SUMMARY
The mammalian telomere-binding protein Rap1 was recently found to have additional nontelomeric functions, acting as a transcriptional cofactor and a regulator of the NF-κB pathway. Here, we assess the effect of disrupting mouse Rap1 in vivo and report on its unanticipated role in metabolic regulation and body-weight homeostasis. Rap1 inhibition causes dysregulation in hepatic as well as adipose function, leading to glucose intolerance, insulin resistance, liver steatosis, and excess fat accumulation. Furthermore, Rap1 appears to play a pivotal role in the transcriptional cascade that controls adipocyte differentiation in vitro. Using a separation-of-function allele, we show that the metabolic function of Rap1 is independent of its recruitment to TTAGGG binding elements found at telomeres and at other interstitial loci. In conclusion, our study underscores an additional function for the most conserved telomere-binding protein, forging a link between telomere biology and metabolic signaling.
INTRODUCTION
Telomeres are specialized nucleoprotein structures that protect the ends of eukaryotic chromosomes from the perils of the DNA double-stranded break repair activities, thereby ensuring genomic stability and cellular viability. In mammalian cells, telomeric DNA is bound by shelterin, a specialized six-subunit complex composed of three proteins with DNA-binding affinities (TRF1, TRF2, and POT1), two anchoring proteins (TIN2 and TPP1), and one additional member (Rap1), which is a TRF2-interacting partner (de Lange, 2005). Rap1, also known as TERF2ip, is unique among the different shelterin subunits in that it is the most conserved telomere binding protein from yeast to humans (Li et al., 2000). The evolutionary conservation of Rap1 is mostly at the level of domain architecture, featuring an N-terminal BRCT domain that is followed by one (mammals) or two (budding yeast) central Myb DNA-binding motifs and a highly conserved protein-protein interaction domain (RCT) at the C terminus (Li et al., 2000). As a result of these multiple protein-protein and protein-DNA interaction modules, Rap1 can act as an adaptor protein, participating in different complexes with various functions (Shore, 1994; Kabir et al., 2010). Accordingly, the function of Rap1 in different organisms is highly diverse and not merely constrained to telomeric chromatin.
Rap1 was first identified in Saccharomyces cerevisiae as a transcriptional regulator that binds to the upstream activation site of the MAT alpha locus and ribosomal protein genes (Shore and Nasmyth, 1987; Kurtz and Shore, 1991). Depending on the context of its DNA binding, it can act either as a transcriptional activator or repressor. Later studies found it to be the primary double-stranded telomere binding protein involved in telomere length regulation (Lustig et al., 1990; Hardy et al., 1992) and transcriptional silencing of subtelomeric genes (Kyrion et al., 1993). Its key function in budding yeast is to protect chromosome ends from end-end fusion by inhibiting the nonhomologous end-joining (NHEJ) repair pathway at telomeres (Pardo and Marcand, 2005). In Schizosaccharomyces pombe, Rap1 performs similar functions but lacks DNA binding activity, thereby relying on its interaction with the telomere-associated protein Taz1 to localize to chromosome ends (Kanoh and Ishikawa, 2001).
Recent in vitro studies have suggested that mammalian Rap1 can bind to DNA directly in a sequence-independent manner (Arat and Griffith, 2012). However, in vivo analyses in both human and mouse cells have demonstrated that Rap1 relies completely on its interaction with TRF2 in order to bind TTAGGG-bearing telomere repeats. Furthermore, the TRF2-Rap1 association is crucial for the stability of endogenous Rap1 protein; as evidenced in the absence of TRF2, where Rap1 levels in the nucleus are greatly diminished (Li et al., 2000; Celli and de Lange, 2005). The chromosome end-protective function of mammalian Rap1 was revealed by loss-of-function analysis in mouse cells, whereby Rap1 was shown to be dispensable for the suppression of DNA damage signaling, as well as DNA double-stranded break repair by NHEJ (Martinez et al., 2010; Sfeir et al., 2010). Instead, in the absence of Ku70/80, Rap1 inhibition was found to unleash homology-directed repair (HDR) at telomeres (Sfeir et al., 2010).
