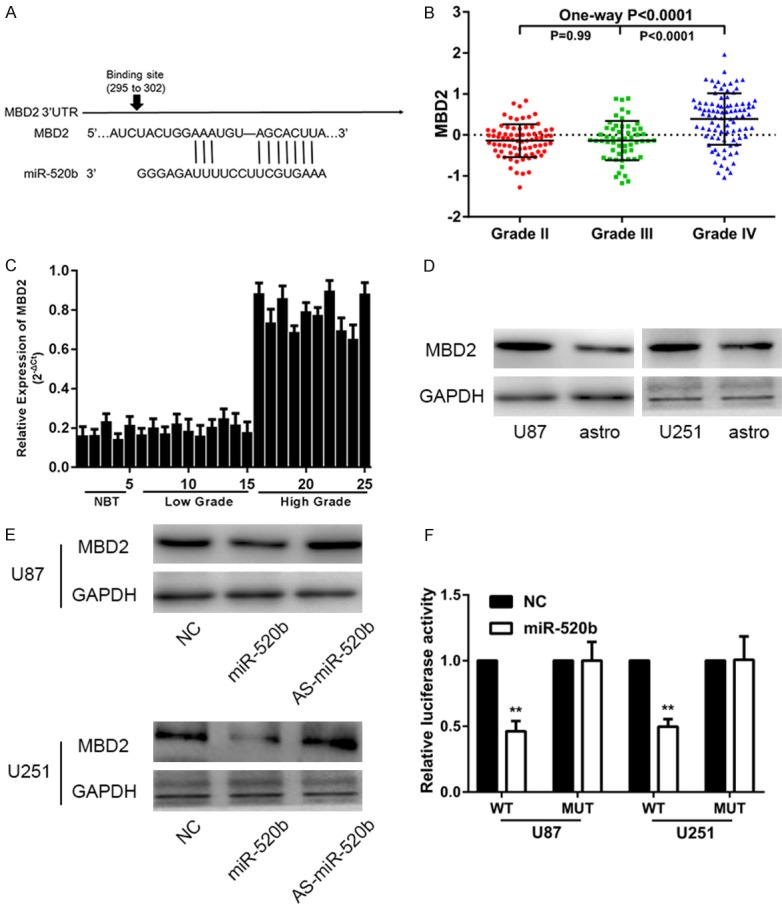
MBD2 is a direct target of miR-520b in glioma cell lines. A. Schematic of the MBD2 3’-UTR including the putative binding sites for miR-520b, as predicted by TargetScan 7.1. B. Expressions of MBD2 in CGGA public database. C. Relative expressions of MBD2 in glioma tissues and normal brain tissues. D. Western blot assay showed the expression of MBD2 in U87, U251 and astrocytic cells. E. Western blot assay showed the expression of MBD2 in U87 and U251 cells transfected with NC, mir-520b mimic or AS-miR-520b. F. miR-520b down-regulated luciferase activity of wild-type MBD 3’-UTR expression vector, but did not reduce expression of a mutant MBD2 3’-UTR (*P < 0.05, **P < 0.01).
MiR-520b inhibits glucose metabolism, invasion, angiogenesis and chemosensitivity by targeting MBD2 in vitro
To determine the role of MBD2 glucose metabolism, invasion, angiogenesis and chemosensitivity in glioma cell lines, we first knocked down the level of MBD2 using chemically synthesized MBD2-targeting siRNA (si-MBD2). In both cells, the transfection efficiency was confirmed by qPCR and Western blot assay (Figure 4A, 4B). We treated both cell lines with increasing concentrations of glucose (0, 10 or 20 mM) for 12 h and found the levels of glucose uptake are dramatically lower in si-MBD2-transfected glioma cells (Figure 4C). Western blotting assay indicated that the protein levels of MBD2 were markedly up-regulated in response to glucose treatment in a dose-dependent manner (Figure 4D). Consistently, knockdown of MBD2 expression significantly inhibited glucose uptake by the glioma cell lines. The invasion of U87 and U251 cells in the si-MBD2 group was decreased compared with the control group (Figure 4E). Additionally, we observed that the down-regulation of MBD2 also decreased the angiogenic capacity of the HUVECs (Figure 4F). As expected, our results showed si-MBD2-transfected glioma cells were more sensitive to TMZ treatment (Figure 4G). Overall, these results suggested that MBD2 functions an onco-gene in glioma.
