Abstract
Transmembrane Protein 39A (TMEM39A) is a member of TMEM family. The understanding about this protein is still limited. The earlier studies indicated that TMEM39A was a key mediator of autoimmune disease. TMEM39A seems to be involved in systemic lupus erythematosus and multiple sclerosis in numerous of populations. All of these works stop at insufficient information by using gene functioning methods such as: Genome-wide association studies (GWASs) and/or follow-up study. It is the fact that the less understood of TMEM39A actually is the attraction to the scientist in near future. In this review the current knowledge about TMEM39A and its possible roles in cell biology, physiology and pathology will be described.
Keywords: Transmembrane proteins family, Multiple sclerosis, Systemic lupus erythematosus, Mitochondria, Mitophagy
INTRODUCTION
A transmembrane protein (TMEM) is a kind of integral membrane protein that exists in the total biological membrane. TMEM family is characterized by a presence of putative transmembrane domains (1). Many TMEMs works as gateways to allow the transport of specific elements through the biological membrane. TMEMs repeatedly experience important conformational changes to transfer a substance through the membrane. The understanding about functions and localization of this family is still unclear at the present moments.
TRANSMEMBRANE PROTEIN FAMILY
TMEMs are predicted to be components of cellular membranes, such as mitochondrial membranes, endoplasmic reticulum (ER), lysosomes and golgi apparatus (2). Indeed, the localization of some protein members was proved by biochemical evidences, including TMEM22 (1) and TMEM45A (3). The functions of several TMEMs in cellular processing were also primarily described. TMEM16A, TMEM16F and TMEM16K were identified as the calcium-activated chloride channel (4,5). Remarkably, TMEM45A firstly was identified as effector of hypoxia condition in 2007 (6). Later on TMEM45A was reported to involve in epidermal keratinization (3). These biological functions implicated that TMEM45A may contribute to cancer development. Indeed, TMEM45A is essential for hypoxia-induced chemo-resistance in breast and liver cancers (7). Consistently, downregulation of TMEM45A suppressed many type of cancers by inhibiting the proliferation, migration and cell invasion. The progression of ductal carcinoma xenografts in situ to invasive breast cancer was dramatically increased by suppressing TMEM45A, which may due to its adhesion regulating effect (8). Similar effects were then observed in ovarian cancer (9) and glioma cells (10). Differential regulation of other TMEM members could be also observed in many cancers, such as lymphomas (TMEM176) (11), colorectal cancer (TMEM25) (12), meningiomas (TMEM30B) (13), renal cell carcinoma (different TMEMs) (2,14–20), paragangliomas and pheochromocytomas (TMEM127) (21), suggesting that TMEM family is prominent group for further cancer research. However there are vast different characteristic and localization between the members of TMEMs. Thus, the biological functions of each candidate member need to be elucidated in detail.
