Abstract
The role of altered redox status and high reactive oxygen species (ROS) is still controversial in cancer development and progression. Intracellular levels of ROS are elevated in cancer cells suggesting a role in cancer initiation and progression; on the contrary, ROS elevated levels may induce programmed cell death and have been associated with cancer suppression. Thus, it is crucial to consider the double-face of ROS, for novel therapeutic strategies targeting redox regulatory mechanisms. In this review, in order to derive cancer-type specific oxidative stress genes' profile and their potential prognostic role, we integrated a publicly available oxidative stress gene signature with patient survival data from the Cancer Genome Atlas database. Overall, we found several genes statistically significant associated with poor prognosis in the examined six tumor types. Among them, FoxM1 and thioredoxin reductase1 expression showed the same pattern in four out of six cancers, suggesting their specific critical role in cancer-related oxidative stress adaptation. Our analysis also unveiled an enriched cellular network, highlighting specific pathways, in which many genes are strictly correlated. Finally, we discussed novel findings on the correlation between oxidative stress and cancer stem cells in order to define those pathways to be prioritized in drug development.
1. Introduction
Reactive oxygen species (ROS) are commonly identified as oxygen reactive molecules associated with a wide variety of physiologic events [1] as well as cancer, diabetes, obesity, neurodegeneration, and other age-related diseases [2, 3]. A reduction-oxidation (redox) reaction concerns the transfer of electrons (reducing power) from a more reduced (nucleophilic) to more oxidized (electrophilic) molecules. ROS can be classified in two groups: (1) free radical ROS containing one or more unpaired electron(s) in their outer molecular orbitals (i.e., superoxide radicals and hydroxyl radicals); (2) nonradical ROS which are chemically reactive and can be converted to radical ROS (i.e., hydrogen peroxide), although they do not have unpaired electron(s). In both cases, ROS can be produced by either enzymatic reactions (i.e., NADPH oxidase, metabolic enzymes such as the cytochrome P450 enzymes, lipoxygenase, and cyclooxygenase) or by nonenzymatic reactions, such as during the mitochondrial respiratory chain. These considerations highlight the concept that the source of ROS is extremely heterogeneous. Indeed, ROS can be found in the environment, as pollutants, tobacco smoke, and iron salts, or generated inside the cells through multiple mechanisms [4]. Within cells, mitochondria, cytosol, single membrane-bound organelles (peroxisomes, endosomes, and phagosomes), or exosomes shed from plasma membranes, as well as extracellular fluids, including plasma, are all involved in ROS generation [3, 5, 6]. Mitochondria are the main ROS producers, principally because they are the site of the respiratory chain when electron leakage can react with molecular oxygen, resulting in the formation of superoxide, which can subsequently be converted to other ROS molecules. Then, generated ROS either can be detoxified or can leave the organelle through channels such as voltage-dependent anion channels (VDAC) or aquaporin, or by small vesicles such as exosomes [3, 5, 7]. However, ROS can also be the product of β-oxidation in peroxisomes, of prostaglandin synthesis and detoxification reactions by cytochrome P450, or of NADPH-mediated reaction in phagocytes [4, 5].
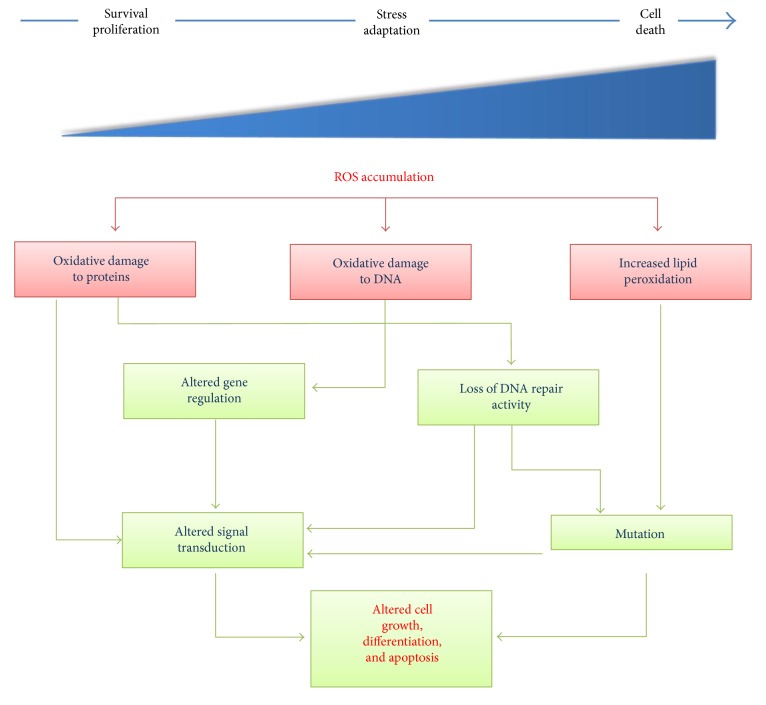
ROS are biologically important in a variety of physiological systems, including adaptation to hypoxia, regulation of autophagy, immunity, differentiation, and longevity. They regulate many signal transduction pathways by directly reacting with proteins and by modulating transcription factors and gene expression [1]. At low levels, ROS promote cellular proliferation, differentiation, and migration as well as cellular stress-responsive survival pathways such as nuclear factor-κB (NF-κB), thus inducing proinflammatory cytokines [4, 8]. Because of ROS' highly reactive potential toward biological molecules, excessive ROS levels can damage cellular components such as DNA, proteins, and lipids. To counteract these effects, cells activate “ROS adaption” mechanisms, involving several antioxidant ROS scavengers, as glutathione peroxidase (GPx), thioredoxin (Trx), catalase (CAT), superoxide-dismutase (SOD), and the nuclear factor erythroid 2 (NRF2) pathway [4, 7]. If a further increase in ROS levels occurs, then the cells undergo apoptotic cell death (Figure 1). Therefore, under physiological conditions, in order to guarantee cellular redox homeostasis, cells regulated intracellular ROS levels by applying a tight regulation of ROS generation and of ROS detoxifying pathways.
Figure 1.
Redox stress activation in physiology. The production of abnormally large amounts of ROS leads to persistent changes in signal transduction and gene expression that, in the last instance, could give to cell death. The steady-state levels of ROS are determined by the rate of ROS production and their clearance by scavenging mechanisms.
In this review, we first summarized the role of oxidative stress molecules in cancer initiation and progression and the proposed oxidative stress-targeted anticancer approaches. Next, in order to derive cancer-type specific oxidative stress gene profiles and their potential prognostic role, we integrated a publicly available oxidative stress gene signature [9] with the data extracted from the Cancer Genome Atlas (TCGA) database. Then, we reviewed some of those genes/pathways correlating with patient's survival, in order to define potential novel anticancer therapeutic targets. Finally, we highlighted novel findings on the correlation between oxidative stress and cancer stem cells (CSC).
2. The Role of Oxidative Stress Molecules in Cancer Initiation and Progression
A link between ROS and cancer progression dates back to 1981 when increased levels of H2O2, induced by insulin were shown to promote tumor cell proliferation. Almost three decades later, several studies sustained this hypothesis, reporting increased levels of oxidative damage products in clinical tumor specimens and plasma as well as in cancer cell lines [5]. Based on these evidences, to date, the idea that altered redox balance and deregulated redox signaling are strongly implicated in any steps of carcinogenesis as well as in the resistance to treatment, by affecting many, if not all, hallmarks of cancer is widely accepted [10, 11]. Indeed, currently, the role of ROS in cancer initiation and progression through the modulation of cell proliferation, apoptosis, angiogenesis, and the alteration of the migration/invasion program is well described [7, 12, 13]. For example, ROS may affect proliferation by a ligand-independent transactivation of different receptor tyrosine kinase via ERK activation and may induce tissue invasion and metastatic dissemination by activation of metalloproteinases. Moreover, the release of vascular endothelial growth factor and angiopoietin induced by ROS promote tumor angiogenesis and anoikis [12, 14].
Nonetheless, the exact origin of ROS generation during cancer development and disease progression and how this event could be druggable remains still unclear. Increasing evidences reported a link between ROS activation and the presence of some oncogenes, such as Ras, c-Myc, or Bcr-Abl [2, 15, 16]. Activation of oncogenic signaling might contribute to the increase of ROS levels, which in turn by promoting genomic instability could affect both nuclear and mitochondrial DNA. The consequent activation of antioxidants' signaling within tumor cells can also promote cancer progression and metastasis [2, 15–18]. Furthermore, cancer cells undergo metabolic changes to counteract the oxidative stress, also contributing to metastatic program [5, 19, 20].
Loss of functional p53 is involved in ROS induction, due to p53 “genome guardian” role in sensing and removing oxidative damage to DNA, thus preventing genetic instability [5, 21]. Anyhow, unlike oncogenes, the role of tumor suppressors in the modulation of ROS is more complex, depending on the specific tumor suppressor itself. For example, ataxia-telangiectasia mutated (ATM) is a cellular damage sensor that by regulating cell cycle and DNA repair preserves genomic integrity. Deficiency of ATM gene, either in patients or in mice, has been shown to produce elevated ROS levels and a chronic oxidative stress status. Recently, cytoplasmic ATM is described to activate a pathway leading to autophagy through repression of mammalian target of rapamycin complex 1 (mTORC1) in response to elevated ROS levels [22, 23]. Another example regards the loss of PTEN that determines AKT hyperactivation and inactivation of the forkhead homeobox type O (FoxO) transcription factor and therefore enhanced susceptibility to oxidative stress [24].
Less evidences are available about the regulation of ROS by microenvironment; however, new efforts have been recently focused in this field [5, 12]. In this regard, Chan et al. demonstrated that cancer-associated fibroblast- (CAF-) derived ROS are able to induce the acquisition of an oxidative CAF-like state on normal fibroblasts. Then, these oxidatively transformed normal fibroblasts promoted the development of aggressive tumors via a TGFβ1-mediated Smad3 signaling, suggesting an important relationship between the extracellular redox state and cancer aggressiveness [25].
3. Targeting Oxidative Stress as Anticancer Therapy
The first approach to prevent or treat cancer, by targeting ROS, was based on the use of antioxidant reagents [11, 15]. In one of the first trials, based on supplementation of selenium, vitamin E and β-carotene on the diet showed a reduction of overall mortality and cancer rates [26]. However, a following trial not only failed to obtain consistent results but also indicated that in certain cases, antioxidants can rather promote cancer initiation and progression. Concordantly, two trials of cancer prevention, the CARET on male smokers, treated with vitamin A and/or β-carotene and the SELECT trial, on older males treated with vitamin E and/or selenium, resulted in an increased incidence of lung and prostate tumors, respectively [27–29]. Similar contradictory results were shown in the trials using antioxidant treatment as adjuvant therapy [30].
Based on these results, almost a decade ago, ROS inducers were proposed as anticancer strategy, in order to overcome the specific threshold of ROS level beyond which cancer cells undergo ROS-mediated cell death [4, 5]. The first agents used are those improving electrons leak from the respiratory complexes in the mitochondria, such as the arsenic trioxide, or conventional chemotherapeutic drugs such as doxorubicin. Indeed, patients treated with those agents showed lipid peroxidation in their plasma as well as low levels of vitamin E, vitamin C, and β-carotene in the blood [4]. The mechanism of action of these agents seems to be related to their ability to generate ROS directly from the mitochondria. Indeed, doxorubicin and arsenic trioxide penetrate in the inner membrane of the mitochondria and induce superoxide radical production by modulating the electron transport chain. Also 5-fluorouracil increases mitochondrial ROS with a different mechanism, mediated by p53 [4, 31]. Ionizing radiations represent other important ROS inducers, because they are able to promote by themselves high level of ROS and also because they might increase NADPH oxidase, an important source of ROS [32]. Moreover, we and others have demonstrated, in different models and in different combination settings, that oxidative injury played a significant functional role in the antitumor effect of histone deacetylase inhibitors (HDACi), a class of epigenetic antitumor compounds currently in clinical practice in haematological malignancies [7, 13, 33–42].