Figure 4.
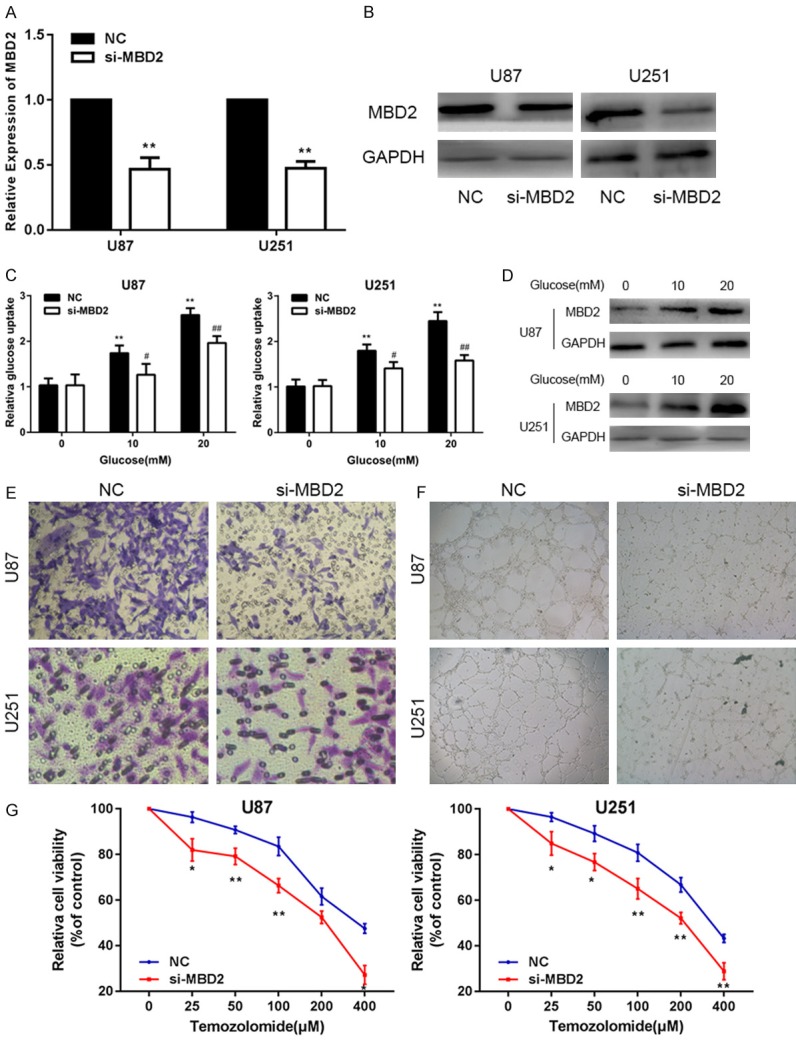
MiR-520b inhibits glucose metabolism, invasion, angiogenesis and chemosensitivity by targeting MBD2 in vitro. A. The expression of MBD2 in U87 and U251 cells transfected with NC or si-MBD2. B. Western blot assay showed the expression of MBD2 in U87 and U251 cells transfected with NC or si-MBD2. C. Relative glucose uptake in U87 and U251 cells transfected with NC or si-MBD2 and treated with the indicated concentrations of glucose (0, 10, or 20 mM). *P < 0.05, **P < 0.01 compared to cells treated with 0 mM glucose, #P < 0.05 compared to control si-MBD2 transfected cells treated with the same glucose concentration. D. Protein levels of MBD2 as determined by Western blot analysis in U87 and u251 cells treated with the indicated concentrations of glucose (0, 10 or 20 mM). E, F. Transwell assay and tube formation assay in U87 and U251 cells also transfected with NC or si-MBD2. G. Cell proliferation was examined in U87 and U251 cells that were expressing NC or si-MBD2 following TMZ treatments at different doses. CCK8 assay was conducted 48 hours after TMZ treatment (*P < 0.05, **P < 0.01, #P < 0.05, ##P < 0.01).