TMEM39A- THE FURTHER FOCUS
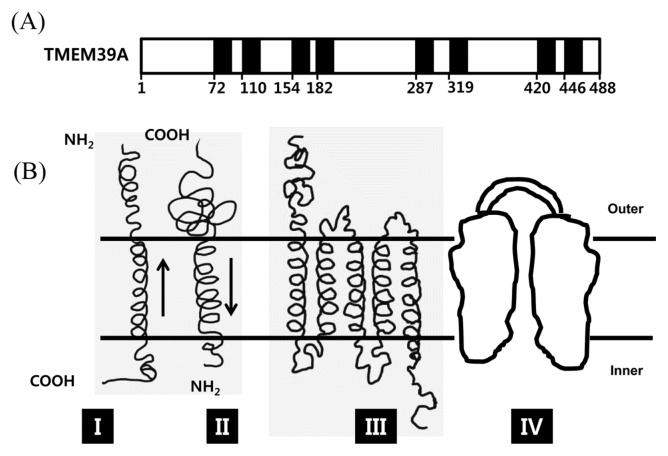
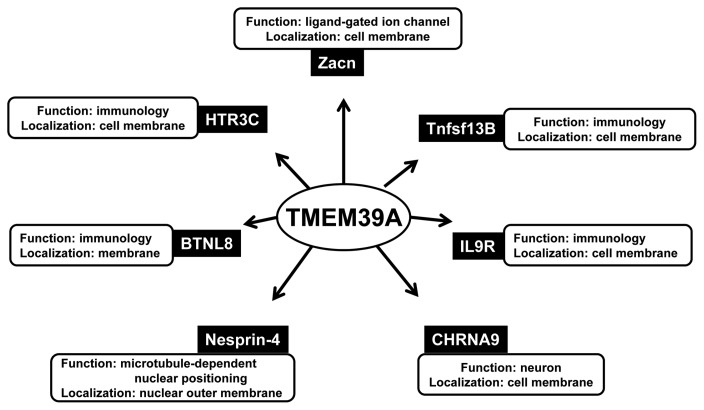
The function of TMEM39A is poorly identified in the member of TMEMs. It contains 488 amino acid and 8 transmembrane helix segments in its sequence (Based on Uniprot database; Fig. 1A). Employing Singer’s classification of proteins, TMEM39A is classified as type 3-transmembrane protein (Fig. 1B) (22). The function of TMEM39A was first revealed in 2010 by the international multiple sclerosis genetics consortium (23). TMEM39A was identified as susceptibility loci of multiple sclerosis. This discovery was again confirmed by Varade et al (24). Multiple sclerosis is the most common neurological disease among young adults, with onset at a mean age of 30 years. It is an autoimmune condition of the central nervous system affecting over 2 million individuals world-wide (23,24). By also using a genome-wide association (GWA) study, recent studies indicated that TMEM39A involved in Erythematosus, which is considered as autoimmune disease (25–27). These implicated that TMEM39A had an important contribution in immune system such as: inflammation, dysregulated type 1 interferon responses, and autoantibodies directed to the nuclear compartment which were characteristics of erythematosus. TMEM39A was found to be bound to seven baits (Fig. 2) (28). Among all the baits, Tumor necrosis factor ligand superfamily member 13B (Tnfsf13B), Butyrophilin-like protein 8 (BTNL8) and 5-hydroxytryptamine receptor 3C (HTR3C), Interleukin-9 receptor (IL9R) were involved in immune system. Thus the further study is required to demonstrate the molecular mechanism of TMEM39A in immune response.
Fig. 1.
TMEM39A is predicted as type III membrane protein. (A) Schematic of TMEM39A topology analyzing in Uniprot database. TMEM39A contains eight transmembrane domains starting at residues: 72, 110, 154, 182, 287, 319, 420, 446. (B) The topography of the four main types (I-IV) of integral membrane proteins. The external and cytoplasmic surfaces of the membrane are designated by outer and inner respectively. (Adopted from (22)).
Fig. 2.
The network showing interactions of TMEM39A with its interactors. TMEM39A was found to be interacted with several protein baits (28). The major function and localization of each interactor were also investigated. Among seven interactor, five proteins including Zacn, Tnfsf13B, IL9R, CHRNA9 and HTR3C are located in cell membrane. Nesprin-4 locates to nuclear outer membrane and BTNL8 is just predicted to be in membrane. Regarding to the functions of those interactors, there are 4 proteins involved in immunology including Tnfsf13B, IL9R, BTNL8, and HTR3C. Other proteins have the following function; Zacn - ion channel, CHRNA9 - neuron development, Nesprin-4 - nuclear positioning. (BTNL8: Butyrophilin-like protein 8, CHRNA9: Neuronal acetylcholine receptor subunit alpha-9, HTR3C: 5-hydroxytryptamine receptor 3C, IL9R: Interleukin-9 receptor, Tnfsf13b: Tumor necrosis factor ligand superfamily member 13B, Zacn: Zinc-activated ligand-gated ion channel).
