Abstract

Halogens are present in a significant number of drugs, contributing favorably to ligand–protein binding. Currently, the contribution of halogens, most notably chlorine and bromine, is largely attributed to halogen bonds involving favorable interactions with hydrogen bond acceptors. However, we show that halogens acting as hydrogen bond acceptors potentially make a more favorable contribution to ligand binding than halogen bonds based on quantum mechanical calculations. In addition, bioinformatics analysis of ligand–protein crystal structures shows the presence of significant numbers of such interactions. It is shown that interactions between halogens and hydrogen bond donors (HBDs) are dominated by perpendicular C–X···HBD orientations. Notably, the orientation dependence of the halogen–HBD (X–HBD) interactions is minimal over greater than 100° with favorable interaction energies ranging from −2 to −14 kcal/mol. This contrasts halogen bonds in that X–HBD interactions are substantially more favorable, being comparable to canonical hydrogen bonds, with a smaller orientation dependence, such that they make significant, favorable contributions to ligand–protein binding and, therefore, should be actively considered during rational ligand design.
Introduction
Halogens are widely known to contribute to ligand–protein interactions, thereby receiving significant attention in drug design. A survey of launched drugs showed that 25% are organohalogens of which organochlorines dominate, composing 57% of halogenated drugs.1 The role of halogens in drugs has focused on the halogen bond, a noncovalent interaction between halogenated ligands and proteins, and its contribution to increased selectivity and binding affinity.2 The physical basis for the halogen bond is generally considered to be due to the presence of a localized positive region on the halogen opposite the C–X covalent bond, termed a σ-hole (Figure S1), making the halogen able to interact favorably with an electronegative atom (e.g., hydrogen bond acceptor) in a highly orientation dependent fashion.2,3 Surveys of biomolecular structures have shed light on how frequently halogen bonds contribute to ligand binding, with the focus on interactions of carbon-bonded halogens (C–X) and the oxygen (O) of carbonyl, hydroxyl, carboxylate, or phosphate groups (C–X···O interactions).3−5
While the halogens are recognized as halogen-bond donors when the C–X···O interaction is approximately linear, halogens can also act as nucleophilic acceptors in directions perpendicular to the C–X bond,2,6,7 yielding a favorable halogen–electrophilic (e.g., hydrogen bond donor) interaction. Murray-West et al. in 1979 showed that the distribution of C–I···O angles was highly populated in the vicinity of 90°8 and these workers subsequently noted the “side-on” interactions of electrophiles with C–X moieties.9 This interaction is due to the electron density being more populated orthogonal to the covalent bond, yielding an electrostatic potential that is more negative in that region (Figure S1) allowing for favorable C–X···H interactions.7 Subsequently, Brammer et al. showed the presence of side-on interactions of halogens when acting as hydrogen-bond acceptors based on surveys of small molecule crystal structures in the Cambridge Structural database10 and on quantum mechanical electrostatic potentials, though no quantitative information was presented concerning the strength of these interactions.11 While such interactions have been noted in the literature, the potential importance of these types of interactions appears to have been underappreciated, as exemplified by Auffinger et al. when they stated “implying the existence of unusual Cl···H–N interactions”.3 More recently, a survey of the PDB and QM calculations by Zhu and co-workers detailed the presence of “side-on” C–X···H interactions, though they conclude, “The C–X···H contacts can be, therefore, considered as secondary interaction contributions to C–X···O halogen bonds that play important roles in conferring specificity and affinity for halogenated ligands.”12 In a subsequent perspective Zhu and co-workers emphasize the importance of C–F···H contributions to ligand–protein interactions while only mentioning, “Furthermore, the mean values of the C–X···H angles for X···H (X = Cl, Br, I) hydrogen bonds are about 100°, whereas the mean C–X···Y angles for halogen bonding interactions in biological system indeed amount to 160° (vide supra).”13 Similarly, another recent study reported that the introduction of Br and I onto aromatic side chains could lead to the formation of both a halogen bond and an C–X···H interaction; however, the role of the C–X···H interaction was largely ignored in the discussion.14 Clearly, the presence of perpendicular, side-on, or lateral interactions of halogens with hydrogen bond donors (HBDs) has been noted, but their contribution to ligand–protein affinity appears to be underappreciated. Such lack of appreciation may, in part, be due to the perception that nonlinear (i.e., side-on, lateral, or perpendicular) interactions may not be favorable enough to significantly contribute to ligand–protein binding combined with a lack of a quantitative evaluation of the strength of these interactions using high-level ab initio quantum mechanical calculations.
In the present work, motivated by results from quantum mechanical (QM) calculations as part of our ongoing empirical force field development efforts (Lin and MacKerell, work in progress),15 we undertook an analysis of the contribution of halogen–HBD (X–HBD) interactions to ligand–protein complexes. This effort involved QM calculations, a survey of X-ray crystallographic structures involving halogenated ligand–protein complexes and analyses of specific halogenated ligand–protein interactions reported in the literature. These analyses indicate that the perpendicular interactions between halogens and HN–, HO–, or HS– hydrogen bond donating groups in proteins make a significant contribution to ligand–protein binding along with halogen bonds, with the former possibly making more favorable contributions, such that they should be explicitly considered during ligand optimization.