Recently, a new ROS inducer compound, Elesclomol (STA-4783), has been developed and tested, both in in vitro and in vivo preclinical studies as well as in clinical trials [5, 43]. Interestingly, the result from a phase II trial using Elesclomol in combination with chemotherapy, in malignant melanoma patients, showed ROS generation and oxidative damage associated with prolonged progression-free survival [44]. Unfortunately, these results were not replicated in a phase III trial, where Elesclomol treatment was suspended due to adverse toxic effects [45]. The reason of this failure could be ascribed, at least in part, to cancer cells' capability to activate ROS adaption mechanisms by increasing levels of ROS scavengers, especially at advanced stages. This event is particularly efficacious in CSC, as described in the last paragraph of this review. To counteract the ROS adaptation mechanisms, a plausible solution could be the combination of ROS inducers either with another ROS inducer or with compounds that suppress cellular antioxidants, to overcome the threshold useful to induce cell death, The latest approach was tested by using an inhibitor of the scavenger SOD2, 2-Me, in combination with arsenic trioxide in lymphocytic leukemia and urothelial carcinoma cells [46, 47]. Similarly, the combination between the inhibitor of the antiapoptotic protein bcl2 ABT-737 and the ROS inducer, N-(4-hydroxyphenyl) retinamide, or the combination between an NRF2 inhibitor and a glutathione-depleting agents, showed increasing therapeutic efficiency compared to single-agent treatment [48, 49]. Based on these data, several clinical trials of combination treatment between ROS inducers and scavenger inhibitors are ongoing, including a multicenter phase II trial with the iron chelator Triapine and gemcitabine in advanced non-small-cell lung cancer [5].
4. Bioinformatics Correlation between Oxidative Stress Gene Expression and Prognosis in Solid Cancer Patients
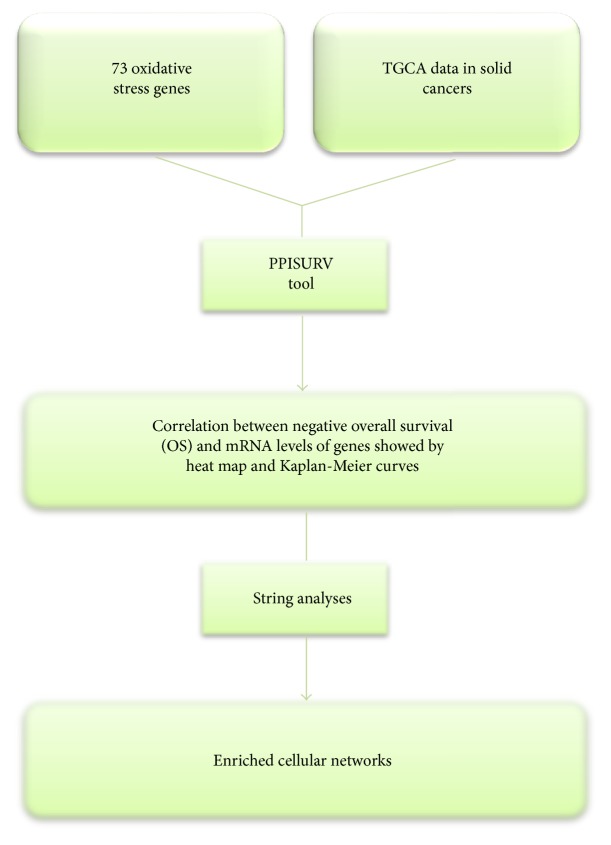
Although the biological role of oxidative stress pathways has been extensively demonstrated, it is still unclear which and how oxidative stress genes predict bad prognosis and if their modulation is cancer-type specific. Here, to address this question, we took advantage of Cancer Genome Atlas (TCGA) database that, by profiling RNA expression levels and DNA mutational status for thousands of genes, has generated comprehensive maps of the key genomic changes in several types of cancer, enabling correlative analysis of critical cellular pathways involved in each type of cancers [50, 51]. In details, we compared cancer patient overall survival (OS) and the mRNA levels of 73 oxidative stress genes, selected from a public available oxidative stress signature [9], in different solid tumors. Specifically, the signature included peroxidases, which are represented by glutathione peroxidases (GPx) and peroxiredoxins (TPx); genes implicated in ROS metabolism (i.e., DUSP1, FoxM1, and HMOX1); and genes involved in superoxide metabolism, such as superoxide dismutase (SOD). Starting from the selection of the 73 oxidative stress genes, bioinformatics investigations were performed as described in Figure 2.
Figure 2.
Bioinformatics analyses. Flow chart reporting step-by-step bioinformatics approach to unveil the most important genes/pathways involved in the correlation between oxidative stress and cancer.
In details, bioinformatics analysis was made by SynTarget online tool (http://www.bioprofiling.de/PPISURV) using the following public datasets: TCGA_PAAD for pancreatic cancer, TCGA_COAD for colon cancer, TCGA_HNSCC for head and neck cancer (HNSCC), GSE31210 for lung cancer, TCGA_PRAD for prostate cancer, and METABRIC for breast cancer [52, 53]. PPISURV automatically derives the currently known interactome for a gene of interest and correlates expression levels of its interactome, with survival outcome in multiple publicly available clinical expression data sets containing microarray expression data set annotated with survival data. In details, as reported by Antonov et al. [54], in the case of the option “single gene survival analyses on a single data set,” the PPISURV program exploits rank information from expression data sets that reflect the relative mRNA expression level. The samples are grouped with respect to expression rank of the gene in order to correlate survival information to the expression level of a gene in a particular data set. The groups are then subdivided in basis to “low expression” and “high expression” where expression rank of the gene is less or more than average expression rank across the data set, respectively. This separation of patients into “low” and “high” groups in the data set along with survival information is then used to find any statistical differences in survival outcome and to draw Kaplan-Meier plot. Hence, PPISURV establishes a correlation of the selected gene with survival and assesses the sign of the effect and if the gene deregulation is associated with positive or negative outcome.
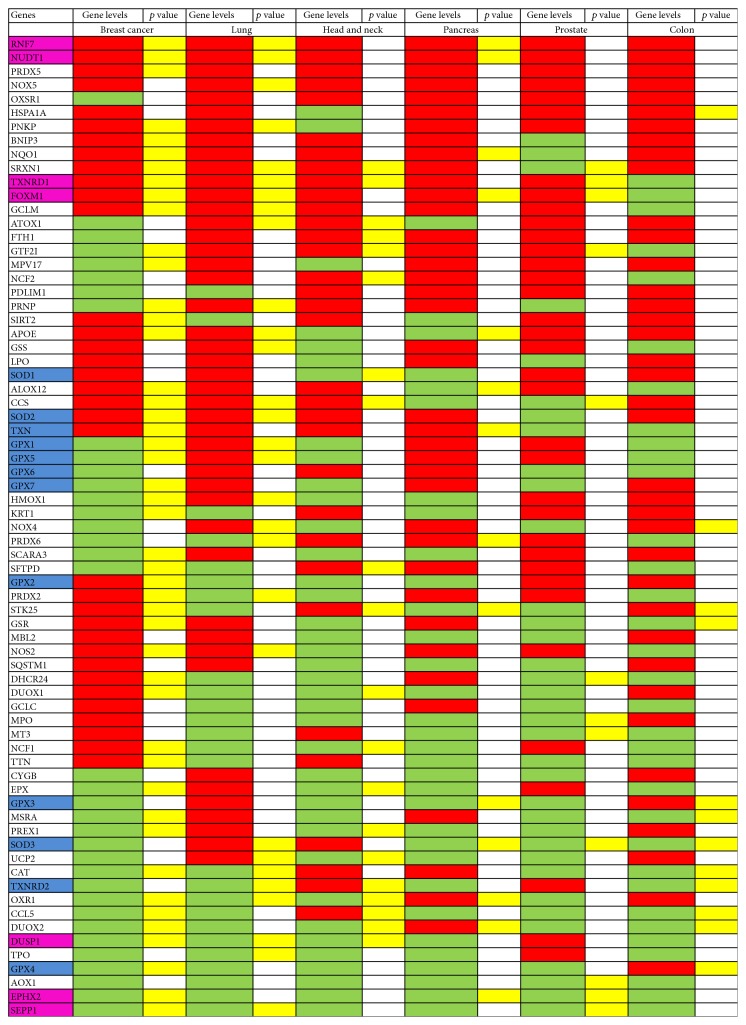
Notably, a significant number of oxidative stress genes were negatively correlated with survival in solid carcinomas, reinforced the idea that oxidative stress plays a crucial role in cancer cells (Figure 3). Furthermore, going deep to our bioinformatics analysis, we observed that breast, lung, and HNSCC cancers were those more susceptible to oxidative stress gene expression fluctuations. To explain these data, one hypothesis could be that all these tumors are more vulnerable to external insults (i.e., pollutants) that, as mentioned above, are an important source of ROS. Furthermore, we speculate that this phenomenon could be also related to the high mutational load of those tumors. Indeed, several studies showed that either breast (particularly triple-negative subgroup) or lung cancer exhibited an elevated mutational load which is closely associated to mutations in DNA damage repair genes as well as to intrinsic genomic instability [55–57]. Similarly, recently it has been demonstrated that the overall mutational load was higher in old HNSCC patients that represent a high percentage of all HNSCC cancers, compared to younger patients [58]. On the contrary, pancreatic, prostate, and even colon (with exception of microsatellite instability (MSI) high subgroup) cancers are described as less hypermutated and thus, we speculate, are also less dependent to the oxidative stress and genomic instability [59–61].
Figure 3.
Bioinformatics correlation between oxidative stress gene expressions and poor prognosis in 6 different tumor types. Heat map in which we report in red or in green if the high or low expression of genes was negatively correlated with survival, respectively. Moreover, we evidenced in yellow when the correlation is statistically significant (with p value <0.05). In the first column, we evidenced in magenta, the genes similar modulated among cancers and in blue those oxidative stress family extracted from STRING analysis. Notably TXNRD1 showed a central role in both analyses.
A further detailed analysis of our correlation between oxidative gene expression signature and OS unveiled that the behavior of modulated genes was different among the cancers examined, with the exception of two genes involved in ROS metabolism, such as FoxM1 and TXNRD1, found as statistically significantly high in poor prognosis patients in four out of six of the tumor types analyzed (Figures 3 and 4). For this reason, those two genes are described below in details in two specific sections of the review. Other five genes, DUSP1, EPHX2, NUDT1, RNF7, and SEPP1, demonstrated a statistically significant modulation in poor prognosis patients in three out of six tumor types (Figure 3 and Suppl. Figure S1 available online at https://doi.org/10.1155/2017/2597581). Briefly, DUSP1 is a dual-specificity phosphatase-1, which is recognized as a key player for inactivating different MAPK isoforms. Recently, a role of DUSP-1 as central redox-sensitive regulator in monocytes has been demonstrated [62]. EPHX2 is a cytosolic epoxide hydrolase, implied in cancer progression and metastasis, in differentially manner based on the stages of carcinogenesis. Indeed, Bracalante et al. demonstrated that in A7 melanotic cells, resembling less aggressive tumor cells, anti-oxidant genes, including EPHX2, were upregulated in response to oxidative stress, while they were downregulated in G10 metastatic melanoma cells [63]. NUDT1, nudix hydrolase 1, is the most prominent mammalian enzyme among other enzymes responsible for hydrolyzing oxidized DNA precursors. NUDT1 is commonly upregulated in a wide variety of tumors to avoid incorporation of oxidized nucleotides that, in turn, induce DNA damage and cell death [64]. RNF7 (RING finger protein-7) acts as a metal chelating protein, a scavenger of ROS at the expense of self-oligomerization. RNF7 was found overexpressed in several tumor types, especially in lung carcinoma, and associated with poor prognosis [65].