The extratelomeric functions of mammalian Rap1 were only recently revealed by two key discoveries. The first was by Martinez et al., who identified Rap1 as a transcriptional repressor/activator and detected several metabolic, cancer-related, and imprinting genes that were deregulated upon deleting Rap1 from mouse embryonic fibroblasts (MEFs). Furthermore, chromatin immunoprecipitation sequencing (ChIP-seq) analysis identified the binding of Rap1 to numerous extratelomeric sites, including regions that are proximal to Rap1-regulated genes (Martinez et al., 2010). Rap1 often associates with nontelomeric sites containing a (TTAGGG)2 consensus motif that is recognized by its interacting partner, TRF2. A second, nontelomeric function for Rap1 was revealed in the cytoplasm, where it acts as a modulator of the NF-κB signaling pathway by partnering with IKB kinase (IKK) to degrade p65, an inhibitor of NF-κB (Teo et al., 2010).
To test the role of Rap1 in vivo, we have analyzed the phenotypes associated with Rap1 loss. Here, we report on an unanticipated function of Rap1 in controlling body weight and regulating metabolism. Rap1 knockout mice display a phenotype characterized by obesity, glucose intolerance, insulin resistance, liver steatosis, and adipose tissue hypertrophy. Dysregulated hepatic function and adipose tissue expansion were preceded by significant alterations in gene expression. Lastly, using a Rap1 mutant that is incapable of binding TRF2, we show that the effect of Rap1 on transcriptional control is independent of its recruitment to TTAGGG-bearing sequences.
RESULTS
Mature-Onset Obesity in Mice Lacking Rap1
Inactivation of Rap1 in vivo was achieved by breeding mice carrying a Rap1 floxed allele with Ella-Cre transgenic mice to generate a Rap1 knockout allele (Sfeir et al., 2010). We assessed the biological function of Rap1 by analyzing offspring from intercrosses of heterozygote mice. Since Rap1+/+ and Rap1−/+ mice displayed no significant difference in phenotype, we pooled their cohorts and compared wild-type (WT) controls (Rap1+/+ and Rap1−/+, referred to as Rap1-WT) to littermate knockout mice (Rap1−/−, referred to Rap1-KO). The loss of Rap1 was confirmed by genotyping PCR (Figure S1A) and western blot analysis (Figure S1B). As we previously reported, mice lacking Rap1 were alive and fertile (Sfeir et al., 2010). Rap1 null mice sustained no telomere dysfunction (Figure S2) and displayed no signs of accelerated aging within the first year of life (data not shown). Surprisingly, Rap1-KO mice displayed a gradual increase in body weight, leading to adult-onset obesity (Figure 1A). To establish a chronology of overweight occurrence in the absence of Rap1, we performed regular weight recordings of individual mice under a standard diet over a 12-month period. By ~22 weeks of age, the body weight of Rap1-deficient females deviated significantly from those of WT littermates (Figure 1B). However, the difference in body weight among males was less pronounced; a durable and significant effect was only established at ~34 weeks of age. Although the underlying basis for the marked sexual dimorphism of weight gain in mice is unknown, this phenotype is similar to what has been previously reported for peroxisome proliferator-activated receptor alpha (PPARα) (Costet et al., 1998) and peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α) (Leone et al., 2005) knockout mice. By comparing the daily food intake, we did not observe an increase in food consumption in Rap1-KO mice when compared to their Rap1-WT counterparts (Figure S1C), thus implicating altered energy expenditure or fuel partitioning as possible causes for the excessive weight gain associated with Rap1 loss. This was supported by careful metabolic assessment indicating a significant increase in fasting blood glucose (Figure S1D) as well as plasma insulin and cholesterol (Figures S1E–S1G) in Rap1-deficient mice as compared to age-matched Rap1-proficient mice. The glucose tolerance test (GTT) and insulin tolerance test (ITT) confirmed that Rap1 deficiency led to glucose intolerance and insulin resistance (Figures 1C–1E). Taken together, these results suggest that Rap1 impairment has adverse metabolic consequences that can lead to obesity in the mouse.
Figure 1. Obese Phenotypes in Rap1-Deficient Mice.