Computational Method
Quantum Mechanical Calculations
Quantum mechanical calculations were performed with the programs Gaussian 0316 and Psi4.17 Geometry optimizations were performed at the MP2 level of theory with the aug-cc-pVDZ basis set18 for all compounds except the brominated species for which the aug-cc-pVDZ-PP basis set19,20 was used. Potential energy scans (PES) were performed with the monomers constrained to the gas phase conformation with only the specified intermolecular degree of freedom varied, with the exception of water which was constrained to the TIP3P intramolecular geometry.21 Single-point interaction energies for the PES were obtained at the RIMP2 level of theory with the cc-pVQZ basis set18 including corrections for the basis set superposition error (BSSE) using the counterpoise correction method.22
PDB Survey
The C–X (X is F, Cl, Br, or I) substructures were used to search for ligand–protein structures in the Protein Data Bank (PDB, January 2017 release)23 that contained halogen atoms. In each structure, the neighboring protein atoms within 4.5 Å of the halogen were identified. Protein atoms were categorized on the basis of their atom names. Hydroxyl moieties on residues Ser, Thr, and Tyr were selected on the basis of atom names, OG, OG1, OH, to evaluate the C–X···O angles. NH moieties in the amide, amino, or guanidinium containing residues Asn, Gln, Lys, Arg, His, and Trp were selected on the basis of the atom names ND2, NE2, NZ, NE, NH1, NH2, ND1, and NE1, respectively, to evaluate the C–X···N angle. SH moieties in Cys residues were selected on the basis of the atom name SG to evaluate the C–X···S angle, where the sulfur atoms involved in disulfide bonds were excluded. To identify C=O and COO– moieties interacting with X atoms via halogen bonds with Asn, Gln, Glu, Asp or the peptide bond selection was based on the atom names OD1, OD2, OE1, OE2, and O1. To ensure that interactions involving noncanonical amino acids that contain atoms with the same names were not included in the survey, the initial search results were verified on the basis of the amino acid names.
Results and Discussion
To systematically investigate potential interactions between protein residues acting as HBDs and halogens, we performed QM analyses of the interactions of model compounds representative of side chains of proteins with monohalogenated analogues of benzene and ethane for F, Cl, and Br. The model compounds were selected to cover the different types of functional groups in protein side chains that act as HBDs, including methanol (MEOH), phenol (PHEN), acetamide (ACEM), imidazole (IMID), indole (INDO), methanthiol (MESH), methylammonium (MAMM), and methylguanidinum (MGUAN), or acetate (ACET) that acts as a HBA. Interaction energies were calculated as a function of the X···HBD distance in the perpendicular, HBD90°, and linear, HBD180°, orientations (Figure 1 and Table 1) and, in the case of the chlorinated species, as a function of C–Cl···HBD angle (Figure 2). Unlike in protein structures where secondary interactions or geometric constraints may impact the halogen–protein interactions, the QM calculations allow for investigation of the interaction energy contribution from the HBD in the model compound itself.
Figure 1.
Quantum mechanical (QM) interaction energies as a function of distance between chlorobenzene and water or model compounds serving as hydrogen bond donors (HBDs) in perpendicular HBD90° and linear HBD180° orientations. Distances are based on the halogen to hydrogen bond donor antecedent (O, N, or S atom). Carbons are colored cyan, hydrogens white, oxygens red, sulfurs yellow, and chlorines green.
Table 1. Minimum Interaction Distances (d(X···HBD), Å) and Energies (Emin,int, kcal/mol) for the Monohalogenated Analogues of Benzene and Ethane with Selected Model Compounds Serving as HBDs or HBAs in Both Perpendicular, C–X···Y = 90° (HBD90°/HBA90°) and Linear C–X···Y = 180° (HBD180°/HBA180°) Orientations, Where X Is F, Cl, or Br and Y Is the Oxygen or Nitrogen of the Model Compounda.
| HBD |
HBA |
|||||
|---|---|---|---|---|---|---|
| model mol. | C–X···HBD angle | d(X···HBD) | Emin,int | d(X···HBA) | Emin,int | Diff. Emin,int |
| FLUBb | ||||||
| MEOH | 90° | 3.17 | –2.05 | |||
| 180° | 3.07 | –1.94 | ||||
| ACEM | 90° | 3.21 | –1.83 | |||
| 180° | 3.21 | –1.66 | ||||
| MAMM | 90° | 2.83 | –12.88 | |||
| 180° | 2.73 | –12.72 | ||||
| ACET | 180° | 5.00c | 3.11 | |||
| FETHb | ||||||
| MEOH | 90° | 2.97 | –3.05 | |||
| 180° | 2.97 | –2.78 | ||||
| ACEM | 90° | 3.21 | –0.95 | |||
| 180° | 3.11 | –2.36 | ||||
| MAMM | 90° | 2.83 | –10.56 | |||
| 180° | 2.73 | –16.01 | ||||
| ACET | 180° | 5.00c | 1.81 | |||
| CHLBb | ||||||
| MEOH | 90° | 3.47 | –2.39 | 3.70 | 0.05 | |
| 180° | 3.77 | –0.60 | 3.10 | –0.83 | –1.56 | |
| ACEM | 90° | 3.51 | –2.23 | 3.60 | 0.22 | |
| 180° | 3.91 | –0.41 | 3.10 | –0.73 | –1.50 | |
| MAMM | 90° | 3.13 | –14.31 | |||
| 180° | 3.23 | –8.65 | ||||
| ACET | 180° | 2.70 | –2.46 | |||
| CLETb | ||||||
| MEOH | 90° | 3.37 | –3.07 | 5.00c | 0.19 | |
| 180° | 3.67 | –1.04 | 3.20 | –0.13 | –2.94 | |
| ACEM | 90° | 3.61 | –1.42 | 5.00c | 0.26 | |
| 180° | 3.81 | –0.71 | 3.30 | –1.38 | –0.04 | |