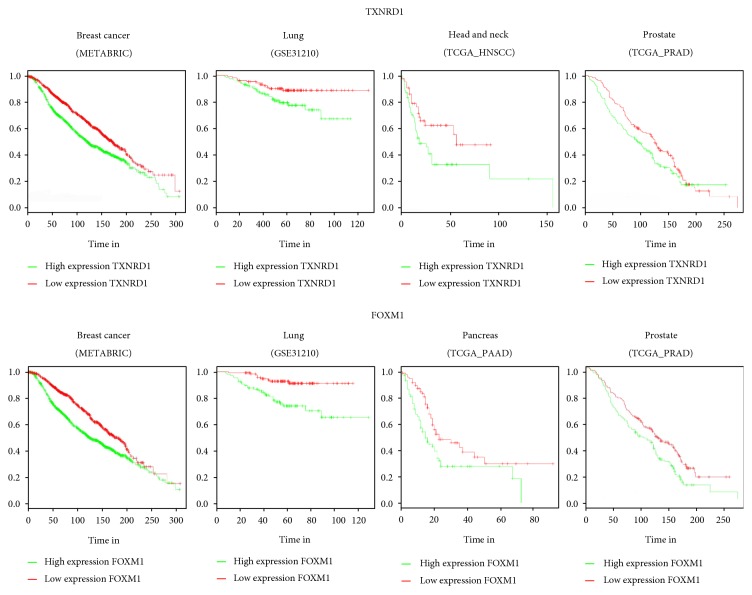
Figure 4.
TXNRD1 and FoxM1 expression related to patient survival. Kaplan-Meier curves showing the survival in the case of high and low expression of TXNRD1 and FOXM1 in solid cancer patients.
SEPP1 is a selenoprotein 1, involved in cellular incorporation of the selenium circulating in the plasma. Moreover, SEPP1 has some antioxidant activity, as target of NRF2 family. In agreement, Bae et al. showed that some antioxidant genes known also as NRF2 targets, including SEPP1, were also transcriptionally modulated by the oncosuppressor BRCA1, thus suggesting that BRCA1 regulates the activity of NRF2 and protects cells against oxidative stress [66].
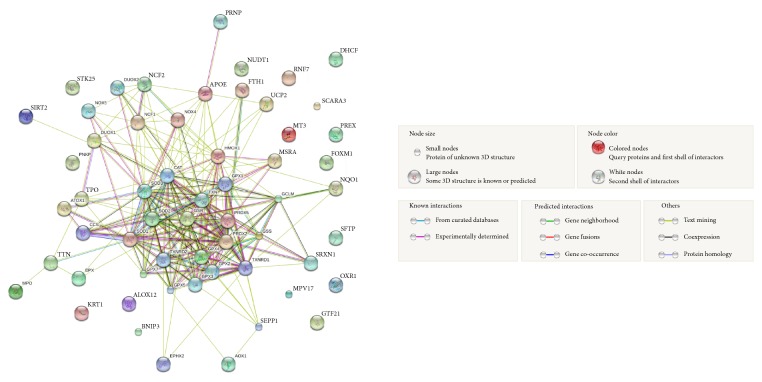
Finally, in order to identify a more relevant oxidative stress family in our setting, we performed an additional bioinformatics analysis where, independently from their trend of expression associated to poor prognosis, all modulated genes were analyzed in the biological database STRING, a resource of known and predicted protein-protein interaction. As shown in Figure 5, our analysis reveals an enriched cellular network, in which many genes, including GPx, SOD, and Trx pathways (the latter including TXNRD1), are strongly correlated, as demonstrated by both experimental studies and text mining (see red and green lines, resp.). Similar analyses were also performed for each tumor type separately, or considering high or low gene expression individually, confirming in almost all tumor types GPx, SOD, and Trx signaling as those predominant (Suppl. Figures S2 and S3).
Figure 5.
STRING analysis of the 58 oxidative stress genes. Association network in STRING analysis shows interactions of glutathione peroxidase, superoxide dismutase, and thioredoxin as principal oxidative stress signaling among six different tumor types.
Based on these analyses, together with TXNRD1, we decided to review the correlated pathways enriched in the network, in details (see below), analyzing their role in cancer and the possible therapeutic strategies to hit them.
5. FoxM1, a Critical Regulator of Oxidative Stress during Tumorigenesis
The highly conserved transcription factor FoxM1 belongs to the forkhead box transcription factor family, similarly to the best known member of FoxO family. However, different from the members of FoxO family, FoxM1 is expressed only in proliferative cells. Indeed, FoxM1 as a target of the cyclinD-CDK4/6 kinases, is reactivated when quiescent cells reenter in the cell cycle and reach a maximal level in S-phase which is maintained throughout G2 and mitosis [67, 68]. Beyond this role on proliferation, FoxM1 regulates metastasis, apoptosis, and DNA damage repair [69–71]. Furthermore, FoxM1 has been shown to prevent oxidative stress-dependent premature senescence. Park et al. showed how ROS themselves are inducers of FoxM1 expression, which in turn is able to stimulate antioxidant genes. The authors proposed the inhibition of FoxM1 as a new therapeutic strategy to kill cancer cells selectively [71]. In agreement, FoxM1 knocking-down was reported to sensitize human pluripotent stem cells to oxidative stress, as a consequence of activated-CAT5 downregulation, a FoxM1 antioxidant target gene [69].
A growing body of evidences reported high FoxM1 as frequently related to poor prognosis in multiple cancers, concordantly with our bioinformatic results [50]. To date, several mechanisms have been proposed to explain the activity of FoxM1 in cancer progression, including the activation of FoxM1 by several oncogenic protein and signalling pathways, such as c-Myc, Ras, and PI3K/AKT [72].
Hereafter, we discussed the role of FoxM1 in the four tumor types where we found statistically significant association of FoxM1 expression and poor prognosis.
The impact of FoxM1 in breast cancer progression is widely demonstrated. Indeed, its high level has been correlated with large tumor size, lymphovascular invasion, lymphnode metastases, and high stage. Two independent studies carried out on ER+ patients, reported that low FoxM1 expression, compared to high FoxM1 expression, is associated to better survival. Another study proved a positive correlation between HER2 status and FoxM1 expression in breast cancer tissue compared to normal breast counterpart [73–75], suggesting that FoxM1 is a downstream target of HER2 and could be used as a marker of HER2 overexpression. However, molecular basis underling the described roles of FoxM1 in cancer progression still needs to be clarified and different mechanisms have been proposed. For example, the induction of EMT by activation of Slug [76], stabilization of Smad3/Smad4 complex, and activation of TGFβ pathway [77] as well as the modulation of extracellular matrix by affecting the levels of uPA, uPAR, MMP-2, MMP-9, and VEGF have been proposed [78, 79]. Moreover, FoxM1 cooperates with survivin and nuclear XIAP in the promotion of chemoresistance [80]. Finally, further studies demonstrated that FoxM1 induces resistance to all the therapeutics tested in breast cancer (including cisplatin, paclitaxel, and trastuzumab) by several mechanisms: (1) acting on DNA-damage repair pathways, (2) promotion of cell cycle progression, (3) inhibition of cell cycle checkpoints, and (4) apoptosis induction [72].
FoxM1 gene is widely described as amplified also in lung cancer, regulating cell proliferation by promoting both G1/S and G2/M transition, differentiation, and transformation [81] as well as inhibition of apoptosis [82]. Recently, a direct link between FoxM1-induced ROS and lung cancer progression has been proposed by Tahmasbpoura et al. Their study showed elevated rate of lung cell proliferation related to high FoxM1 expression in patients exposed to sulfur mustard, a well known agent able to induce ROS [83].
Beyond the mechanisms described, the molecular basis of FoxM1 dysregulation has been also related to the capability of vitamin D receptor (VDR)/FoxM1 axis to affect cell stemness and to induce an invasive and metastatic phenotypes in pancreatic cancer. Indeed, the authors observed that VDR activation reduced the levels of FoxM1, inducing nuclear accumulation of β-catenin [84].
In prostate cancer (PCa), only few studies focused on the role of FoxM1; for instance, FoxM1 and its target CENPF, a structural protein of kinetochore, have been both proposed as critical drivers of PCa development and as prognostic markers of poor survival [85]. Concordantly, Lin et al. unveiled different miRNAs regulating FoxM1-CENPF axis taking advantage of miRNA expression profile available in Taylor dataset of prostate specimens (normal, localized, and metastatic tissues) [86]. Notably, since CENPF regulates several genes important for metastasis, including MMP2, MMP9, LOX, CXCR4, and CXCL12, dysregulation of the miRNA-COUP-TFII-FoxM1-CENPF axis can inhibit also PCa metastatization [86].
Overall, these considerations identified FoxM1 as a potential anticancer therapeutic target. Unfortunately, the druggability of FoxM1 remains a big challange because of the lack of substrate-binding pockets and hydrophobic surfaces [72, 87]. Several in vitro studies proposed RNA interference (RNAi) as a strategy to knockdown FoxM1, either alone or in combination with ROS inducers, in order to provoke ROS-mediated cell death [82]. Some studies reported that proteasome inhibitors, including bortezomib or thiostrepton, directly reduce both FoxM1 expression and its transcriptional activity with the same efficacy as that obtained by FoxM1 silencing [82, 88]. This latter approach is very promising, considering that bortezomib is already in clinical practice to treat multiple myeloma, and that RNAi treatment, so far, is not a reasonable therapeutic approach in patients [50, 82, 88]. Thus, bortezomib treatment has been proposed as effective therapeutic strategy in highly expressing FoxM1 solid tumor, also in association with ROS inducers.
6. Thioredoxin, Glutathione Peroxidase, and Superoxide Dismutase Families as Mediators of Carcinogenesis
Thioredoxin system, composed of thioredoxin reductase (TrxR), thioredoxin (Trx), and NADPH, senses and responds to oxidative stress and modulates the redox status by scavenging ROS and by regulating several redox enzymes and signaling proteins. Mammalian genomes encode two main Trx systems: Trx1 and Trx reductase (TrxR) 1, which together constitutes the cytosolic system; Trx2 and TrxR2, which are localized in mitochondria (a Trx3 isoform has been also reported, as a testis-specific form, mainly expressed in male germ cells and associated to reproductive disorders) [89]. Trx1 reducing power allows the transfer of two electrons from its dithiol motif to an acceptor, then the oxidized disulfide form of the enzyme is recycled to the dithiol form by TrxR1, thereby oxidizing one molecule of NADPH.