To further confirm that miR-520b exerted its function by directly targeting MBD2 in the glioma cell lines, we transfected miR-520b together with an MBD2-expression plasmid into the two cell lines, followed by a series of functional assays. First, we used RT-qPCR and western blot assay to determine whether MBD2 was involved in changes induced by miR-520b expression (Figure 5A, 5B). RT-qPCR and western blot assay proved that compared with the two other groups, MBD2 was significantly downregulated in the miR-520b-transfected group. Afterwards, we also confirmed that the restoration of MBD2 expression was capable of partially counteracting the effects of miR-520b on glucose metabolism, invasion, angiogenesis and chemosensitivity in U87 and U251 cell lines (Figure 5C-F). These data suggested that miR-520b inhibited glucose metabolism, invasion, angiogenesis and chemosensitivity in glioma cell lines, partly by directly targeting MBD2.
Figure 5.
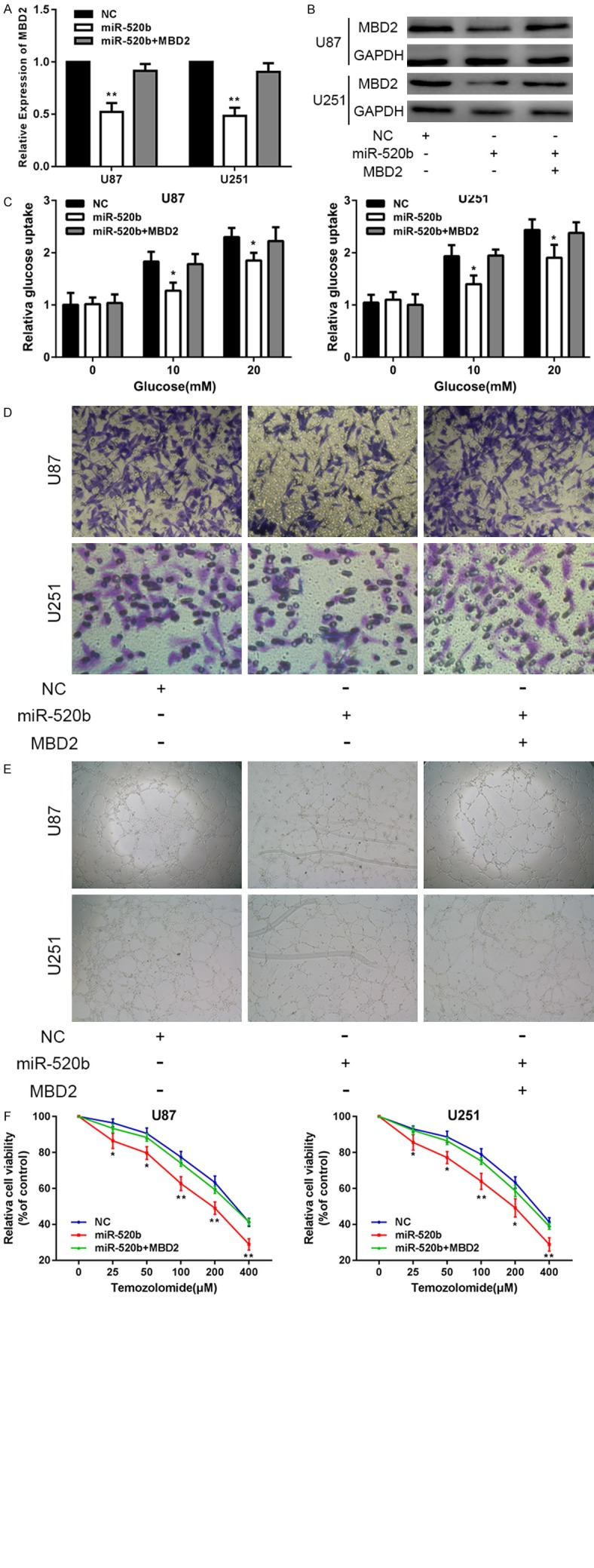
MiR-520b inhibits glucose metabolism, invasion, angiogenesis and chemosensitivity by targeting MBD2 in vitro. A, B. The expression of MBD2 in U87 and U251 cells transfected with NC, miR-520b or miR-520b+MBD2. C. Relative glucose uptake in U87 and U251 cells transfected with NC, miR-520b or miR-520b+MBD2 and treated with the indicated concentrations of glucose (0, 5, or 20 mM). *P < 0.05, **P < 0.01 compared to cells treated with miR-520b+MBD2. D. Transwell assay in U87 and U251 cells transfected with NC, miR-520b or miR-520b+MBD2. E. Tube formation assay in U87 and U251 cells transfected with NC, miR-520b or miR-520b+MBD2. F. Cell proliferation was examined in U87 and U251 cells that were expressing NC, miR-520b or miR-520b+MBD2 following TMZ treatments at different doses. CCK8 assay was conducted 48 hours after TMZ treatment (*P < 0.05, **P < 0.01).