TMEM39A was significantly changed in sclerosis-a neurological disease. Cholinergic receptor nicotinic alpha 9 subunit (CHRNA9), an interactor of TMEM39A (29), also functioned in neuronal activity. In brain, the functions, dynamics and homeostasis of mitochondria are really important and associating with several neuronal disorder diseases. Mitochondria dose not only provide ATP but also perform a variety of roles in processes such as the transduction of metabolic and stress signals. Thereby mitochondria was a factor deciding the cell fate. The quality and quantity controls of mitochondrial thus were highlighted. Mitophagy, a key process to remove damaged mitochondria, was well known to be regulated by Pink/Parkin pathway which linked to the neurodegenerative diseases. Mutations in these protein encoded-genes defected mitophagy leading to accumulation of damage mitochondria, resulting in neuronal cell death (30). Taken these evidences raised a question that: is there any contribution of TMEM39A in mitochondria, mitophagy and other molecular signaling in neuron? Indeed, image-based genome-wide siRNA screen study showed that TMEM39 was potential factor involved in Parkin-mediated mitophagy (31). However the molecular mechanism for this phenomenon has not been provided yet. In addition, it has been found that TMEM39A was mutated in breast cancer (32). Moreover, mitophagy and mitochondria were considered as the important modulators of tumorigenesis and development (33–35). Note that in TMEM family, TMEM59 was known as regulator of neuron stem cell during differentiation (36) as well as an Alzheimer’s related protein (37,38). Interestingly, Emilio et al. showed that TMEM59 binds to ATG16L and promotes local activation LC3 (39). This indicated for a connection between a trans-membrane protein with autophagy and neuronal disease that can also be happened in case of TMEM39A. From these all information, the relationship between TMEM39A and brain tumor-GBM, autophagy may become a good point for further study.
In other aspect, the discovery of TMEM39A role in mitochondria pointed that intracellular location of TMEM39A is now also in needed and mitochondria might be a possibility. However five of seven interactors identified by Hunttlin et al. (28) were cell membrane proteins or were predicted to be cell membrane proteins (Fig. 2). This implicates that TMEM39A may be in cell membrane and thus may have more change to interact with those interactors.
CONCLUSION REMARKS
The function of TMEM39A is poorly understood. There is no biochemical evidence to bring this protein into the brightness. However, the genetic studies have discovered that TMEM39A may contribute to Multiple Sclerosis and Erythematosus, the autoimmune diseases. Base on all of our knowledge about TMEM39A, this review has described and connected its characteristics with cellular biology topics, therefore providing some promising points for TMEM39A study in future:
The molecular mechanism of TMEM39A in regulating the autoimmune diseases. Investigation of some of TMEM39A interactors such as Tnfsf13B, BTNL8 and HTR3C, IL9R might provide the interesting results with TMEM39A in autoimmune diseases.
The function of TMEM39A in mitochondria and mitophagy has been suggested. Thus, the further study on the effects of TMEM39A on mitophagy in neurobiology and brain cancer will be required.
The exact localization of TMEM39A is urgently required to investigate the function of TMEM39A. TMEM39A might localize to plasma membrane since most of its interactors are plasma membrane proteins. However mitochondrial localization of TMEM39A is also possible, due to the effects of TMEM39A on mitophagy.
ACKNOWLEDGMENTS
This work was financially supported by research fund of Chungnam National University in 2014 (Seon-Hwan Kim) and by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MEST) (NRF-2012M3A9B6055302, NRF-2014R1A1A3050752, NRF-2015R1A2A2A01003597, NRF-2015R1D1A3A01015694).
REFERENCES
- 1.Dobashi S, Katagiri T, Hirota E, Ashida S, Daigo Y, Shuin T, Fujioka T, Miki T, Nakamura Y. Involvement of TMEM22 overexpression in the growth of renal cell carcinoma cells. Oncol Rep. 2009;21:305–312. [PubMed] [Google Scholar]