| MAMM | 90° | 3.13 | –12.23 | |||
| 180° | 3.23 | –10.92 | ||||
| ACET | 180° | 2.90 | 1.28 | |||
| BROBb | ||||||
| MEOH | 90° | 3.57 | –2.34 | 3.80 | 0.09 | |
| 180° | 4.07 | –0.37 | 3.10 | –1.55 | –0.79 | |
| ACEM | 90° | 3.71 | –2.22 | 3.70 | 0.22 | |
| 180° | 4.21 | –0.23 | 3.10 | –1.52 | –0.70 | |
| MAMM | 90° | 3.33 | –14.39 | |||
| 180° | 3.43 | –7.68 | ||||
| ACET | 180° | 2.70 | –5.41 | |||
| BRETb | ||||||
| MEOH | 90° | 3.57 | –2.93 | 5.00c | 0.21 | |
| 180° | 3.87 | –0.75 | 3.20 | –0.81 | –2.12 | |
| ACEM | 90° | 3.71 | –1.48 | 5.00c | 0.27 | |
| 180° | 4.01 | –0.47 | 3.20 | –0.57 | –0.91 | |
| MAMM | 90° | 3.33 | –12.56 | |||
| 180° | 3.33 | –9.79 | ||||
| ACET | 180° | 2.80 | –1.36 | |||
The distance is measured between the halogen and the oxygen or nitrogen atom of the model compounds. Diff. Emin,int is the energy differences of Emin,int (HBD90°–HBA180°) indicating the relative strength of perpendicular halogen–hydrogen bond donor (X–HBD) versus halogen bonds.
Fluorobenzene (FLUB), fluoroethane (FETH), chlorobenzene (CHLB), chloroethane (CLET), bromobenzene (BROB), and bromoethane (BRET).
Repulsive interaction with no minimum <5 Å. Energies correspond to 5 Å.
Figure 2.
Quantum mechanical (QM) interaction energies as a function of angle between chlorobenzene and water or model compounds representative of hydrogen bond donors (HBDs) in proteins as a function of the C–Cl···HBD angle at Cl···HBD non-hydrogen-atom distances of 3.5 and 4.0 Å. Distances and angles are based on the hydrogen bond donor antecedents (O, N, or S atom). Carbons are colored cyan, hydrogens white, oxygens red, sulfurs yellow, and chlorines green.
Analysis of the interaction energy surfaces as a function of distance shows the perpendicular HBD90° interaction energies to be systematically more favorable than the HBD180° interactions, with minima in the range from 3.2 to 4.0 Å (Figure 1). This trend is maintained for the F and Br species (Table 1). Interaction energy surfaces for the Cl species were calculated for the C–Cl···HBD angle, based on the HBD non-hydrogen atom, at Cl···HBD distances of 3.5 and 4.0 Å (Figure 2). Results show the interaction energies to be most favorable in the vicinity of the perpendicular, HBD90° orientation. These interactions are 2–3 kcal/mol more favorable than the linear HBD180° orientation with the neutral model compounds and by up to 6 kcal/mol with the positively charged compounds, with similar differences for Br for a subset of the model compounds (Table 1). The magnitudes of the interactions themselves range from −2 to −4 kcal/mol with the neutral species and −10 and −14 kcal/mol for the model compounds representing the positively charged side chains of Arg (MGUAN) and Lys (MAMM). While the interactions are most favorable in the perpendicular orientation, the favorable interactions range from ∼40 to 180° (Figure 2) with the energy remaining favorable in most cases for the linear interaction, indicating the angular dependence to be minimal. The results for fluorinated species in Table 1 show the interaction energies of the perpendicular and linear orientations to be similar for fluorobenzene consistent with the lack of a σ-hole such that fluorine still acts as a hydrogen bond acceptor in the linear orientation.4
To put the X–HBD interactions in context with halogen bonds, QM minimum interaction distances and energies were obtained with the Cl and Br benzene and ethane analogues for selected model compounds acting as hydrogen bond acceptors (HBA) (Table 1). Consistent with previous studies,2−4,12,13 interactions of Cl and Br with the acceptor atoms in the linear HBA180° orientation are favorable. However, when comparing the halogen bond with the X–HBD interactions (HBD90°), the latter interactions are equivalent or more favorable by up to −3 kcal/mol for the neutral model compounds. Analysis of the halogen bond interactions was extended to include a negatively charged acceptor using acetate (ACET) as a model compound (Table 1). The halogen bond with the negatively charged ACET has a more favorable halogen bond interaction than the neutral halogen bond interactions. However, the interaction is significantly less favorable than the interactions of charged hydrogen bond donors with Cl and Br in X–HBD90° interactions. For example, the most favorable halogen bond interaction involving ACET is with bromobenzene with an energy of −5.4 kcal/mol at 2.7 Å, which is significantly less favorable than the X–HBD90° interaction of −14.4 kcal/mol with MAMM (Table 1). Notably, the perpendicular X–HBD interactions are more favorable with the Cl and Br aliphatic species than any of the interactions involving either HBA90° or HBA180° interactions with those molecules. The size of those interactions ranges from approximately −1.5 to −3.1 kcal/mol, with the HBA180° halogen bonds ranging from −0.1 to −1.6 kcal/mol. Overall, these results indicate that X–HBD interactions have the potential to make significant contributions to ligand–protein interactions, with those favorable interactions involving halogens in both aromatic and aliphatic systems, versus halogen bonds being primarily limited to halogenated aromatic species.