MiR-520b suppressed tumor growth in nude mice by inhibiting MBD2
Considering the remarkable glioblastoma-inhibiting effects of miR-520b in vitro, we extended our investigation to examine if miR-520b could impede glioma growth in vivo using nude mice. To investigate the role of miR-520b on the tumor growth in vivo, we implanted U87 cells stably expressing miR-520b in the subcutaneous tissue of nude mice. The tumor growth was evaluated every 7 days for 21 days (Figure 6A). The tumor growth kinetics showed that the tumors derived from the U87 cells transfected with miR-520b mimics exhibited slower growth than the tumors derived from the control cells. The mice were sacrificed after 21 days. Subsequently, the tumors were dissected and weighed (Figure 6B, 6C). The average tumor weight of miR-520b mimics-treated group was significantly lower than that of the control. In addition, we also measured MBD2 expression in tumor tissues (Figure 6D). Therefore, these results demonstrated that increased levels of miR-520b could inhibit the development of glioma in vivo.
Figure 6.
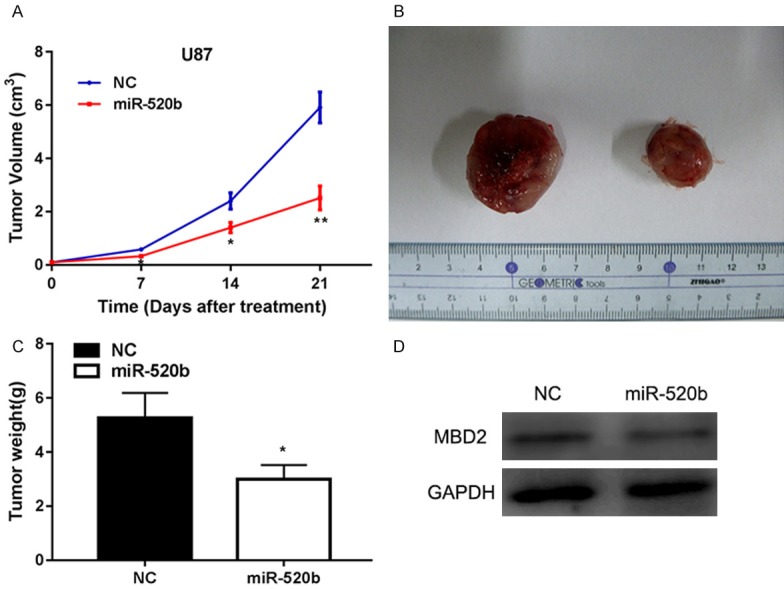
MiR-520b suppressed tumor growth in nude mice by inhibiting MBD2. A. Tumor growth curves were established by measuring tumor volume every 7 days for 21 days after injection. B. Tumor formation was performed in nude mice and the excision tumor of U87 xenografts. C. Tumor weight isolated from nude mice in each treatment group on day 21 after injection. D. Western blot assay showed the expression of MBD2 in the tumor tissues (*P < 0.05, **P < 0.01).