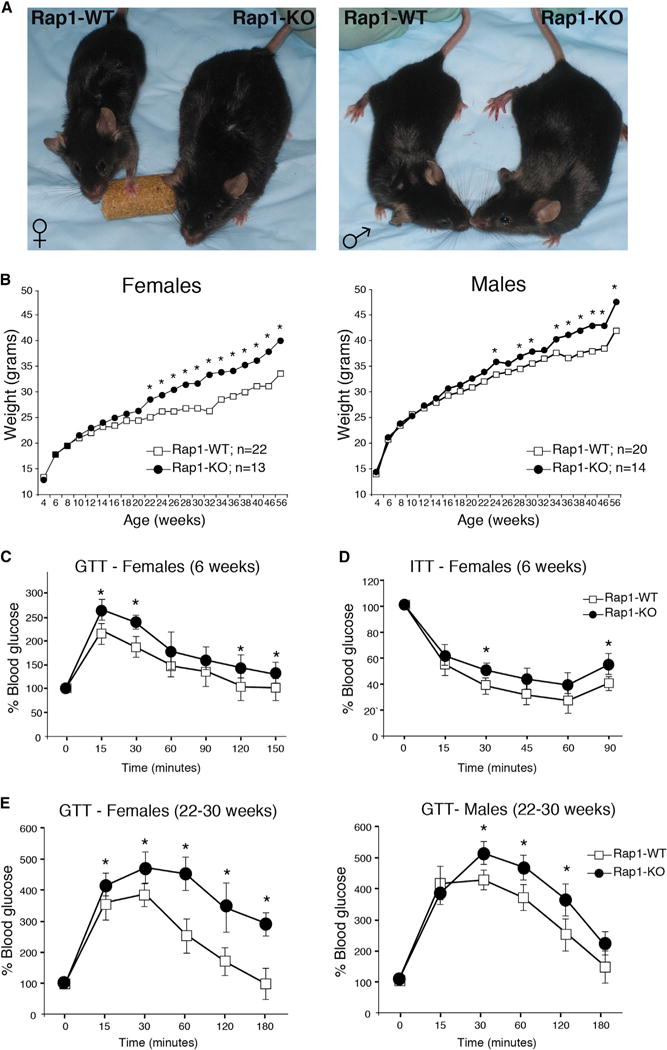
(A) Rap1-KO mice display morphological signs of obesity when compared to Rap1-WT controls. Shown are representative photographs of 24-week-old female (left) and male (right) mice.
(B) Body-weight curves of Rap1-WT (white boxes) and Rap-KO (black circles) mice over a period of 56 weeks. Animals were fed a normal chow diet and weighed once every 2 weeks. Data represent mean value, and p values were calculated with the Student’s t test (*p < 0.05).
(C) Plasma glucose levels during glucose tolerance test (GTT) performed on 6- to 8-week-old female mice. Closed circles represent Rap1-KO mice (n = 5) and open boxes represent WT control mice (n = 5). Values were normalized to t = 0 time point. Data represent mean ±SEM. All p values were calculated with the Student’s t test (*p < 0.05).
(D) Plasma glucose levels during Insulin tolerance test (ITT) performed on 6-8 week-old female mice (n = 5 for each group). p values were calculated by Student’s t test and (*) indicates p < 0.05.
(E) GTT performed on 24–32 week-old females (right panel) and males (left panel) mice. Closed circles represent Rap1-KO mice (n = 15 for females and n = 14 for males) and open boxes represent WT control mice (n = 18 for females and n = 20 for males). Data represent mean ±SEM. All p values were calculated with the Student’s t test (*p < 0.05).