To verify the potential importance of X–HBD interactions to ligand–protein interactions, we performed a survey of protein crystallographic structures in the Protein Data Bank (PDB)23 that contain halogenated ligands. The survey determined the distribution of C–X···HBD angles, where X is the halogen and the HBDs are the oxygen, nitrogen, or sulfur atoms of protein side chain hydrogen bond donors. Results from the survey for all crystal structures with a resolution better than 3.0 Å are shown in Figure 3 for X = Cl and in Figures S2, S3, and S4 for the X = F, Br, and I, respectively. The probability distributions of the C–X···HBD angles were binned into 10° windows at search radii of less than 3.5, 3.5–4.0, and 4.0–4.5 Å based on the X to HBD O, N, or S atom distances. Evident in the survey data is the maxima in the plots in the range from 80 to 110°. This is consistent with the QM interaction energy surfaces showing energies to be more favorable in the vicinity of 90° as compared to linear interactions in the vicinity of 180° (Figure 2). The second maximum in the survey data occurs at 140–150° with the exception of X = F. This is primarily due to the oxygen or sulfur associated HBDs also being able to act as HBAs, thereby interacting with X via a halogen bond. Separation of the HBD survey data into contributions from N, O, and S HBDs (panels b, c, and d in Figure 3 and Figures S3 and S4, respectively) shows the 140° maxima to be prominent with oxygens and sulfurs for X = Cl, Br, and I, while only a small increase is observed with nitrogens. The small peak in the nitrogen distribution is due to the nitrogen HBA in the neutral His side chain. Further support for the peak in the vicinity of 140° being a halogen bond was the omission of the peak with X = F (Figure S2) and from a survey of C–X···O angles for protein carbonyl groups (Figure S5). For Cl, Br, and I, there are clear maxima present in the regions of 130–180° especially for X···O distances less than 3.5 Å (Figure S5).
Figure 3.

Probability distribution of C–Cl···HBD angles from a survey of ligand–protein complexes in the Protein Data Bank. HBD is defined by the oxygen, nitrogen, or sulfur atom of the protein hydrogen bond donor functional group. Part a shows the combined normalized distribution of C–Cl···O/N/S. Parts b, c, and d are the respective normalized probability distributions for C–Cl···N, C–Cl···O, and C–Cl···S. Search radii for the survey were <3.5, 3.5–4.0, and 4.0–4.5 Å for the Cl to HBD distance. The numbers of interactions identified in the survey are shown in Table S1 and comparisons of the number of possible X-HBD interactions versus halogen bonds are presented in Table S2 and S3 along with discussion of the analyses.
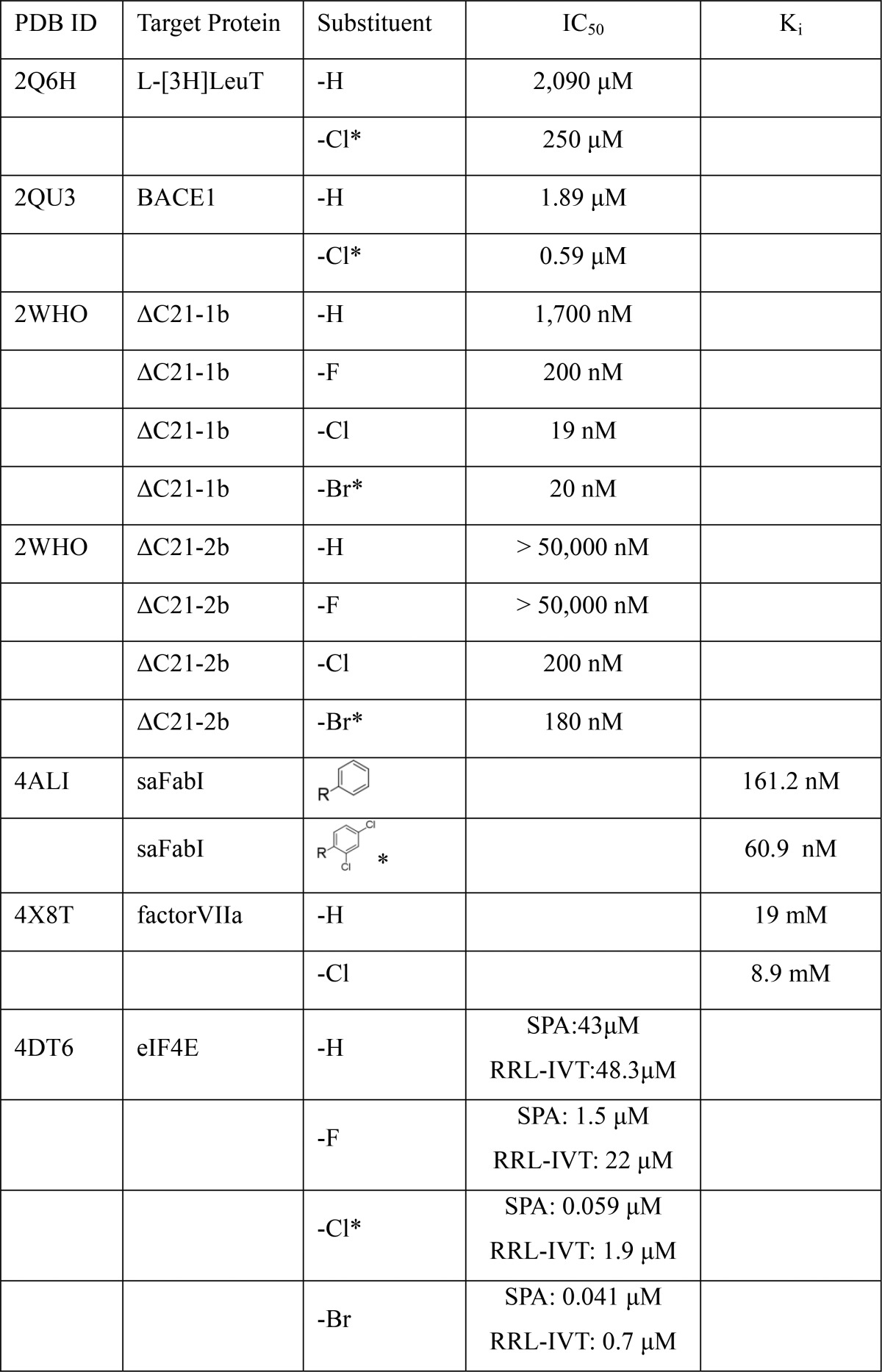
To further verify the role of X–HBD interactions in ligand–protein binding, selected crystal structures from the survey were identified for which it has been experimentally shown that adding a Cl or Br atom to the ligand improves the binding affinity. Images from six example structures are shown in Figure 4.24−29 In all cases, the X···HBD distances are 3.5 Å or less, including an interaction between Br and Arg of 3.3 Å in Figure 4c. The C–X···HBD angles range from 81 to 96°. Published biochemical data for the ligands shown in Figure 4 and