Discussion
MiRNAs act as posttranscriptional gene regulators in the etiology of various pathological events. Abnormal expression of miRNAs has been considered to participate in cancer progression. Several studies have shown that miR-520b functions as a biomarker and has significant roles in many types of cancers [17,18]. However, there has not been any study that on the role of miR-520b in glioma so far. In our study, we discovered that miR-520b is down-regulated in gliomas and that it acts as a novel tumor suppressor in glioma. Meanwhile, up-regulation of miR-520b significantly suppressed glucose metabolism, invasion and angiogenesis and enhanced the chemosensitivity of glioma cell lines. Lastly, we demonstrated that miR-520b acts as a tumor suppresser by suppressing its target MBD2.
The MBD family members MBD1, MBD2, MBD4 and MeCP2 bind to methylated DNA via methyl-binding domain [14]. Therefore, MBD proteins are often described as ‘readers’ of DNA methylation [19]. Moreover, MBD proteins also have a role in gene repression. The MBD family proteins are vital players in determining the transcriptional state of the epigenome [19]. In recent years, the associations of MBD2 with human cancers have been reported. Current studies have emphasized the tumorigenic role of MBD2 in prostate cancer, breast cancer and human oral squamous cell carcinoma [11,20,21]. However, the effect of MBD2 on glioma is still poorly understood. In the present study, we showed that compared with that in Low Grade Glioma, the expression of MBD2 in High Grade Glioma was significantly higher. The down-regulation MBD2 markedly repressed glucose metabolism, invasion and angiogenesis and enhanced the chemosensitivity of glioma cell lines. The results above showed that MBD2 contributed to glioma progression in both U87 and U251 cells. Moreover, our findings also provide the first evidence that MBD2 is a predominant mediator of miR-520b-induced tumor-suppressive function.
In conclusion, our present study showed that miR-520b was down-regulated and played the role of a tumor suppressor in glioma by directly targeting MBD2. In addition, we demonstrated, for the first time, that the importance of the miR-520b/MBD2 axis in glioma progression. Moreover, miR-520b rendered glioma cells more sensitive to TMZ treatment and promoted TMZ-induced apoptosis in glioma cells via suppression of its target gene, MBD2. Thus, the induction of miR-520b in combination with TMZ treatment could be a useful therapeutic strategy for suppressing glioma growth. Nevertheless, further studies are needed to determine the exact mechanism of decreased miR-520b expression during the progression of glioma and to further explore other possible targets of miR-520b in glioma. Additionally, a large cohort study, incorporating MBD2 expression and function should also be further investigated.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81502168, No. 81302180), Program Funding for Health Youth Talent Training Project of Nanjing (QRX11027), Scientific and Technologic Development Program of Nanjing City (No. 20160525), the Key Project supported by Medical Science and Technology Development Foundation, Nanjing Department of Health (Grant No. YKK16139 and YKK12139), Science and Technology Development Foundation of Nanjing Medical University (2015NJMU049). We thank General Clinical Research Center, Nanjing First Hospital, for technical support.
Disclosure of conflict of interest
None.