Interestingly, our analysis revealed that TXNRD1, the gene encoding TrxR1, is upregulated and correlates with bad prognosis in pancreatic, colon, HNSCC, lung, prostate, and breast cancers. Trx1 enzyme has been shown to regulate NF-κB, playing opposite roles, depending to its intracellular localization: overexpression of Trx in cytoplasm reduced NF-κB activity, blocking the degradation of the NF-κB inhibitor IκB; in the nucleus, Trx directly reduces the cysteine(s) of NF-κB allowing the NF-κB-dependent gene expression [90]. Following NF-κB stimuli, such as UVB irradiation and TNFα treatment, Trx quickly translocates from the cytoplasm into the nucleus. Trx1 has also been reported as a secreted protein by normal and neoplastic cells [91], but not via exosomes [92]. Notably, Trx-increased secretion contributed to high ROS production in cisplatin-resistant lung tumors, both in vitro and in vivo [93].
Trx1 itself is regulated both by hypoxia and by oxidative stress conditions via binding of NRF2 to an antioxidant responsive element in the Trx promotor [94]. Moreover, Trx1 complex functions as a molecular switch turning the cellular redox state into kinase signaling. Thus, the system is able to regulate DNA synthesis, cell proliferation [95, 96], apoptosis, and transcription. In details, the reduced form of Trxs binds to apoptosis signal-regulating kinase 1 (ASK1) and inhibits its activity to prevent stress- and cytokine-induced apoptosis; when Trx is oxidized, it dissociates from ASK1 and apoptosis is stimulated [97–100]. The impact of Trx1 intracellular localization on its role may be taken into account especially in tumors (as colon and prostate) where a low expression of TXNRD1 correlates to poor patient outcome (as described in Figure 3()). In fact, although increased Trx1 protein expression has been associated to hypoxic regions of certain tumours, tumor grade and chemoresistence, for instance by scavenging ROS species generated by various anticancer agents [101, 102], its localization and activity have to be both taken into account. In prostate cancer, Shan and colleagues identified constitutive nuclear and transiently increased cytoplasmic Trx1 oxidation by androgen but decreased Trx1 activities with the progression of prostate cancer, despite high levels of Trx1 protein expression in cancer cells [103]. The role of TrxR1 in dysplastic transformation has been pointed out in human breast epithelial cells, triggered by chronic oxidative stress [104]. In addition, Trx1 has been proposed as serum biomarker for either early diagnosis or prognosis of breast cancer in association with CEA and CA15-3 [105]. In non-small-cell lung cancer, Trx1 is able to modulate transcription of cyclooxygenase-2 via hypoxia-inducible factor- (HIF-) 1α [106]. It is actually worth to mention that many human cancers have low levels of thioredoxin-binding protein-2 (TBP-2), a Trx regulator which is able to bind Trx, blocking its reducing activity. These mechanisms have been identified as druggable: histone deacetylase inhibitors (HDACi) have been demonstrated to upregulate TBP-2 in various transformed cells, associated with a decrease in Trx levels [102].
Recently, Park and colleagues observed that TrxR2 is a novel binding protein for ribonucleotide reductase small subunit p53R2, which is involved in nuclear and mitochondrial DNA replication and repair, stimulating the enzymatic activity of TrxR in vitro. Their findings also suggest that p53R2 acts as a positive regulator of TrxR2 activity in the mitochondria both under normal physiological conditions and during the cellular response to DNA damage [107].
Although STRING analyses highlighted glutathione peroxidases (GPx) as one of the main family involved in oxidative stress adaptation, we found high heterogeneity in the disregulation of GPx family members among the tumor types we have investigated (Figure 5). GPx reduces either free hydrogen peroxide to water or lipid hydroperoxides to their corresponding alcohols. So far, eight different isoforms of GPx, 1 to 8, have been identified in humans, carrying different affinities for their substrates and different localizations. GPx1, found in the cytoplasm of mammalian cells, is mainly able to target the hydrogen peroxide, while GPx4 showed high affinity for lipid hydroperoxides. GPx2 is an intestinal and extracellular enzyme, while GPx3 is extracellularly secreted [99].
GPx1 allelic loss or polymosphisms have been known for years to contribute to both lung [108] and breast cancers [109]. Interestingly, in HNSCC cancer, almost all the isoforms showed low expression (Figure 3). In agreement, a decrease in GPx activity accompanied by SOD and CAT decrease as well as higher levels of oxidative DNA damage was found in HNSCC patients compared to healthy donors [110].
An increase of both Trx and GSH metabolism is a mechanism widely implicated in the resistance of cancer cells to chemotherapy. Loss of TXNRD1 makes tumors highly susceptible to pharmacological GSH deprivation, and concomitant inhibition of both GSH and TxrR systems was recently proposed as an anticancer strategy [18, 111]. Recently, Rodman and colleagues demonstrated that depletion of GSH and inhibition of TrxR activity enhanced radiation responses in human breast cancer stem cells by a mechanism involving thiol-dependent oxidative stress [112]. Furthermore, Scarbrough and colleagues reported that simultaneous GSH/Trx inhibition sensitizes human breast and prostate cancer cells to 2DG + 17AAG-mediated killing [113].
Among the most important antioxidant enzymes, it is also important to highlight the role of SOD. SOD is able to convert the superoxide (O2−) radical into either oxygen (O2) or the less reactive hydrogen peroxide (H2O2) which can then be removed by CAT, GPx, or TPx. Among the three major families of SOD, those we single out in humans are the copper and zinc (Cu-Zn) SOD1, whose localization is in cytosol, nucleus, peroxisome, and intermembrane space of the mitochondria [114], the mitochondrial enzyme manganese SOD2 (MnSOD), and the (Cu-Zn) extracellular SOD3. SOD enzymes are able to exert a strong antioxidant activity. In a recent study, Elchuri and colleagues observed that mice deficient in CuZn SOD1 (which contributes to the majority of cellular SOD activity [115]) showed a reduced lifespan and increased incidence of neoplastic changes in the liver [116]. Conversely, it has been also observed by several authors that SOD1 overexpression makes tumor cells resistant to oxidative stress and chemotherapy [117]. Increased expression and activity of MnSOD has been correlated with cancer aggressiveness in several tumors and through different pathways [118]. Recently, dysregulation of MnSOD function has been linked to an acetylation-mediated impairment [119, 120] which triggers an increase in oxidative stress, leading to AKT activation via oxidative inactivation of PTEN [119]. MnSOD acetylation (and activity) is regulated by the deacetylase Sirt3, a mitochondrial fidelity protein. Interestinlgly, Zou et al. showed that loss of Sirt3 results in endocrine therapy resistance of human luminal B breast cancer [120]. In agreement, we and others demonstrated that HDAC inhibition increases MnSOD protein expression in both solid and haematological diseases [121, 122].
Overall, similar to FoxM1, the described antioxidant systems represent putative good targets to improve therapeutical oxidative stress-dependent strategies. In details, several recent efforts have focused on the targeting of Trx/TrxR system [123–130]. Moreover, increasing evidences on a putative key role of HDAC inhibitors in the modulation of these pathways may deserve further investigations. In this regard, our recent study on the effect of HDACi in regulating NRF2/Keap1 pathway is of interest, considering the interplay between this pathway and thioredoxin [7].
7. Oxidative Stress and Cancer Stem Cells
In the multitude of morphological, functional, and responsive cancer cells, a subset of the so-called “cancer stem cells” (CSC), carrying peculiar features, was identified almost ten years ago in solid cancers [131]. However, the name CSC is not referred to an origin from normal stem counterpart but rather represents a specific population that displays some exceptional properties normally attributed to stem cells. Specific features, like hierarchical differentiation, self-renewal, enhanced invasive capacity, metastatic proficiency, and tumorigenicity, make CSC critical for tumor initiation and growth [132], while CSC elevated apoptosis resistance, drug-efflux pumps, enhanced DNA repair efficiency, detoxification enzyme expression, and quiescence are all identified as prosurvival mechanisms associated with resistance to chemotherapy and tumor relapse [133].
Few studies reported the behavior of cancer stem cells in oxidative stress condition, but notably in contrast to their normal stem cell counterparts, cancer stem cells are characterized by increased ROS levels, reduced oxidative damage, and thus longer survival [134, 135]. For example, Im and colleagues showed that significantly higher ROS levels were observed in the supernatant of glioblastoma cells, grown in serum-free sphere medium, either in polystyrene-treated tissue culture plates or in nonadherent plates. Moreover, it has been also shown that ROS is a critical factor for maintaining stemness, regulating the expression of the transcription factor SOX-2 [136]. This can be due to a combination of mechanisms that arise in the tumor, such as modulation of (1) multiple antioxidative enzyme systems [137] or (2) redox-sensitive signaling pathways, as NRF2, NF-κB, c-Jun, and HIFs, leading to the increased expression of antioxidant molecules [5].
The higher ROS levels in CSC could be associated with lower basal expression of ROS-scavenging systems, such as SODs, CAT, GPx, and TPx, compared to normal stem cells. In this regard, Yang et al. published those nonglioma stem cells which displayed significantly lower basal GPx1 expression and activity than glioma stem cells and that miR-153/NRF2/GPx1 pathway plays an important role in regulating radiosensitivity and stemness of glioma stem cells via ROS [138].
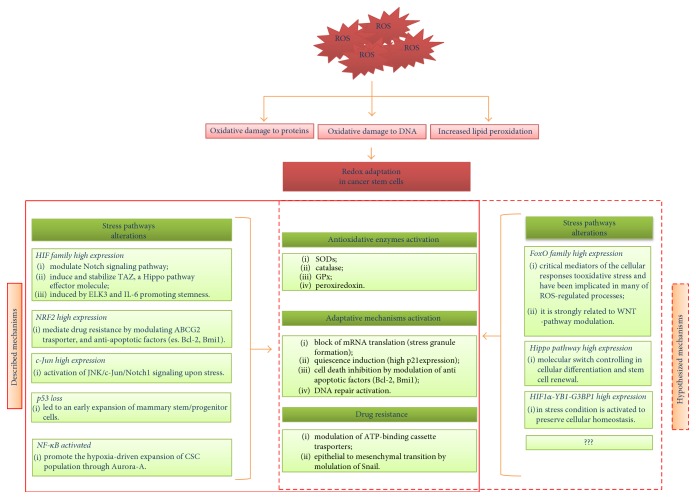
Due to the growing body of studies focused on the differential modulation of redox-sensitive signaling pathways (as summarized in Figure 6) in CSC subpopulation, compared to cancer cells or normal stem cells, in this paraghraph we discuss the relevance of the ROS-related pathways modulated in CSC phenotype.
Figure 6.
Redox stress in cancer stem cells. The persistent production of abnormally large amounts of ROS induced the mechanism of redox adaptation that, in turn, is translated in a various alteration in stress signaling. Here, we reported both known and hypothesized modulated pathways.
In hypoxic environments, limited amount of oxygen leads to metabolic switches in both normal and malignant cells by HIFs. Paradoxically, recent studies have shown that CSC exhibit high HIF activity in normoxic environments and that HIF activity is critical in the maintenance of CSC as well as in the differentiation [139]. In agreement, Wang et al. found that overexpression of stem cell factor in hepatocellular carcinoma is regulated by hypoxic conditions through a selective HIF2α-dependent mechanism which promotes metastasis [140].
Several studies showed that HIF factors can enhance CSC population growth by modulating Notch signaling pathway in glioma [141], Hippo pathway through direct stabilization of TAZ in breast cancer [142], Ras-ERK-ELK3 in hepatocellular cancer, hypoxia-NOTCH1-SOX2 in ovarian cancer [143], and IL6-HIF1α in non-small-cell lung cancer [144]. Additionally, Yang et al. established that gastric CSC exhibited a marked increase in HIF1α expression and increased migration and invasion capabilities compared with the normoxic control upon hypoxia treatment. Also HIF-1α was responsible for activating EMT via increased expression of the transcription factor Snail in gastric CSC [145].