analogues are presented in Table 2. An additional five examples of X–HBD interactions for which both structural and biochemical data is available are included in Figure S6 and Table S4. The contributions of the X–HBD interactions are evident, with the changes in affinity ranging from a factor of 2 to over 250-fold with PDB ID 2WHO. The over 250-fold change involves the interaction of an Arg side chain with a bromophenyl moiety, indicating that a significant contribution from the charged guanidinium moiety with the Br atom is occurring. In PDB ID 4DT6, replacement of hydrogen with a F leads to a 2–40-fold increase in binding with Cl and Br substitutions yielding up to 100-fold improvements in affinity. While these results represent a small sample of possible X–HBD interactions, in combination with the survey data, they indicate that these types of interactions are common in ligand–protein complexes and further support the significant role X–HBD interactions contribute to ligand–protein binding affinities.
Figure 4.
Examples of crystal structures that are observed to have X–HBD interactions including (PDBIDs) (a) 2Q6H,24 (b) 2QU3,25 (c) 2WHO,26 (d) 4ALI,27 (e) 4X8T,28 and (f) 4DT6.29 Carbons are colored in cyan, hydrogens white, oxygens red, sulfurs yellow, chlorines green, and bromine is colored in dark red in panel c.
Table 2. Impact of Halogen Substitution on Affinities in Selected Ligand–Protein Complexes Participating in X–HBD Interactionsa.
Conclusion
In the present study, we have investigated the contribution of interactions between halogens and protein side-chain hydrogen-bond donors, termed X–HBD interactions, to ligand–protein binding. While a few studies have reported that the inclusion of a halogen, particularly Cl, improved ligand affinity due to an interaction with a hydrogen bond donor group in the protein,24,30−33 the phenomenon has not previously been systematically studied. QM data based on model compounds representative of hydrogen bond donors in proteins shows the X–HBD interactions, which are most favorable in the perpendicular orientation, to have favorable energies from −2 kcal/mol with thiol groups to more than −14 kcal/mol with the positively charged ammonium groups on the side chain of Lys. The interaction energy is not highly sensitive to orientation, as defined by the C–X···HBD angle. In all cases, there is a minimum close to the perpendicular orientation of approximately 90°; however, the change in energy is relatively small as a function of angle with favorable interactions still occurring at 180°. These properties are in contrast to halogen bonds, associated with the σ-hole and favorable interactions with hydrogen-bond acceptors, where the interaction is linear in nature (i.e., C–X···HBA angle is ∼180°), the interaction energies based on neutral model compounds are in the range of −2 kcal/mol or less, of up to −5.4 kcal/mol with negatively charged acetate, and the interaction energies rapidly become less favorable as the C–X···HBA angle deviates below 150°.13,15 In addition, the predominance of X–HBD interactions in the perpendicular orientation contrasts the orientation dependence of canonical hydrogen bonds,34 such that X–HBD interactions, where X is Cl, Br, or I, would not be considered to be a standard or canonical hydrogen bond.