Rap1 Deficiency Leads to Dysregulated White Adipose Tissue Function
Obesity is often coupled to excessive expansion of lipid storing white adipose tissue (WAT), whereas the energy dissipating brown adipose tissue (BAT) inversely correlates with body mass index (Gesta et al., 2007). Surgical dissection of Rap1-KO mice revealed an excess accumulation of subcutaneous as well as intra-abdominal fat (Figures 2A and 3A). To confirm these observations, we measured body composition using a dual-energy X-ray absorption (DEXA) scan and noted a significant increase in the percentage of fat mass in 24- to 32-week-old Rap1-KO mice relative to age-matched controls (Figure 2B). Furthermore, morphological analysis of intra-abdominal WAT from 6-week-old Rap1-KO mice displayed an increase in the size of white adipose cells as compared to WAT from age-matched control mice (Figure 2C). On the other hand, hematoxylin and eosin (H&E) staining of interscapular BAT (iBAT) revealed no difference in the morphology or extent of lipid accumulation in the iBAT of Rap1-KO mice when compared to WT controls (Figure S3A). In addition, 6- to 8-week-old Rap1-KO mice were fully capable of maintaining body temperature when subjected to cold challenge (Figure S3B). Furthermore, consistent with the fact that the thermogenic capacity of mice is unaffected by Rap1, the induction of the Ucp1 gene in iBAT of Rap1-KO mice exposed to cold temperature (4°C) was similar to that of age-matched control mice (Figure S3C).
Figure 2. White Adipose Tissue Dysregulation in the Absence of Rap1.
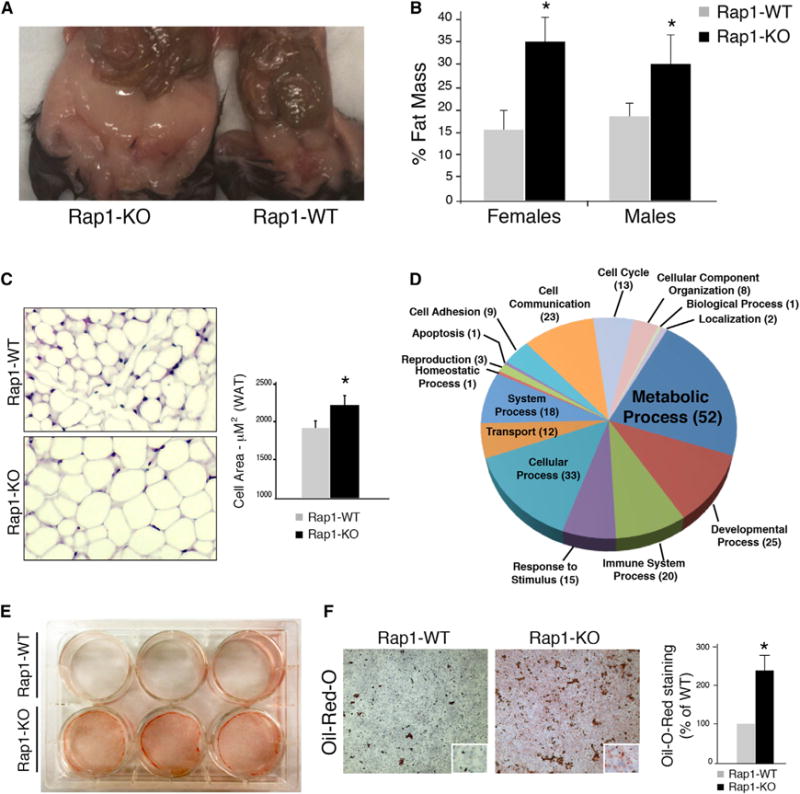
(A) Representative photograph of 40-week-old Rap1-KO (left) and Rap1-WT (right) female mice displaying WAT accumulation.
(B) A DEXA scan was performed on 24- to 32-week-old Rap1-WT mice. Fat percentage is represented as average values ±SEM. The Student’s t test was performed for significance and (*p < 0.05).
(C) Shown are representative sections of intra-abdominal WAT from 6-week-old Rap1-WT and Rap1-KO mice stained with H&E. Morphometric analysis of white fat cells was carried out using MetaMorph. Cell area was quantified for each genotype and values represent mean ±SEM.
(D) Gene Ontology analysis was performed to classify differentially expressed genes from intra-abdominal WAT of 6-week-old Rap1-KO and Rap1-WT female mice into biological processes.
(E) Transdifferentiation of MEFs isolated from Rap1-KO and Rap1-WT mice into adipocytes. Lipid accumulation is visualized using oil red O staining.
(F) Shown is a representative image of a transdifferentiated adipocyte stained with oil red O. Lipid accumulation was quantified and normalized to WT levels. Values are mean ±SEM. A Student’s t test was performed for significance and (*p < 0.05).
Figure 3. Rap1 Deficiency Alters Liver Metabolism, Leading to Steatosis.