NF-κB is also related to hypoxia and HIF1α induction. It has been shown that inhibition of NFκB signaling promoted a significant reduction in the hypoxia-driven expansion of CD44+CD24−/low CSC which was due to increased CD24 expression in breast cancer models [146]. Similarly, Aurora A kinase which can activate NF-κB pathway has been found highly expressed in ovarian CSC [147].
NRF2 represents another antioxidant system involved in the maintenance of quiescence as well as in the determination of differentiation fate in normal stem cells, as described and reviewed by Ryoo et al. [148]. For example, NRF2-deficient mice showed defective stem cell function. Indeed, haematopoietic stem cell, derived from those mice, dispayed lower levels of prosurvival cytochines and exibited spontaneous apoptosis related to wild-type cells [149].
Recently, several studies showed that high levels of NRF2 are related to CSC survival and anticancer drug resistance in HNSCC, cervical, breast, and ovarian cancers [150–153]. Notably, it was reported that NRF2 overexpression is related to an induction of ATP-binding cassette trasporters and thus drug resistance mechanisms. Other described redox-signaling pathway implicated in redox regulation in CSC could be c-Jun and/or p53 and NF-κB and FoxO family. In details, Chiche et al. showed that the loss of p53 in K5ΔNβcat (βcat activated) mice led to an early expansion of mammary stem/progenitor cells and accelerated the formation of triple-negative breast cancers. In particular, p53-deficient tumors expressed high levels of integrins and extracellular matrix components and were enriched in cancer stem cells [154].
Moreover, Xie et al. found that knockdown of JNK1 or JNK2 or treatment with JNK-IN-8, an adenosine triphosphate-competitive irreversible pan-JNK inhibitor, significantly reduced cell proliferation, the ALDH1+ and CD44+/CD24- CSC subpopulations, and mammosphere formation, indicating that JNK family promotes CSC self-renewal and maintenance in triple-negative breast cancer [155].
However, other factors could be implicated in CSC capability to adapt high level of intracellular ROS and would be very interesting to better define them as potential therapeutic targets, mostly because many anticancer drugs increase intracellular ROS levels.
In this regard, the transcription factors FoxO1, FoxO3a, and FoxO4 are critical mediators of the cellular responses to oxidative stress and have been implicated in many of ROS-regulated processes [156]. It is also known that FoxO competes with TCF for the same binding site of β-catenin and suppresses β-catenin-TCF signaling toward proliferation, thus attenuating WNT-mediated signaling activities. Also, FoxO factors reduce mitochondrial output to prevent excess ROS production through inhibition of c-Myc function and alter the hypoxia response [157].
Another candidate is the Hippo pathway, which acts as a molecular switch controlling in cellular differentiation and stem cell renewal but is also modulated in stress condition and is described as highly mutated in cancer. Lehtinen and colleagues elegantly demonstrated the activation of Mst1, a serine/threonine kinase activated in the Hippo cascade, upon oxidative stress induced by exposure to increasing concentrations of exogenous H2O2. This was accompanied by phosphorylation of the transcription factor FoxO3a at S207, thereby disrupting its association with 14-3-3 binding protein and leading to its nuclear localization and transcriptional activation of the BH3- only Bcl-2 protein, Bim, which triggered neuronal apoptosis [158].
One of the first mechanisms modulated upon stress condition is messenger RNA translation, likely as a mean to limit energy demanding protein synthesis, leading to stress granule (SG) formation in cancer cells. Many evidences suggest that altered mRNA translational control is a critical factor in cancer progression, and in this regard, a new axis has been described. In details, Somasekharan et al. showed that under stress condition, a YB1, nuclease-sensitive element-binding protein 1, facilitates tumor metastasis through two mechanisms: first, it directly binds to HIF1α that drives stress adaptation and metastatic capacity in vivo; second, YB1 mediates formation of cytosolic SGs through translational activation of G3BP1, a SG nucleator [159]. Accumulating evidences suggest that SG formation is protective against stress-induced cell damage and death [160], and few studies suggested SG implication in cancer biology [161, 162].
8. Conclusions and Future Perspectives
The idea that the oxidative stress modulation has a crucial role in cancer cells to promote proliferation, adaptation, and resistance to therapy is now widely accepted [7, 12, 13]. Thus, modulating redox regulatory mechanisms represents an attractive therapeutic strategy. However, to date, the oxidative stress-related therapeutic strategies evaluated in preclinical and clinical studies did not produce homogenous results, due to several variables associated to ROS generation and redox adaptation mechanisms.
Furthermore, the identification of tumor-type specific oxidative stress gene profiles and how they could predict prognosis still represent critical challenges. Thanks to the increasing availability of cancer gene expression profile, mutation, epigenetic, and survival data from the TGCA dataset, it was possible to use bioinformatics to screen the role of oxidative stress genes from a publicly available signature in large cohorts of several solid cancer patients [52, 53].
The TCGA database provides correlative evidences suggesting the involvement of the FoxM1, thioredoxin, superoxide-dismutase, and glutathione pathways as principally and commonly modulated in breast, lung, HNSCC, pancreatic, prostate, and colon cancers. The differential expression levels of each gene observed in different settings revealed a precise spatial context where redox alterations may promote genome instability or redox adaptation.
For this reason, tumors should be classified into subclasses based on different oxidative stress alterations occurring in redox homeostasis genes, to guarantee the development of precision medicine-based approaches in selected subgroups of cancer patients. Further mechanistic studies are needed to identify either new compounds or molecules to be repositioned, in order to target the described redox pathways.
Supplementary Material
Supplementary Figure S1. Kaplan Meier curves showing the survival in the case of high and low expression of DUSP1, EPHX2, NUDT1, RNF7 and SEPP1 in solid cancers patients. Supplementary Figure S2 STRING analysis of modulated oxidative stress genes in six different type of cancer. Supplementary Figure S3 STRING analysis of oxidative stress genes in six different type of cancer: high vs low network.
Acknowledgments
This study was partially supported by the following Research Grant to Alfredo Budillon: Italian Ministry of Health (RF-2011-02346914).
Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Sena L. A., Chandel N. S. Physiological roles of mitochondrial reactive oxygen species. Molecular Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill J. G., Piskounova E., Morrison S. J. Cancer, oxidative stress, and metastasis. Cold Spring Harbor Symposia on Quantitative Biology. 2016;81: 163–175. doi: 10.1101/sqb.2016.81.030791. [DOI] [PubMed] [Google Scholar]
- 3.Tafani M., Sansone L., Limana F., et al. The interplay of reactive oxygen species, hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxidative Medicine and Cellular Longevity. 2016;2016:18. doi: 10.1155/2016/3907147.3907147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorrini C., Harris I. S., Mak T. W. Modulation of oxidative stress as an anticancer strategy. Nature Reviews Drug Discovery. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 5.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature Reviews Drug Discovery. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez-Garcia D., Wood C. D., Castro-Obregón S., Covarrubias L. Reactive oxygen species: a radical role in development? Free Radical Biology and Medicine. 2010;49(2):130–143. doi: 10.1016/j.freeradbiomed.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Leone A., Roca M. S., Ciardiello C., et al. Vorinostat synergizes with EGFR inhibitors in NSCLC cells by increasing ROS via up-regulation of the major mitochondrial porin VDAC1 and modulation of the c-Myc-NRF2-KEAP1 pathway. Free Radical Biology and Medicine. 2015;89:287–299. doi: 10.1016/j.freeradbiomed.2015.07.155. [DOI] [PubMed] [Google Scholar]
- 8.Waypa G. B., Marks J. D., Guzy R., et al. Hypoxia triggers subcellular compartmental redox signaling in vascular smooth muscle cells. Circulation Research. 2010;106(3):526–535. doi: 10.1161/CIRCRESAHA.109.206334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. https://www.qiagen.com/de/shop/pcr/primer-sets/rt2-profiler-pcr-arrays/?catno=PAHS-065Z#geneglobe.
- 10.Hanahan D., Weinberg R. A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Panieri E., Santoro M. M. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death & Disease. 2016;7(6, article e2253) doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiaschi T., Chiarugi P. Oxidative stress, tumor microenvironment, and metabolic reprogramming: a diabolic liaison. International Journal of Cell Biology. 2012;2012:8. doi: 10.1155/2012/762825.762825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruzzese F., Pucci B., Milone M. R., et al. Panobinostat synergizes with zoledronic acid in prostate cancer and multiple myeloma models by increasing ROS and modulating mevalonate and p38-MAPK pathways. Cell Death & Disease. 2013;4, article e878 doi: 10.1038/cddis.2013.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues S. F., Granger D. N. Blood cells and endothelial barrier function. Tissue Barriers. 2015;3(1-2, article e978720) doi: 10.4161/21688370.2014.978720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J., Kim J., Bae J. S. ROS homeostasis and metabolism: a critical liaison for cancer therapy. Experimental & Molecular Medicine. 2016;48(11, article e269) doi: 10.1038/emm.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabharwal S. S., Schumacker P. T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nature Reviews Cancer. 2014;14(11):709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeNicola G. M., Karreth F. A., Humpton T. J., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris I. S., Treloar A. E., Inoue S., et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27(2):211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Kuehne A., Emmert H., Soehle J., et al. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Molecular Cell. 2015;59(3):359–371. doi: 10.1016/j.molcel.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Sukhatme V. P., Chan B. Glycolytic cancer cells lacking 6-phosphogluconate dehydrogenase metabolize glucose to induce senescence. FEBS Letters. 2012;586(16):2389–2395. doi: 10.1016/j.febslet.2012.05.052. [DOI] [PubMed] [Google Scholar]
- 21.Wormann S. M., Song L., Ai J., et al. Loss of P53 function activates JAK2-STAT3 signaling to promote pancreatic tumor growth, stroma modification, and gemcitabine resistance in mice and is associated with patient survival. Gastroenterology. 2016;151(1, article e12):180–193. doi: 10.1053/j.gastro.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Alexander A., Cai S. L., Kim J., et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander A., Kim J., Walker C. L. ATM engages the TSC2/mTORC1 signaling node to regulate autophagy. Autophagy. 2010;6(5):672–673. doi: 10.4161/auto.6.5.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCabe N., Walker S. M., Kennedy R. D. When the guardian becomes the enemy: targeting ATM in PTEN-deficient cancers. Molecular and Cellular Oncology. 2016;3(1, article e1053595) doi: 10.1080/23723556.2015.1053595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan J. S., Tan M. J., Sng M. K., et al. Cancer-associated fibroblasts enact field cancerization by promoting extratumoral oxidative stress. Cell Death & Disease. 2017;8(1, article e2562) doi: 10.1038/cddis.2016.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao Y. L., Dawsey S. M., Kamangar F., et al. Total and cancer mortality after supplementation with vitamins and minerals: follow-up of the Linxian general population nutrition intervention trial. Journal of the National Cancer Institute. 2009;101(7):507–518. doi: 10.1093/jnci/djp037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omenn G. S., Goodman G., Thornquist M., et al. Chemoprevention of lung cancer: the beta-carotene and retinol efficacy trial (CARET) in high-risk smokers and asbestos-exposed workers. IARC Scientific Publications. 1996;136:67–85. [PubMed] [Google Scholar]