With respect to the relative strength of the perpendicular X–HBD interactions (C–X···HBD90°) relative to canonical hydrogen bonds, the overall interaction energies are similar. For example, published QM hydrogen bond interactions of small molecules at the MP2/6-311++G** model chemistry show hydrogen bonds between a hydroxyl group and a carbonyl oxygen or a sp2 nitrogen to have interaction energies of −3.7 and −4.6 kcal/mol, respectively,35 which are somewhat more favorable than the X–HBD interactions involving MEOH (Table 1, −2.4 kcal/mol with chlorobenzene). However, the C–X···HBD90° interaction of the hydroxyl of phenylalanine with chlorobenzene is −3.4 kcal/mol (Figure 1) comparable with or even more favorable than hydrogen bonds of the hydroxyl with a carbonyl oxygen (−3.7 kcal/mol), an ether oxygen (−2.9 kcal/mol), or a hydroxyl group oxygen (−2.7 kcal/mol).35 For neutral NH groups serving as HBD in C–X···HBD90° interactions involving imidazole (IMID) or indole (INDO), the interaction energies are −3.7 and −4.1 kcal/mol, respectively, which are comparable to or more favorable than the hydrogen bond between a sp2 nitrogen (serving as a HBD) and a carbonyl oxygen (−3.0 kcal/mol), , an ether oxygen (−4.1 kcal/mol), or a hydroxyl group oxygen (−4.0 kcal/mol).35 Thus, the presented X–HBD interactions are comparable to canonical hydrogen bonds with respect to interaction energies and offer the additional advantage of a limited angular dependency (Figure 2).
The survey of the PDB23 performed in this study revealed the X–HBD interactions to be quite common with the C–X···HBD angular distribution consistent with the interaction energies as a function of orientation observed in the model compound QM calculations. In addition, for selected ligands for which crystal structures and affinity data are available, it is shown how the presence of a specific X–HBD interaction can improve the binding affinity by up to 250-fold.
In practical terms, given the wide presence of halogens in drugs, awareness of the importance of halogens to ligand–protein affinity is not new. However, in order to more rationally exploit the use of halogens in rational ligand design, it is necessary to quantify the detailed nature of those interactions including their energetic contributions as well as their orientation dependence. The relatively “flat” orientation dependence of the X–HBDs is particularly noteworthy, indicating that the insertion of halogens into ligands can produce favorable interactions with HBDs without the well-known orientation dependence associated with halogen bonds and, importantly, standard hydrogen bonds in general.36 Indeed, the wide presence of halogens in drugs is likely, in large part, due to the presence of the X–HBD interactions quantified in the present study.
As a final point with respect to rationally exploiting X–HBD interactions in ligand design is their ability to be accurately treated by molecular mechanics force fields. Traditionally, halogens are treated as having a small partial negative atomic charge with relatively large favorable dispersion contributions.37 This treatment assures that the halogen will have favorable interactions with HBDs and those interactions will be relatively insensitive to orientation, as shown in the present study. More recent force field models of halogens have included a lone pair carrying a positive charge to mimic the σ-hole,15,38−40 thereby modeling the halogen bond. Importantly, in these models, the halogen atom still carries a negative charge, allowing it to continue to interact favorably with HBDs. However, it is the magnitude of these interactions that are likely incorrectly treated, largely due to a lack of QM data on the orientation dependence of X–HBD for use as target data for force field optimization. The results reported in the present study, as well as a more extensive data set to be presented in a forthcoming publication (Lin and MacKerell, work in progress), will allow that limitation to be overcome and lead to the improved exploitation of halogens both qualitatively by the ligand designer being cognitive of their existence as well as quantitatively by their more accurate treatment in empirical force fields.
Acknowledgments
This work was supported by National Institutes of Health grants GM070855 and GM072558. The University of Maryland Computer-Aided Drug Design Center and XSEDE are acknowledged for their generous allocations of computer time.
Glossary
Abbreviations
- methanol
(MEOH)
- phenol
(PHEN)
- acetamide
(ACEM)
- imidazole
(IMID)
- indole
(INDO)
- methanthiol
(MESH)
- methylammonium
(MAMM)
- methylguanidinum
(MGUAN)
- acetate
(ACET)
- fluorobenzene
(FLUB)
- fluoroethane
(FETH)
- chlorobenzene
(CHLB)
- chloroethane
(CLET)
- bromobenzene
(BROB)
- bromoethane
(BRET)
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.7b04198.
Figures showing molecular electrostatic potentials of halogen molecules and additional PDB survey results (PDF)
Author Contributions
A.D.M. conceived the study, F.-Y.L. performed the calculations, and F.-Y.L. and A.D.M. analyzed the data and wrote the manuscript.
The authors declare the following competing financial interest(s): A.D.M. is co-founder and CSO of SilcsBio LLC.