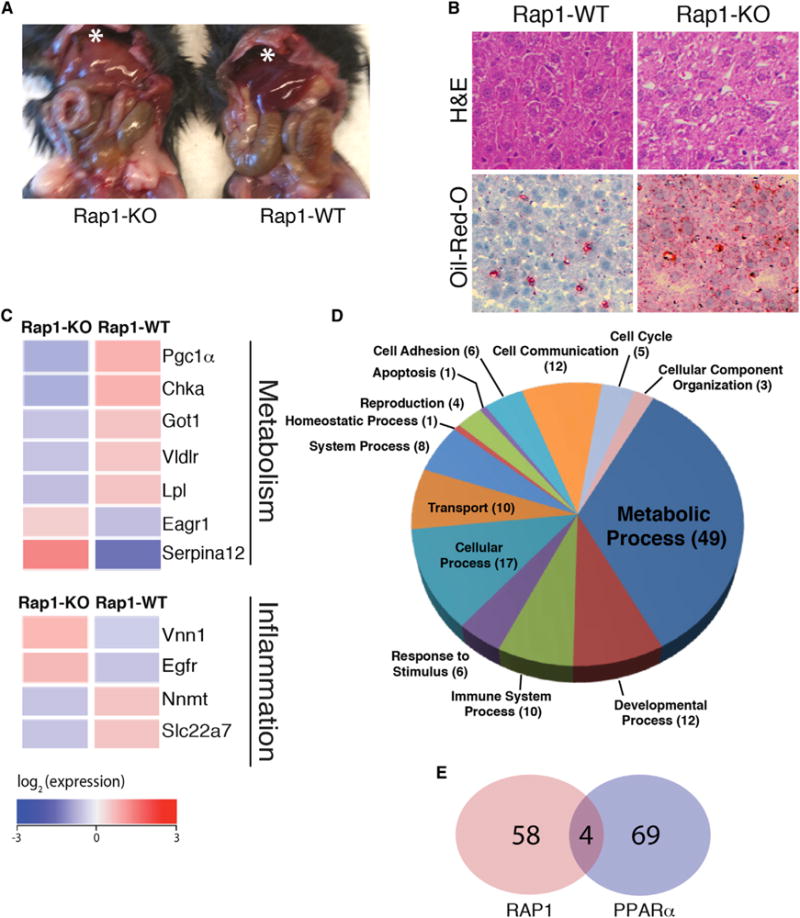
(A) Shown is a representative photograph of 24-week-old Rap1-KO and Rap1-WT mice, displaying pale/fatty liver (marked by asterisk) in the absence of Rap1.
(B) Representative H&E (upper) and oil red O (bottom) staining liver sections from 20-week-old WT and Rap1-KO mice. The red stain is an indicator for accumulation of neutral lipids.
(C) Microarray analysis of messenger RNA from the livers of6-week-old Rap1-KOand Rap1-WT female mice. Heat map representation of differentially expressed metabolism and inflammation genes. Average gene expression is colored proportionally to signal intensity with blue denoting a lower expression and red representing high expression.
(D) Gene Ontology analysis and classification of differentially expressed genes into biological processes.
(E) A Venn diagram showing the overlap of Rap1-regulated genes (red) with the PPARα pathway (blue) genes identified using GSEA analysis(p = 0.00028; Fisher’s exact test).
To further delineate the impact of Rap1 loss on WAT function, we assessed gene expression patterns of intra-abdominal WAT using Affymetrix GeneChip microarray. Animals were analyzed at 6 weeks of age, prior to when the effect of Rap1 deletion on body weight is evident. Of the total 39,000 transcripts analyzed, 118 WAT genes were differentially expressed between Rap1-WT and Rap1-KO mice (p < 0.05; 93 downregulated and 35 upregulated genes) (Tables S1, S2, and S3). Gene set enrichment analysis (GSEA) and Gene Ontology (GO) analysis on all differentially expressed genes indicated a significant deregulation in genes with a negative impact on metabolism as well as in genes involved in the developmental process, inflammation, and cell adhesion (Figure 2D; Tables S1, S2, and S3).