- 28.Omenn G. S., Goodman G. E., Thornquist M. D., et al. Risk factors for lung cancer and for intervention effects in CARET, the beta-carotene and retinol efficacy trial. Journal of the National Cancer Institute. 1996;88(21):1550–1559. doi: 10.1093/jnci/88.21.1550. [DOI] [PubMed] [Google Scholar]
- 29.Klein E. A., Thompson I. M., Tangen C. M., et al. Vitamin E and the risk of prostate cancer: the selenium and vitamin E cancer prevention trial (SELECT) JAMA. 2011;306(14):1549–1556. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs C., Hutton B., Ng T., Shorr R., Clemons M. Is there a role for oral or intravenous ascorbate (vitamin C) in treating patients with cancer? A systematic review. The Oncologist. 2015;20(2):210–223. doi: 10.1634/theoncologist.2014-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longley D. B., Latif T., Boyer J., Allen W. L., Maxwell P. J., Johnston P. G. The interaction of thymidylate synthase expression with p53-regulated signaling pathways in tumor cells. Seminars in Oncology. 2003;30(3, Supplement 6):3–9. doi: 10.1016/s0093-7754(03)00119-2. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida T., Goto S., Kawakatsu M., Urata Y., Li T. S. Mitochondrial dysfunction, a probable cause of persistent oxidative stress after exposure to ionizing radiation. Free Radical Research. 2012;46(2):147–153. doi: 10.3109/10715762.2011.645207. [DOI] [PubMed] [Google Scholar]
- 33.Bruzzese F., Rocco M., Castelli S., Di Gennaro E., Desideri A., Budillon A. Synergistic antitumor effect between vorinostat and topotecan in small cell lung cancer cells is mediated by generation of reactive oxygen species and DNA damage-induced apoptosis. Molecular Cancer Therapeutics. 2009;8(11):3075–3087. doi: 10.1158/1535-7163.MCT-09-0254. [DOI] [PubMed] [Google Scholar]
- 34.Rosato R. R., Almenara J. A., Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer Research. 2003;63(13):3637–3645. [PubMed] [Google Scholar]
- 35.Pei X. Y., Dai Y., Grant S. Synergistic induction of oxidative injury and apoptosis in human multiple myeloma cells by the proteasome inhibitor bortezomib and histone deacetylase inhibitors. Clinical Cancer Research. 2004;10(11):3839–3852. doi: 10.1158/1078-0432.CCR-03-0561. [DOI] [PubMed] [Google Scholar]
- 36.Yu C., Friday B. B., Lai J. P., et al. Abrogation of MAPK and Akt signaling by AEE788 synergistically potentiates histone deacetylase inhibitor-induced apoptosis through reactive oxygen species generation. Clinical Cancer Research. 2007;13(4):1140–1148. doi: 10.1158/1078-0432.CCR-06-1751. [DOI] [PubMed] [Google Scholar]
- 37.Bhalla S., Balasubramanian S., David K., et al. PCI-24781 induces caspase and reactive oxygen species-dependent apoptosis through NF-kappaB mechanisms and is synergistic with bortezomib in lymphoma cells. Clinical Cancer Research. 2009;15(10):3354–3365. doi: 10.1158/1078-0432.CCR-08-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koshikawa N., Hayashi J. I., Nakagawara A., Takenaga K. Reactive oxygen species-generating mitochondrial DNA mutation up-regulates hypoxia-inducible factor-1alpha gene transcription via phosphatidylinositol 3-kinase-Akt/protein kinase C/histone deacetylase pathway. The Journal of Biological Chemistry. 2009;284(48):33185–33194. doi: 10.1074/jbc.M109.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf I. M., Fan Z., Rauh M., et al. Histone deacetylases inhibition by SAHA/Vorinostat normalizes the glioma microenvironment via xCT equilibration. Scientific Reports. 2014;4:p. 6226. doi: 10.1038/srep06226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hui K. F., Yeung P. L., Chiang A. K. Induction of MAPK- and ROS-dependent autophagy and apoptosis in gastric carcinoma by combination of romidepsin and bortezomib. Oncotarget. 2016;7(4):4454–4467. doi: 10.18632/oncotarget.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao P. H., Hsu H. H., Chen T. S., et al. Phosphorylation of cofilin-1 by ERK confers HDAC inhibitor resistance in hepatocellular carcinoma cells via decreased ROS-mediated mitochondria injury. Oncogene. 2016 doi: 10.1038/onc.2016.357. [DOI] [PubMed] [Google Scholar]
- 42.Makena M. R., Koneru B., Nguyen T. H., Kang M. H., Reynolds C. P. Reactive oxygen species-mediated synergism of fenretinide and romidepsin in preclinical models of T-cell lymphoid malignancies. Molecular Cancer Therapeutics. 2017 doi: 10.1158/1535-7163.MCT-16-0749. [DOI] [PubMed] [Google Scholar]
- 43.Kirshner J. R., He S., Balasubramanyam V., et al. Elesclomol induces cancer cell apoptosis through oxidative stress. Molecular Cancer Therapeutics. 2008;7(8):2319–2327. doi: 10.1158/1535-7163.MCT-08-0298. [DOI] [PubMed] [Google Scholar]
- 44.O'Day S., Gonzalez R., Lawson D., et al. Phase II, randomized, controlled, double-blinded trial of weekly elesclomol plus paclitaxel versus paclitaxel alone for stage IV metastatic melanoma. Journal of Clinical Oncology. 2009;27(32):5452–5458. doi: 10.1200/JCO.2008.17.1579. [DOI] [PubMed] [Google Scholar]
- 45.O'Day S. J., Eggermont A. M., Chiarion-Sileni V., et al. Final results of phase III SYMMETRY study: randomized, double-blind trial of elesclomol plus paclitaxel versus paclitaxel alone as treatment for chemotherapy-naive patients with advanced melanoma. Journal of Clinical Oncology. 2013;31(9):1211–1218. doi: 10.1200/JCO.2012.44.5585. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi S. Combination therapy with arsenic trioxide for hematological malignancies. Anti-Cancer Agents in Medicinal Chemistry. 2010;10(6):504–510. doi: 10.2174/1871520611009060504. [DOI] [PubMed] [Google Scholar]
- 47.Kuo K. L., Lin W. C., Ho I. L., et al. 2-methoxyestradiol induces mitotic arrest, apoptosis, and synergistic cytotoxicity with arsenic trioxide in human urothelial carcinoma cells. PloS One. 2013;8(8, article e68703) doi: 10.1371/journal.pone.0071342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruno S., Ghiotto F., Tenca C., et al. N-(4-hydroxyphenyl)retinamide promotes apoptosis of resting and proliferating B-cell chronic lymphocytic leukemia cells and potentiates fludarabine and ABT-737 cytotoxicity. Leukemia. 2012;26(10):2260–2268. doi: 10.1038/leu.2012.98. [DOI] [PubMed] [Google Scholar]
- 49.Lee H. R., Cho J. M., Shin D. H., et al. Adaptive response to GSH depletion and resistance to L-buthionine-(S,R)-sulfoximine: involvement of Nrf2 activation. Molecular and Cellular Biochemistry. 2008;318(1-2):23–31. doi: 10.1007/s11010-008-9853-y. [DOI] [PubMed] [Google Scholar]
- 50.Rotblat B., Grunewald T. G., Leprivier G., Melino G., Knight R. A. Anti-oxidative stress response genes: bioinformatic analysis of their expression and relevance in multiple cancers. Oncotarget. 2013;4(12):2577–2590. doi: 10.18632/oncotarget.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuzu O. F., Noory M. A., Robertson G. P. The role of cholesterol in cancer. Cancer Research. 2016;76(8):2063–2070. doi: 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amelio I., Tsvetkov P. O., Knight R. A., Lisitsa A., Melino G., Antonov A. V. SynTarget: an online tool to test the synergetic effect of genes on survival outcome in cancer. Cell Death and Differentiation. 2016;23(5):p. 912. doi: 10.1038/cdd.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antonov A. V. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Research. 2011;39:W323–W327. doi: 10.1093/nar/gkr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antonov A. V., Krestyaninova M., Knight R. A., Rodchenkov I., Melino G., Barlev N. A. PPISURV: a novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene. 2014;33(13):1621–1628. doi: 10.1038/onc.2013.119. [DOI] [PubMed] [Google Scholar]
- 55.Budczies J., Bockmayr M., Denkert C., et al. Classical pathology and mutational load of breast cancer - integration of two worlds. Journal of Pathology: Clinical Research. 2015;1(4):225–238. doi: 10.1002/cjp2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chabanon R. M., Pedrero M., Lefebvre C., Marabelle A., Soria J. C., Postel-Vinay S. Mutational landscape and sensitivity to immune checkpoint blockers. Clinical Cancer Research. 2016;22(17):4309–4321. doi: 10.1158/1078-0432.CCR-16-0903. [DOI] [PubMed] [Google Scholar]
- 57.Jamieson N. B., Maker A. V. Gene-expression profiling to predict responsiveness to immunotherapy. Cancer Gene Therapy. 2016 doi: 10.1038/cgt.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meucci S., Keilholz U., Tinhofer I., Ebner O. A. Mutational load and mutational patterns in relation to age in head and neck cancer. Oncotarget. 2016;7(43):69188–69199. doi: 10.18632/oncotarget.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy S. J., Hart S. N., Lima J. F., et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology. 2013;145(5):1098–1109. doi: 10.1053/j.gastro.2013.07.049. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCrow J. P., Petersen D. C., Louw M., et al. Spectrum of mitochondrial genomic variation and associated clinical presentation of prostate cancer in South African men. Prostate. 2016;76(4):349–358. doi: 10.1002/pros.23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gelsomino F., Barbolini M., Spallanzani A., Pugliese G., Cascinu S. The evolving role of microsatellite instability in colorectal cancer: a review. Cancer Treatment Reviews. 2016;51:19–26. doi: 10.1016/j.ctrv.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Kim H. S., Ullevig S. L., Zamora D., Lee C. F., Asmis R. Redox regulation of MAPK phosphatase 1 controls monocyte migration and macrophage recruitment. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(41):E2803–E2812. doi: 10.1073/pnas.1212596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bracalente C., Ibañez I. L., Berenstein A., et al. Reprogramming human A375 amelanotic melanoma cells by catalase overexpression: upregulation of antioxidant genes correlates with regression of melanoma malignancy and with malignant progression when downregulated. Oncotarget. 2016;7(27):41154–41171. doi: 10.18632/oncotarget.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waz S., Nakamura T., Hirata K., et al. Structural and kinetic studies of the human nudix hydrolase MTH1 reveal the mechanism for its broad substrate specificity. The Journal of Biological Chemistry. 2017;292(7):2785–2794. doi: 10.1074/jbc.M116.749713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Y., Li H. Functional characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and an E3 ubiquitin ligase. Protein & Cell. 2013;4(2):103–116. doi: 10.1007/s13238-012-2105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bae I., Fan S., Meng Q., et al. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Research. 2004;64(21):7893–7909. doi: 10.1158/0008-5472.CAN-04-1119. [DOI] [PubMed] [Google Scholar]