- 2.Wrzesinski T, Szelag M, Cieslikowski WA, Ida A, Giles R, Zodro E, Szumska J, Pozniak J, Kwias Z, Bluyssen HA, Wesoly J. Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer. 2015;15:518. doi: 10.1186/s12885-015-1530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayez A, Malaisse J, Roegiers E, Reynier M, Renard C, Haftek M, Geenen V, Serre G, Simon M, de Rouvroit CL, Michiels C, Poumay Y. High TMEM45A expression is correlated to epidermal keratinization. Exp Dermatol. 2014;23:339–344. doi: 10.1111/exd.12403. [DOI] [PubMed] [Google Scholar]
- 4.Ferrera L, Caputo A, Galietta LJ. TMEM16A protein: A new identity for Ca2+-dependent Cl− channels. Physiology (Bethesda) 2010;25:357–363. doi: 10.1152/physiol.00030.2010. [DOI] [PubMed] [Google Scholar]
- 5.Ishihara K, Suzuki J, Nagata S. Role of Ca2+ in the Stability and Function of TMEM16F and 16K. Biochemistry. 2016;55:3180–3188. doi: 10.1021/acs.biochem.6b00176. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Rendon E, Hale SJ, Ryan D, Baban D, Forde SP, Roubelakis M, Sweeney D, Moukayed M, Harris AL, Davies K, Watt SM. Transcriptional profiling of human cord blood CD133+ and cultured bone marrow mesenchymal stem cells in response to hypoxia. Stem Cells. 2007;25:1003–1012. doi: 10.1634/stemcells.2006-0398. [DOI] [PubMed] [Google Scholar]
- 7.Flamant L, Roegiers E, Pierre M, Hayez A, Sterpin C, De Backer O, Arnould T, Poumay Y, Michiels C. TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer. 2012;12:391. doi: 10.1186/1471-2407-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, Medina D, Allred DC. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 2012;72:4574–4586. doi: 10.1158/0008-5472.CAN-12-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J, Chen L, Luo N, Yang W, Qu X, Cheng Z. Inhibition of TMEM45A suppresses proliferation, induces cell cycle arrest and reduces cell invasion in human ovarian cancer cells. Oncol Rep. 2015;33:3124–3130. doi: 10.3892/or.2015.3902. [DOI] [PubMed] [Google Scholar]
- 10.Sun W, Qiu G, Zou Y, Cai Z, Wang P, Lin X, Huang J, Jiang L, Ding X, Hu G. Knockdown of TMEM45A inhibits the proliferation, migration and invasion of glioma cells. Int J Clin Exp Pathol. 2015;8:12657–12667. [PMC free article] [PubMed] [Google Scholar]
- 11.Cuajungco MP, Podevin W, Valluri VK, Bui Q, Nguyen VH, Taylor K. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 2012;114:705–712. doi: 10.1016/j.acthis.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrasovec S, Hauptman N, Glavac D, Jelenc F, Ravnik-Glavac M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis Markers. 2013;34:93–104. doi: 10.1155/2013/427890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Magan E, Campos-Martin Y, Mur P, Fiano C, Ribalta T, Garcia JF, Rey JA, Rodriguez de Lope A, Mollejo M, Melendez B. Genetic alterations associated with progression and recurrence in meningiomas. J Neuropathol Exp Neurol. 2012;71:882–893. doi: 10.1097/NEN.0b013e31826bf704. [DOI] [PubMed] [Google Scholar]
- 14.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WG, Jr, Signoretti S. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cifola I, Spinelli R, Beltrame L, Peano C, Fasoli E, Ferrero S, Bosari S, Signorini S, Rocco F, Perego R, Proserpio V, Raimondo F, Mocarelli P, Battaglia C. Genome-wide screening of copy number alterations and LOH events in renal cell carcinomas and integration with gene expression profile. Mol Cancer. 2008;7:6. doi: 10.1186/1476-4598-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, Sun LZ, Ahlquist DA, Wood CG, Copland JA. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 17.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, Jonas D, Libermann TA. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 18.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ, Copland JA. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS ONE. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Roche O, Yan MS, Finak G, Evans AJ, Metcalf JL, Hast BE, Hanna SC, Wondergem B, Furge KA, Irwin MS, Kim WY, Teh BT, Grinstein S, Park M, Marsden PA, Ohh M. Regulation of endocytosis via the oxygen-sensing pathway. Nat Med. 2009;15:319–324. doi: 10.1038/nm.1922. [DOI] [PubMed] [Google Scholar]
- 20.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O’Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abermil N, Guillaud-Bataille M, Burnichon N, Venisse A, Manivet P, Guignat L, Drui D, Chupin M, Josseaume C, Affres H, Plouin PF, Bertherat J, Jeunemaitre X, Gimenez-Roqueplo AP. TMEM127 screening in a large cohort of patients with pheochromocytoma and/or paraganglioma. J Clin Endocrinol Metab. 2012;97:E805–E809. doi: 10.1210/jc.2011-3360. [DOI] [PubMed] [Google Scholar]