Supplementary Material
References
- Xu Z.; Yang Z.; Liu Y.; Lu Y.; Chen K.; Zhu W. Halogen Bond: Its Role beyond Drug–Target Binding Affinity for Drug Discovery and Development. J. Chem. Inf. Model. 2014, 54 (1), 69–78. 10.1021/ci400539q. [DOI] [PubMed] [Google Scholar]
- Cavallo G.; Metrangolo P.; Milani R.; Pilati T.; Priimagi A.; Resnati G.; Terraneo G. The Halogen Bond. Chem. Rev. 2016, 116 (4), 2478–2601. 10.1021/acs.chemrev.5b00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P.; Hays F. A.; Westhof E.; Ho P. S. Halogen Bonds in Biological Molecules. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (48), 16789–16794. 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholfield M. R.; Zanden C. M. V.; Carter M.; Ho P. S. Halogen Bonding (X-Bonding): A Biological Perspective. Protein Sci. Publ. Protein Soc. 2013, 22 (2), 139–152. 10.1002/pro.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisini E.; Metrangolo P.; Pilati T.; Resnati G.; Terraneo G. Halogen Bonding in Halocarbon–protein Complexes: A Structural Survey. Chem. Soc. Rev. 2011, 40 (5), 2267–2278. 10.1039/c0cs00177e. [DOI] [PubMed] [Google Scholar]
- Zhou P.-P.; Qiu W.-Y.; Liu S.; Jin N.-Z. Halogen as Halogen-Bonding Donor and Hydrogen-Bonding Acceptor Simultaneously in Ring-Shaped H3N·X(Y)·HF (X = Cl, Br and Y = F, Cl, Br) Complexes. Phys. Chem. Chem. Phys. 2011, 13 (16), 7408–7418. 10.1039/c1cp00025j. [DOI] [PubMed] [Google Scholar]
- Politzer P.; Murray J. S.; Clark T. Halogen Bonding and Other σ-Hole Interactions: A Perspective. Phys. Chem. Chem. Phys. 2013, 15 (27), 11178–11189. 10.1039/c3cp00054k. [DOI] [PubMed] [Google Scholar]
- Murray-Rust P.; Motherwell W. D. S. Computer Retrieval and Analysis of Molecular Geometry. 4. Intermolecular Interactions. J. Am. Chem. Soc. 1979, 101 (15), 4374–4376. 10.1021/ja00509a056. [DOI] [Google Scholar]
- Ramasubbu N.; Parthasarathy R.; Murray-Rust P. Angular Preferences of Intermolecular Forces around Halogen Centers: Preferred Directions of Approach of Electrophiles and Nucleophiles around Carbon-Halogen Bond. J. Am. Chem. Soc. 1986, 108 (15), 4308–4314. 10.1021/ja00275a012. [DOI] [Google Scholar]
- Groom C. R.; Bruno I. J.; Lightfoot M. P.; Ward S. C. The Cambridge Structural Database. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016, 72 (2), 171–179. 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brammer L.; Bruton E. A.; Sherwood P. Understanding the Behavior of Halogens as Hydrogen Bond Acceptors. Cryst. Growth Des. 2001, 1 (4), 277–290. 10.1021/cg015522k. [DOI] [Google Scholar]
- Lu Y.; Wang Y.; Xu Z.; Yan X.; Luo X.; Jiang H.; Zhu W. C–X···H Contacts in Biomolecular Systems: How They Contribute to Protein–Ligand Binding Affinity. J. Phys. Chem. B 2009, 113 (37), 12615–12621. 10.1021/jp906352e. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Wang Y.; Zhu W. Nonbonding Interactions of Organic Halogens in Biological Systems: Implications for Drug Discovery and Biomolecular Design. Phys. Chem. Chem. Phys. 2010, 12 (18), 4543–4551. 10.1039/b926326h. [DOI] [PubMed] [Google Scholar]
- Scholfield M. R.; Ford M. C.; Carlsson A.-C. C.; Butta H.; Mehl R. A.; Ho P. S. Structure–Energy Relationships of Halogen Bonds in Proteins. Biochemistry 2017, 56 (22), 2794–2802. 10.1021/acs.biochem.7b00022. [DOI] [PubMed] [Google Scholar]
- Soteras Gutiérrez I.; Lin F.-Y.; Vanommeslaeghe K.; Lemkul J. A.; Armacost K. A.; Brooks C. L. III; MacKerell A. D. Jr. Parametrization of Halogen Bonds in the CHARMM General Force Field: Improved Treatment of Ligand–protein Interactions. Bioorg. Med. Chem. 2016, 24 (20), 4812–4825. 10.1016/j.bmc.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Montgomery J. A. Jr.; Vreven T.; Kudin K. N.; Burant J. C.; et al. Gaussian 03, revision C.02; Gaussian, Inc.: Wallingford, CT, 2004.
- Turney J. M.; Simmonett A. C.; Parrish R. M.; Hohenstein E. G.; Evangelista F. A.; Fermann J. T.; Mintz B. J.; Burns L. A.; Wilke J. J.; Abrams M. L.; et al. Psi4: An Open-Source Ab Initio Electronic Structure Program. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2 (4), 556–565. 10.1002/wcms.93. [DOI] [Google Scholar]
- Woon D. E.; Dunning T. H. Jr. Gaussian Basis Sets for Use in Correlated Molecular Calculations. III. The Atoms Aluminum through Argon. J. Chem. Phys. 1993, 98 (2), 1358–1371. 10.1063/1.464303. [DOI] [Google Scholar]
- Peterson K. A.; Figgen D.; Goll E.; Stoll H.; Dolg M. Systematically Convergent Basis Sets with Relativistic Pseudopotentials. II. Small-Core Pseudopotentials and Correlation Consistent Basis Sets for the Post-d Group 16–18 Elements. J. Chem. Phys. 2003, 119 (21), 11113–11123. 10.1063/1.1622924. [DOI] [Google Scholar]