- 67.Mowla S. N., Lam E. W., Jat P. S. Cellular senescence and aging: the role of B-MYB. Aging Cell. 2014;13(5):773–779. doi: 10.1111/acel.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh M., Igarashi M., Fukuda H., Nakagama H., Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Letters. 2013;328(2):198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 69.Kwok C. T., Leung M. H., Qin J., et al. The forkhead box transcription factor FOXM1 is required for the maintenance of cell proliferation and protection against oxidative stress in human embryonic stem cells. Stem Cell Research. 2016;16(3):651–661. doi: 10.1016/j.scr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 70.Wierstra I. The transcription factor FOXM1 (forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Advances in Cancer Research. 2013;118:97–398. doi: 10.1016/B978-0-12-407173-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 71.Park H. J., Carr J. R., Wang Z., et al. FoxM1, a critical regulator of oxidative stress during oncogenesis. The EMBO Journal. 2009;28(19):2908–2918. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saba R., Alsayed A., Zacny J. P., Dudek A. Z. The role of forkhead box protein M1 in breast cancer progression and resistance to therapy. International Journal of Breast Cancer. 2016;2016:8. doi: 10.1155/2016/9768183.9768183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahn H., Sim J., Abdul R., et al. Increased expression of forkhead box M1 is associated with aggressive phenotype and poor prognosis in estrogen receptor-positive breast cancer. Journal of Korean Medical Science. 2015;30(4):390–397. doi: 10.3346/jkms.2015.30.4.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bergamaschi A., Madak-Erdogan Z., Kim Y. J., Choi Y. L., Lu H., Katzenellenbogen B. S. The forkhead transcription factor FOXM1 promotes endocrine resistance and invasiveness in estrogen receptor-positive breast cancer by expansion of stem-like cancer cells. Breast Cancer Research. 2014;16(5):p. 436. doi: 10.1186/s13058-014-0436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Francis R. E., Myatt S. S., Krol J., et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. International Journal of Oncology. 2009;35(1):57–68. doi: 10.3892/ijo_00000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang C., Chen H., Tan G., et al. FOXM1 promotes the epithelial to mesenchymal transition by stimulating the transcription of slug in human breast cancer. Cancer Letters. 2013;340(1):104–112. doi: 10.1016/j.canlet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 77.Xue J., Lin X., Chiu W. T., et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. The Journal of Clinical Investigation. 2014;124(2):564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahmad A., Wang Z., Kong D., et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Research and Treatment. 2010;122(2):337–346. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- 79.Karadedou C. T., Gomes A. R., Chen J., et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene. 2012;31(14):1845–1858. doi: 10.1038/onc.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Moraes G. N., Delbue D., Silva K. L., et al. FOXM1 targets XIAP and survivin to modulate breast cancer survival and chemoresistance. Cellular Signalling. 2015;27(12):2496–2505. doi: 10.1016/j.cellsig.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 81.Yang D. K., Son C. H., Lee S. K., Choi P. J., Lee K. E., Roh M. S. Forkhead box M1 expression in pulmonary squamous cell carcinoma: correlation with clinicopathologic features and its prognostic significance. Human Pathology. 2009;40(4):464–470. doi: 10.1016/j.humpath.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 82.Halasi M., Pandit B., Wang M., Nogueira V., Hay N., Gartel A. L. Combination of oxidative stress and FOXM1 inhibitors induces apoptosis in cancer cells and inhibits xenograft tumor growth. The American Journal of Pathology. 2013;183(1):257–265. doi: 10.1016/j.ajpath.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tahmasbpour E., Ghanei M., Qazvini A., Vahedi E., Panahi Y. Gene expression profile of oxidative stress and antioxidant defense in lung tissue of patients exposed to sulfur mustard. Mutation Research, Genetic Toxicology and Environmental Mutagenesis. 2016;800-801:12–21. doi: 10.1016/j.mrgentox.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 84.Li Z., Gao Y., Xie D., et al. Activation of vitamin D receptor signaling downregulates the expression of nuclear FOXM1 protein and suppresses pancreatic cancer cell stemness. Clinical Cancer Research. 2015;21(4):844–853. doi: 10.1158/1078-0432.CCR-14-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aytes A., Mitrofanova A., Lefebvre C., et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25(5):638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin S. C., Kao C. Y., Lee H. J., et al. Dysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancer. Nature Communications. 2016;7, article 11418 doi: 10.1038/ncomms11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halasi M., Gartel A. L. Targeting FOXM1 in cancer. Biochemical Pharmacology. 2013;85(5):644–652. doi: 10.1016/j.bcp.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 88.Gartel A. L. Suppression of the oncogenic transcription factor FOXM1 by proteasome inhibitors. Scientifica. 2014;2014:5. doi: 10.1155/2014/596528.596528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jimenez A., Zu W., Rawe V. Y., et al. Spermatocyte/spermatid-specific thioredoxin-3, a novel Golgi apparatus-associated thioredoxin, is a specific marker of aberrant spermatogenesis. The Journal of Biological Chemistry. 2004;279(33):34971–34982. doi: 10.1074/jbc.M404192200. [DOI] [PubMed] [Google Scholar]
- 90.Hirota K., Murata M., Sachi Y., et al. Distinct roles of thioredoxin in the cytoplasm and in the nucleus. A two-step mechanism of redox regulation of transcription factor NF-kappaB. The Journal of Biological Chemistry. 1999;274(39):27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 91.Rubartelli A., Bajetto A., Allavena G., Wollman E., Sitia R. Secretion of thioredoxin by normal and neoplastic cells through a leaderless secretory pathway. The Journal of Biological Chemistry. 1992;267(34):24161–24164. [PubMed] [Google Scholar]
- 92.Manabe Y., Takagi M., Nakamura-Yamada M., et al. Redox proteins are constitutively secreted by skeletal muscle. The Journal of Physiological Sciences. 2014;64(6):401–409. doi: 10.1007/s12576-014-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wangpaichitr M., Sullivan E. J., Theodoropoulos G., et al. The relationship of thioredoxin-1 and cisplatin resistance: its impact on ROS and oxidative metabolism in lung cancer cells. Molecular Cancer Therapeutics. 2012;11(3):604–615. doi: 10.1158/1535-7163.MCT-11-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim Y. C., Masutani H., Yamaguchi Y., Itoh K., Yamamoto M., Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. The Journal of Biological Chemistry. 2001;276(21):18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 95.Song J. S., Cho H. H., Lee B. J., Bae Y. C., Jung J. S. Role of thioredoxin 1 and thioredoxin 2 on proliferation of human adipose tissue-derived mesenchymal stem cells. Stem Cells and Development. 2011;20(9):1529–1537. doi: 10.1089/scd.2010.0364. [DOI] [PubMed] [Google Scholar]
- 96.Prigge J. R., Eriksson S., Iverson S. V., et al. Hepatocyte DNA replication in growing liver requires either glutathione or a single allele of txnrd1. Free Radical Biology and Medicine. 2012;52(4):803–810. doi: 10.1016/j.freeradbiomed.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsuzawa A., Ichijo H. Redox control of cell fate by MAP kinase: physiological roles of ASK1-MAP kinase pathway in stress signaling. Biochimica et Biophysica Acta. 2008;1780(11):1325–1336. doi: 10.1016/j.bbagen.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 98.Saitoh M., Nishitoh H., Fujii M., et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. The EMBO Journal. 1998;17(9):2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanschmann E. M., Hanschmann E. M., Godoy J. R., Berndt C., Hudemann C., Lillig C. H. Thioredoxins, glutaredoxins, and peroxiredoxins—molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxidants & Redox Signaling. 2013;19(13):1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu J., Holmgren A. Thioredoxin system in cell death progression. Antioxidants & Redox Signaling. 2012;17(12):1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 101.Mukherjee A., Martin S. G. The thioredoxin system: a key target in tumour and endothelial cells. The British Journal of Radiology. 2008;81(1):S57–S68. doi: 10.1259/bjr/34180435. [DOI] [PubMed] [Google Scholar]
- 102.Marks P. A. Thioredoxin in cancer--role of histone deacetylase inhibitors. Seminars in Cancer Biology. 2006;16(6):436–443. doi: 10.1016/j.semcancer.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shan W., Zhong W., Zhao R., Oberley T. D. Thioredoxin 1 as a subcellular biomarker of redox imbalance in human prostate cancer progression. Free Radical Biology and Medicine. 2010;49(12):2078–2087. doi: 10.1016/j.freeradbiomed.2010.10.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dong C., Zhang L., Sun R., et al. Role of thioredoxin reductase 1 in dysplastic transformation of human breast epithelial cells triggered by chronic oxidative stress. Scientific Reports. 2016;6, article 36860 doi: 10.1038/srep36860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park B. J., Cha M. K., Kim I. H. Thioredoxin 1 as a serum marker for breast cancer and its use in combination with CEA or CA15-3 for improving the sensitivity of breast cancer diagnoses. BMC Research Notes. 2014;7:p. 7. doi: 10.1186/1756-0500-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Csiki I., Yanagisawa K., Haruki N., et al. Thioredoxin-1 modulates transcription of cyclooxygenase-2 via hypoxia-inducible factor-1alpha in non-small cell lung cancer. Cancer Research. 2006;66(1):143–150. doi: 10.1158/0008-5472.CAN-05-1357. [DOI] [PubMed] [Google Scholar]
- 107.Park S. J., Kim H. B., Piao C., et al. p53R2 regulates thioredoxin reductase activity through interaction with TrxR2. Biochemical and Biophysical Research Communications. 2017;482(4):706–712. doi: 10.1016/j.bbrc.2016.11.099. [DOI] [PubMed] [Google Scholar]
- 108.Moscow J. A., Schmidt L., Ingram D. T., Gnarra J., Johnson B., Cowan K. H. Loss of heterozygosity of the human cytosolic glutathione peroxidase I gene in lung cancer. Carcinogenesis. 1994;15(12):2769–2773. doi: 10.1093/carcin/15.12.2769. [DOI] [PubMed] [Google Scholar]
- 109.Hu Y. J., Diamond A. M. Role of glutathione peroxidase 1 in breast cancer: loss of heterozygosity and allelic differences in the response to selenium. Cancer Research. 2003;63(12):3347–3351. [PubMed] [Google Scholar]
- 110.Milonski J., Zielinska-Blizniewska H., Olszewski J., Majsterek I., Mrowicka M. DNA damage and oxidant-antioxidant status in blood of patients with head and neck cancer. DNA and Cell Biology. 2015;34(3):213–219. doi: 10.1089/dna.2014.2706. [DOI] [PubMed] [Google Scholar]
- 111.Mandal P. K., Schneider M., Kölle P., et al. Loss of thioredoxin reductase 1 renders tumors highly susceptible to pharmacologic glutathione deprivation. Cancer Research. 2010;70(22):9505–9514. doi: 10.1158/0008-5472.CAN-10-1509. [DOI] [PubMed] [Google Scholar]
- 112.Rodman S. N., Spence J. M., Ronnfeldt T. J., et al. Enhancement of radiation response in breast cancer stem cells by inhibition of thioredoxin- and glutathione-dependent metabolism. Radiation Research. 2016;186(4):385–395. doi: 10.1667/RR14463.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scarbrough P. M., Mapuskar K. A., Mattson D. M., Gius D., Watson W. H., Spitz D. R. Simultaneous inhibition of glutathione- and thioredoxin-dependent metabolism is necessary to potentiate 17AAG-induced cancer cell killing via oxidative stress. Free Radical Biology and Medicine. 2012;52(2):436–443. doi: 10.1016/j.freeradbiomed.2011.10.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang S., Xue J., Zheng J., et al. The superoxide dismutase 1 3'UTR maintains high expression of the SOD1 gene in cancer cells: the involvement of the RNA-binding protein AUF-1. Free Radical Biology and Medicine. 2015;85:33–44. doi: 10.1016/j.freeradbiomed.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Crapo J. D., Oury T., Rabouille C., Slot J. W., Chang L. Y. Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(21):10405–10409. doi: 10.1073/pnas.89.21.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Elchuri S., Oberley T. D., Qi W., et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24(3):367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 117.Brown D. P., Chin-Sinex H., Nie B., Mendonca M. S., Wang M. Targeting superoxide dismutase 1 to overcome cisplatin resistance in human ovarian cancer. Cancer Chemotherapy and Pharmacology. 2009;63(4):723–730. doi: 10.1007/s00280-008-0791-x. [DOI] [PubMed] [Google Scholar]