References
- 1.Yan S, Tao T, Ning L, Luan WK, Jin Q, Rui L, Qi H, Yan W, Zhang J, You Y. PPARα, a predictor of patient survival in glioma, inhibits cell growth through the E2F1/miR-19a feedback loop. Oncotarget. 2016;7:84623–84633. doi: 10.18632/oncotarget.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, Cui S, Zhang R, Shi Y, Luo L. MiR-421 inhibits the malignant phenotype in glioma by directly targeting MEF2D. Am J Cancer Res. 2017;7:857–868. [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, Qian J, Liu N, You Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang K, Zhi T, Xu W, Xu X, Wu W, Yu T, Nie E, Zhou X, Bao Z, Jin X, Zhang J, Wang Y, Liu N. MicroRNA-1468-5p inhibits glioma cell proliferation and induces cell cycle arrest by targeting RRM1. Am J Cancer Res. 2017;7:784–800. [PMC free article] [PubMed] [Google Scholar]
- 5.Labak CM, Wang PY, Arora R, Guda MR, Asuthkar S, Tsung AJ, Velpula KK. Glucose transport: meeting the metabolic demands of cancer, and applications in glioblastoma treatment. Am J Cancer Res. 2016;6:1599–608. [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Zhang G, Shi Y, Qiu R, Khan AA. Neuropilin-1 (NRP-1) and magnetic nanoparticles, a potential combination for diagnosis and therapy of gliomas. Curr Pharma Des. 2015;21:5434–5449. doi: 10.2174/1381612821666150917092658. [DOI] [PubMed] [Google Scholar]
- 7.Jiang R, Zhang C, Liu G, Gu R, Wu H. MicroRNA-101 inhibits proliferation, migration and invasion in osteosarcoma cells by targeting ROCK1. Am J Cancer Res. 2017;7:88–97. [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Zhang H, Ning T, Wang X, Liu R, Yang H, Han Y, Deng T, Zhou L, Zhang L, Bai M, Wang X, Ge S, Ying G, Ba Y. MiR-520b/e regulates proliferation and migration by simultaneously targeting EGFR in gastric cancer. Cell Physiol Biochem. 2016;40:1303–15. doi: 10.1159/000453183. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang Q, Cai N, Wang H, Liu F, Ye L, Zhang X. Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP. Mol Cancer. 2014;13:128. doi: 10.1186/1476-4598-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu N, Zhang J, Cui W, Kong G, Zhang S, Yue L, Bai X, Zhang Z, Zhang W, Zhang X, Ye L. miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J Biol Chem. 2011;286:13714–13722. doi: 10.1074/jbc.M110.204131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stirzaker C, Song JZ, Ng W, Du Q, Armstrong NJ, Locke WJ, Statham AL, French H, Pidsley R, Valdesmora F, Zotenko E, Clark SJ. Methyl-CpGbinding protein MBD2 plays a key role in maintenance and spread of DNA methylation at CpG islands and shores in cancer. Oncogene. 2017;36:1328–1338. doi: 10.1038/onc.2016.297. [DOI] [PubMed] [Google Scholar]
- 12.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434–9. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- 15.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmc enriched within active genes and accessible chromatin in the nervous system. Cell. 2013;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barr H, Hermann A, Berger J, Tsai HH, Adie K, Prokhortchouk A, Hendrich B, Bird A. Mbd2 contributes to DNA methylation-directed repression of the Xist gene. Mol Cell Biol. 2007;27:3750–3757. doi: 10.1128/MCB.02204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Pang W, Zuo Z, Zhang W, He W. MicroRNA-520b suppresses proliferation, migration, and invasion of spinal osteosarcoma cells via downregulation of Frizzled-8. Oncol Res. 2017 doi: 10.3727/096504017X14873430389189. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Liu Q, Bai X, Li H, Zhang Y, Zhao Y, Zhang X, Ye L. The oncoprotein HBXIP upregulates Lin28B via activating TF II D to promote proliferation of breast cancer cells. Int J Cancer. 2013;133:1310–1322. doi: 10.1002/ijc.28154. [DOI] [PubMed] [Google Scholar]
- 19.Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2017;7:1–23. doi: 10.2217/epi.15.39. [DOI] [PubMed] [Google Scholar]
- 20.Desai MA, Webb HD, Sinanan LM, Scarsdale JN, Walavalkar NM, Ginder GD, Williams DC Jr. An intrinsically disordered region of methyl-CpG binding domain protein 2 (MBD2) recruits the histone deacetylase core of the NuRD complex. Nucleic Acids Res. 2015;43:3100–13. doi: 10.1093/nar/gkv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He S, Lai R, Chen D, Yan W, Zhang Z, Liu Z, Ding X, Chen Y. Downregulation of miR-221 inhibits cell migration and invasion through targeting methyl-CpG binding domain protein 2 in human oral squamous cell carcinoma cells. Biomed Res Int. 2015;2015:751672. doi: 10.1155/2015/751672. [DOI] [PMC free article] [PubMed] [Google Scholar]