- 22.Singer SJ. The structure and insertion of integral proteins in membranes. Annu Rev Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- 23.International Multiple Sclerosis Genetics Consortium (IMSGC) Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum Mol Genet. 2010;19:953–962. doi: 10.1093/hmg/ddp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varade J, Comabella M, Ortiz MA, Arroyo R, Fernandez O, Pinto-Medel MJ, Fedetz M, Izquierdo G, Lucas M, Gomez CL, Rabasa AC, Alcina A, Matesanz F, Alloza I, Antiguedad A, Garcia-Barcina M, Otaegui D, Olascoaga J, Saiz A, Blanco Y, Montalban X, Vandenbroeck K, Urcelay E. Replication study of 10 genes showing evidence for association with multiple sclerosis: validation of TMEM39A, IL12B and CBLB [correction of CLBL] genes. Mult Scler. 2012;18:959–965. doi: 10.1177/1352458511432741. [DOI] [PubMed] [Google Scholar]
- 25.Sheng YJ, Xu JH, Wu YG, Zuo XB, Gao JP, Lin Y, Zhu ZW, Wen LL, Yang C, Liu L, Cheng YY, Chang Y, Yang LL, Zhou FS, Tang XF, Zheng XD, Yin XY, Tang HY, Sun LD, Cui Y, Yang S, Zhang XJ. Association analyses confirm five susceptibility loci for systemic lupus erythematosus in the Han Chinese population. Arthritis Res Ther. 2015;17:85. doi: 10.1186/s13075-015-0602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You Y, Zhai ZF, Chen FR, Chen W, Hao F. Autoimmune risk loci of IL12RB2, IKZF1, XKR6, TMEM39A and CSK in Chinese patients with systemic lupus erythematosus. Tissue Antigens. 2015;85:200–203. doi: 10.1111/tan.12522. [DOI] [PubMed] [Google Scholar]
- 27.Lessard CJ, Adrianto I, Ice JA, Wiley GB, Kelly JA, Glenn SB, Adler AJ, Li H, Rasmussen A, Williams AH, Ziegler J, Comeau ME, Marion M, Wakeland BE, Liang C, Ramos PS, Grundahl KM, Gallant CJ, Alarcon-Riquelme ME, Alarcon GS, Anaya JM, Bae SC, Boackle SA, Brown EE, Chang DM, Cho SK, Criswell LA, Edberg JC, Freedman BI, Gilkeson GS, Jacob CO, James JA, Kamen DL, Kimberly RP, Kim JH, Martin J, Merrill JT, Niewold TB, Park SY, Petri MA, Pons-Estel BA, Ramsey-Goldman R, Reveille JD, Scofield RH, Song YW, Stevens AM, Tsao BP, Vila LM, Vyse TJ, Yu CY, Guthridge JM, Kaufman KM, Harley JB, Wakeland EK, Langefeld CD, Gaffney PM, Montgomery CG, Moser KL BIOLUPUS Network; GENLES Network. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. American Journal of Human Genetics. 2012;90:648–660. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. The bioplex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lips KS, Pfeil U, Kummer W. Coexpression of alpha 9 and alpha 10 nicotinic acetylcholine receptors in rat dorsal root ganglion neurons. Neuroscience. 2002;115:1–5. doi: 10.1016/S0306-4522(02)00274-9. [DOI] [PubMed] [Google Scholar]
- 30.Son JH, Shim JH, Kim KH, Ha JY, Han JY. Neuronal autophagy and neurodegenerative diseases. Exp Mol Med. 2012;44:89–98. doi: 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 33.Palikaras K, Lionaki E, Tavernarakis N. Mitophagy: In sickness and in health. Mol Cell Oncol. 2016;3:e1056332. doi: 10.1080/23723556.2015.1056332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senft D, Ronai ZA. Regulators of mitochondrial dynamics in cancer. Curr Opin Cell Biol. 2016;39:43–52. doi: 10.1016/j.ceb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernardini JP, Lazarou M, Dewson G. Parkin and mitophagy in cancer. Oncogene. 2016;36:1315–1327. doi: 10.1038/onc.2016.302. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Ju X, Cheng Y, Guo X, Wen T. Identifying Tmem59 related gene regulatory network of mouse neural stem cell from a compendium of expression profiles. BMC Syst Biol. 2011;5:152. doi: 10.1186/1752-0509-5-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ullrich S, Munch A, Neumann S, Kremmer E, Tatzelt J, Lichtenthaler SF. The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J Biol Chem. 2010;285:20664–20674. doi: 10.1074/jbc.M109.055608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guttula SV, Allam A, Gumpeny RS. Analyzing microarray data of Alzheimer’s using cluster analysis to identify the biomarker genes. Int J Alzheimers Dis. 2012;2012:649456. doi: 10.1155/2012/649456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boada-Romero E, Letek M, Fleischer A, Pallauf K, Ramon-Barros C, Pimentel-Muinos FX. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 2013;32:566–582. doi: 10.1038/emboj.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]