- 118.Piecuch A., Brzozowa-Zasada M., Dziewit B., et al. Immunohistochemical assessment of mitochondrial superoxide dismutase (MnSOD) in colorectal premalignant and malignant lesions. Przeglad Gastroenterologiczny. 2016;11(4):239–246. doi: 10.5114/pg.2016.57943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vidimar V., Gius D., Chakravarti D., Bulun S. E., Wei J. J., Kim J. J. Dysfunctional MnSOD leads to redox dysregulation and activation of prosurvival AKT signaling in uterine leiomyomas. Science Advances. 2016;2(11, article e1601132) doi: 10.1126/sciadv.1601132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zou X., Santa-Maria C. A., O'Brien J., Gius D., Zhu Y. Manganese superoxide dismutase acetylation and dysregulation, due to loss of SIRT3 activity, promote a luminal B-like breast carcinogenic-permissive phenotype. Antioxidants & Redox Signaling. 2016;25(6):326–336. doi: 10.1089/ars.2016.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bianchi L., Bruzzese F., Leone A., et al. Proteomic analysis identifies differentially expressed proteins after HDAC vorinostat and EGFR inhibitor gefitinib treatments in Hep-2 cancer cells. Proteomics. 2011;11(18):3725–3742. doi: 10.1002/pmic.201100092. [DOI] [PubMed] [Google Scholar]
- 122.Rosato R. R., Kolla S. S., Hock S. K., et al. Histone deacetylase inhibitors activate NF-kappaB in human leukemia cells through an ATM/NEMO-related pathway. The Journal of Biological Chemistry. 2010;285(13):10064–10077. doi: 10.1074/jbc.M109.095208. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Raninga P. V., Di Trapani G., Vuckovic S., Bhatia M., Tonissen K. F. Inhibition of thioredoxin 1 leads to apoptosis in drug-resistant multiple myeloma. Oncotarget. 2015;6(17):15410–15424. doi: 10.18632/oncotarget.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xie L., Luo Z., Zhao Z., Chen T. Anticancer and antiangiogenic iron(II) complexes that target thioredoxin reductase to trigger cancer cell apoptosis. Journal of Medicinal Chemistry. 2017;60(1):202–214. doi: 10.1021/acs.jmedchem.6b00917. [DOI] [PubMed] [Google Scholar]
- 125.Hwang-Bo H., Cheong J., Kim W. J., Yoo Y. H., Choi Y. H. Auranofin, an inhibitor of thioredoxin reductase, induces apoptosis in hepatocellular carcinoma Hep3B cells by generation of reactive oxygen species. General Physiology and Biophysics. 2017 doi: 10.4149/gpb_2016043. [DOI] [PubMed] [Google Scholar]
- 126.Ng H. L., Ma X., Chew E. H., Chui W. K. Design, synthesis, and biological evaluation of coupled bioactive scaffolds as potential anticancer agents for dual targeting of dihydrofolate reductase and thioredoxin reductase. Journal of Medicinal Chemistry. 2017;60(5):1734–1745. doi: 10.1021/acs.jmedchem.6b01253. [DOI] [PubMed] [Google Scholar]
- 127.Zhu J. Y., Zhang C. Y., Dai J. J., Rahman K., Zhang H. Diterpenoids with thioredoxin reductase inhibitory activities from Jatropha multifida. Natural Product Research. 2017:1–6. doi: 10.1080/14786419.2017.1315575. [DOI] [PubMed] [Google Scholar]
- 128.Duan D., Zhang J., Yao J., Liu Y., Fang J. Targeting thioredoxin reductase by parthenolide contributes to inducing apoptosis of HeLa cells. The Journal of Biological Chemistry. 2016;291(19):10021–10031. doi: 10.1074/jbc.M115.700591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang J., Peng S., Li X., Liu R., Han X., Fang J. Targeting thioredoxin reductase by plumbagin contributes to inducing apoptosis of HL-60 cells. Archives of Biochemistry and Biophysics. 2017;619:16–26. doi: 10.1016/j.abb.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 130.Schroeder A., Warnken U., Röth D., et al. Targeting thioredoxin-1 by dimethyl fumarate induces ripoptosome-mediated cell death. Scientific Reports. 2017;7, article 43168 doi: 10.1038/srep43168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Medema J. P. Cancer stem cells: the challenges ahead. Nature Cell Biology. 2013;15(4):338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 132.Visvader J. E., Lindeman G. J. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 133.Colak S., Medema J. P. Cancer stem cells—important players in tumor therapy resistance. The FEBS Journal. 2014;281(21):4779–4791. doi: 10.1111/febs.13023. [DOI] [PubMed] [Google Scholar]
- 134.Trachootham D., Zhang H., Zhang W., et al. Effective elimination of fludarabine-resistant CLL cells by PEITC through a redox-mediated mechanism. Blood. 2008;112(5):1912–1922. doi: 10.1182/blood-2008-04-149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lagadinou E. D., Sach A., Callahan K., et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12(3):329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Im C. N., Yun H. H., Yoo H. J., Park M. J., Lee J. H. Enhancement of SOX-2 expression and ROS accumulation by culture of A172 glioblastoma cells under non-adherent culture conditions. Oncology Reports. 2015;34(2):920–928. doi: 10.3892/or.2015.4021. [DOI] [PubMed] [Google Scholar]
- 137.Liu J., Wang Z. Increased oxidative stress as a selective anticancer therapy. Oxidative Medicine and Cellular Longevity. 2015;2015:12. doi: 10.1155/2015/294303.294303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang W., Shen Y., Wei J., Liu F. MicroRNA-153/Nrf-2/GPx1 pathway regulates radiosensitivity and stemness of glioma stem cells via reactive oxygen species. Oncotarget. 2015;6(26):22006–22027. doi: 10.18632/oncotarget.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Peng G., Liu Y. Hypoxia-inducible factors in cancer stem cells and inflammation. Trends in Pharmacological Sciences. 2015;36(6):374–383. doi: 10.1016/j.tips.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang X., Dong J., Jia L., et al. HIF-2-dependent expression of stem cell factor promotes metastasis in hepatocellular carcinoma. Cancer Letters. 2017 doi: 10.1016/j.canlet.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 141.Li Z., Bao S., Wu Q., et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xiang L., Gilkes D. M., Hu H., et al. Hypoxia-inducible factor 1 mediates TAZ expression and nuclear localization to induce the breast cancer stem cell phenotype. Oncotarget. 2014;5(24):12509–12527. doi: 10.18632/oncotarget.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seo E. J., Kim D. K., Jang I. H., et al. Hypoxia-NOTCH1-SOX2 signaling is important for maintaining cancer stem cells in ovarian cancer. Oncotarget. 2016;7(34):55624–55638. doi: 10.18632/oncotarget.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang F., Duan S., Tsai Y., et al. Cisplatin treatment increases stemness through upregulation of hypoxia-inducible factors by interleukin-6 in non-small cell lung cancer. Cancer Science. 2016;107(6):746–754. doi: 10.1111/cas.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang S. W., Zhang Z. G., Hao Y. X., et al. HIF-1alpha induces the epithelial-mesenchymal transition in gastric cancer stem cells through the snail pathway. Oncotarget. 2017;8(6):9535–9545. doi: 10.18632/oncotarget.14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Iriondo O., Rábano M., Domenici G., et al. Distinct breast cancer stem/progenitor cell populations require either HIF1alpha or loss of PHD3 to expand under hypoxic conditions. Oncotarget. 2015;6(31):31721–31739. doi: 10.18632/oncotarget.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chefetz I., Holmberg J. C., Alvero A. B., Visintin I., Mor G. Inhibition of aurora-a kinase induces cell cycle arrest in epithelial ovarian cancer stem cells by affecting NFkB pathway. Cell Cycle. 2011;10(13):2206–2214. doi: 10.4161/cc.10.13.16348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ryoo I. G., Lee S. H., Kwak M. K. Redox modulating NRF2: a potential mediator of cancer stem cell resistance. Oxidative Medicine and Cellular Longevity. 2016;2016:14. doi: 10.1155/2016/2428153.2428153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Merchant A. A., Singh A., Matsui W., Biswal S. The redox-sensitive transcription factor Nrf2 regulates murine hematopoietic stem cell survival independently of ROS levels. Blood. 2011;118(25):6572–6579. doi: 10.1182/blood-2011-05-355362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lu B. C., Li J., Yu W. F., Zhang G. Z., Wang H. M., Ma H. M. Elevated expression of Nrf2 mediates multidrug resistance in CD133+ head and neck squamous cell carcinoma stem cells. Oncology Letters. 2016;12(6):4333–4338. doi: 10.3892/ol.2016.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jia Y., Chen J., Zhu H., Jia Z. H., Cui M. H. Aberrantly elevated redox sensing factor Nrf2 promotes cancer stem cell survival via enhanced transcriptional regulation of ABCG2 and Bcl-2/Bmi-1 genes. Oncology Reports. 2015;34(5):2296–2304. doi: 10.3892/or.2015.4214. [DOI] [PubMed] [Google Scholar]
- 152.Ryoo I. G., Choi B. H., Kwak M. K. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget. 2015;6(10):8167–8184. doi: 10.18632/oncotarget.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mizuno T., Suzuki N., Makino H., et al. Cancer stem-like cells of ovarian clear cell carcinoma are enriched in the ALDH-high population associated with an accelerated scavenging system in reactive oxygen species. Gynecologic Oncology. 2015;137(2):299–305. doi: 10.1016/j.ygyno.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 154.Chiche A., Moumen M., Romagnoli M., et al. p53 deficiency induces cancer stem cell pool expansion in a mouse model of triple-negative breast tumors. Oncogene. 2016 doi: 10.1038/onc.2016.396. [DOI] [PubMed] [Google Scholar]
- 155.Xie X., Kaoud T. S., Edupuganti R., et al. c-Jun N-terminal kinase promotes stem cell phenotype in triple-negative breast cancer through upregulation of Notch1 via activation of c-Jun. Oncogene. 2016 doi: 10.1038/onc.2016.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Storz P. Forkhead homeobox type O transcription factors in the responses to oxidative stress. Antioxidants & Redox Signaling. 2011;14(4):593–605. doi: 10.1089/ars.2010.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ferber E. C., Peck B., Delpuech O., Bell G. P., East P., Schulze A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death and Differentiation. 2012;19(6):968–979. doi: 10.1038/cdd.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lehtinen M. K., Yuan Z., Boag P. R., et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125(5):987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 159.Somasekharan S. P., El-Naggar A., Leprivier G., et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. The Journal of Cell Biology. 2015;208(7):913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nature Cell Biology. 2008;10(11):1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 161.Moeller B. J., Cao Y., Li C. Y., Dewhirst M. W. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5(5):429–441. doi: 10.1016/S1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 162.Fournier M. J., Gareau C., Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell International. 2010;10:p. 12. doi: 10.1186/1475-2867-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Kaplan Meier curves showing the survival in the case of high and low expression of DUSP1, EPHX2, NUDT1, RNF7 and SEPP1 in solid cancers patients. Supplementary Figure S2 STRING analysis of modulated oxidative stress genes in six different type of cancer. Supplementary Figure S3 STRING analysis of oxidative stress genes in six different type of cancer: high vs low network.