- Peterson K. A.; Shepler B. C.; Figgen D.; Stoll H. On the Spectroscopic and Thermochemical Properties of ClO, BrO, IO, and Their Anions. J. Phys. Chem. A 2006, 110 (51), 13877–13883. 10.1021/jp065887l. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79 (2), 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Boys S. F.; Bernardi F. The Calculation of Small Molecular Interactions by the Differences of Separate Total Energies. Some Procedures with Reduced Errors. Mol. Phys. 1970, 19 (4), 553–566. 10.1080/00268977000101561. [DOI] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000, 28 (1), 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K.; Yamashita A.; Gouaux E. Antidepressant Binding Site in a Bacterial Homologue of Neurotransmitter Transporters. Nature 2007, 448 (7156), 952–956. 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- Fobare W. F.; Solvibile W. R.; Robichaud A. J.; Malamas M. S.; Manas E.; Turner J.; Hu Y.; Wagner E.; Chopra R.; Cowling R.; et al. Thiophene Substituted Acylguanidines as BACE1 Inhibitors. Bioorg. Med. Chem. Lett. 2007, 17 (19), 5353–5356. 10.1016/j.bmcl.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Ontoria J. M.; Rydberg E. H.; Di Marco S.; Tomei L.; Attenni B.; Malancona S.; Martin Hernando J. I.; Gennari N.; Koch U.; Narjes F.; et al. Identification and Biological Evaluation of a Series of 1H-Benzo[de]Isoquinoline-1,3(2H)-Diones as Hepatitis C Virus NS5B Polymerase Inhibitors. J. Med. Chem. 2009, 52 (16), 5217–5227. 10.1021/jm900517t. [DOI] [PubMed] [Google Scholar]
- Schiebel J.; Chang A.; Lu H.; Baxter M. V.; Tonge P. J.; Kisker C. Staphylococcus Aureus FabI: Inhibition, Substrate Recognition, and Potential Implications for in Vivo Essentiality. Structure 2012, 20 (5), 802–813. 10.1016/j.str.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney D. L.; Bozarth J. M.; Metzler W. J.; Morin P. E.; Mueller L.; Newitt J. A.; Nirschl A. H.; Rendina A. R.; Tamura J. K.; Wei A.; et al. Discovery of Novel P1 Groups for Coagulation Factor VIIa Inhibition Using Fragment-Based Screening. J. Med. Chem. 2015, 58 (6), 2799–2808. 10.1021/jm501982k. [DOI] [PubMed] [Google Scholar]
- Chen X.; Kopecky D. J.; Mihalic J.; Jeffries S.; Min X.; Heath J.; Deignan J.; Lai S.; Fu Z.; Guimaraes C.; et al. Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the EIF4E MRNA–Cap Interaction. J. Med. Chem. 2012, 55 (8), 3837–3851. 10.1021/jm300037x. [DOI] [PubMed] [Google Scholar]
- Tomar D.; Khan T.; Singh R. R.; Mishra S.; Gupta S.; Surolia A.; Salunke D. M. Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites. PLoS One 2012, 7 (9), e43522. 10.1371/journal.pone.0043522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K. H. G.; Seljée F.; Rozeboom H. J.; Kalk K. H.; Dijkstra B. W. Crystallographic Analysis of the Catalytic Mechanism of Haloalkane Dehalogenase. Nature 1993, 363 (6431), 693–698. 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- Tipparaju S. K.; Mulhearn D. C.; Klein G. M.; Chen Y.; Tapadar S.; Bishop M. H.; Yang S.; Chen J.; Ghassemi M.; Santarsiero B. D.; et al. Design and Synthesis of Aryl Ether Inhibitors of the Bacillus Anthracis Enoyl-ACP Reductase. ChemMedChem 2008, 3 (8), 1250–1268. 10.1002/cmdc.200800047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone V.; Chung R.; Endo S.; Hara A.; El-Kabbani O. Structure of Aldehyde Reductase in Ternary Complex with Coenzyme and the Potent 20α-Hydroxysteroid Dehydrogenase Inhibitor 3,5-Dichlorosalicylic Acid: Implications for Inhibitor Binding and Selectivity. Arch. Biochem. Biophys. 2008, 479 (1), 82–87. 10.1016/j.abb.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Panigrahi S. K.; Desiraju G. R. Strong and Weak Hydrogen Bonds in the Protein–ligand Interface. Proteins: Struct., Funct., Genet. 2007, 67 (1), 128–141. 10.1002/prot.21253. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Wang G.; Li Z.; Wang R. Geometrical Preferences of the Hydrogen Bonds on Protein–Ligand Binding Interface Derived from Statistical Surveys and Quantum Mechanics Calculations. J. Chem. Theory Comput. 2008, 4 (11), 1959–1973. 10.1021/ct800267x. [DOI] [PubMed] [Google Scholar]
- Morozov A. V.; Kortemme T.; Tsemekhman K.; Baker D. Close Agreement between the Orientation Dependence of Hydrogen Bonds Observed in Protein Structures and Quantum Mechanical Calculations. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (18), 6946–6951. 10.1073/pnas.0307578101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Hatcher E.; Acharya C.; Kundu S.; Zhong S.; Shim J.; Darian E.; Guvench O.; Lopes P.; Vorobyov I.; et al. CHARMM General Force Field (CGenFF): A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31 (4), 671–690. 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M. A. A. Molecular Mechanical Study of Halogen Bonding in Drug Discovery. J. Comput. Chem. 2011, 32 (12), 2564–2574. 10.1002/jcc.21836. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Schyman P. Treatment of Halogen Bonding in the OPLS-AA Force Field: Application to Potent Anti-HIV Agents. J. Chem. Theory Comput. 2012, 8 (10), 3895–3901. 10.1021/ct300180w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X.; Wang Q.; Wang L.-P.; Fried S. D.; Piquemal J.-P.; Dalby K. N.; Ren P. Modeling Organochlorine Compounds and the σ-Hole Effect Using a Polarizable Multipole Force Field. J. Phys. Chem. B 2014, 118 (24), 6456–6465. 10.1021/jp411671a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.