Rap1 Loss Promotes Adipocyte Differentiation of Immortalized MEFs In Vitro
The discernible hypertrophy of WAT (Figure 2C), together with the significant deregulation of gene expression (Figure 2D), prompted us to ask whether Rap1 affects the differentiation of preadipocytes into WAT. To directly address this question, we tested the transdifferentiation capacity of fibroblasts into adipocytes using a tissue culture model. SV40-LgT immortalized MEFs from Rap1 null mice and WT controls were subject to well-established adipocyte differentiation protocols (Nakae et al., 2003). Following hormonal induction with insulin, dexamethasone, and IBMX, both cell lines displayed morphological features consistent with adipocyte differentiation. Interestingly, Rap1-KO MEFs generated significantly more lipid droplets as indicated by oil red O staining (Figures 2E and 2F). These results suggest that Rap1 deficiency alters adipogenesis in a cell-autonomous manner, leading to increased white fat accumulation.
Rap1 Deficiency Alters Hepatic Metabolism and Leads to Liver Steatosis
We next turned our attention to the liver due to its central role in regulating glucose homeostasis, a major pathway that was significantly impacted in Rap1 null mice (Figures 1C–1E; Figure S1D). We found that Rap1 deficiency resulted in liver steatosis (Figure 3A), a common feature of obesity. This observation was confirmed by histological analysis that revealed excess fat droplets accumulation in liver tissue section from Rap1-KO mice (Figure 3B). Moreover, oil red O staining confirmed the accumulation of lipid droplets in liver of Rap1 null mice as compared to their WT counterparts (Figure 3B).
To investigate the biological processes underlying this altered hepatic phenotype, we assessed the effects of Rap1 inhibition on gene transcription in the liver of 6-week-old mice. Differential gene expression analysis revealed 62 genes that were significantly deregulated (false discovery rate [FDR] < 0.2) in Rap1 null mice as compared to age-matched controls (Figure 3C; Figure S4; Tables S4, S5, and S6). The list comprised metabolic and inflammation-related genes including PGC1-α, very low-density lipoprotein (Vldlr), and nicotinamide N-methyltransferase (Nnmt). GO classification of the deregulated genes highlighted several biological processes that were impacted by the loss of Rap1, including metabolic and developmental processes as well as the inflammation gene network (Figure 3D). Furthermore, GSEA analysis identified a number of pathways that were significantly deregulated in the context of Rap1 deficiency, the most notable being the PPAR-α signaling pathway genes (Figure 3E; Table S6), thereby highlighting a potential connection between the gene networks of Rap1 and PPAR-α in controlling hepatic function.
Rap1 Effect on Metabolic Gene Transcription Is Independent of TRF2-Mediated Recruitment to TTAGGG Sequence
Previous ChIP-seq analysis revealed that Rap1 occupies thousands of extratelomeric sites, many of which are associated with genes (Martinez et al., 2010; Yang et al., 2011). Motif discovery tools were utilized to identify “TTAGGG2” as the highest ranking motif for Rap1 binding to DNA, thereby implicating TRF2-mediated recruitment of Rap1 to interstitial telomere repeats in the regulation of gene transcription. However, ~75% of Rap1-associated genes were devoid of the TTAGGG consensus motif and the majority were not bound by TRF2 (Martinez et al., 2010; Yang et al., 2011). Moreover, upon matching gene expression profiles for Rap1-KO versus Rap1-WT MEF data sets (Table S7) with the Rap1-binding peaks identified by ChIP-seq analysis (Martinez et al., 2010), we found that 25% of the differentially regulated genes were bound by Rap1 (47 of 187; p = 0.0058, Fisher’s exact test), yet only eight genes contained the TTAGGG consensus motif (p = 0.25) (Figure S5; Tables S7, S8, and S9). This lack of enrichment for the TTAGGG motif within Rap1-regulated genes prompted us to ask whether the metabolic function of Rap1 is independent of its TRF2-mediated localization to DNA sequences at telomeres or elsewhere in the genome. To address this question, we generated an allele of mouse Rap1 carrying an isoleucine to arginine mutation at amino acid 312, a conserved residue in the RCT domain that was previously shown to promote the interaction between human Rap1 and TRF2 (Figure 4A) (Chen et al., 2011). Coimmunoprecipitation of cotransfected Myc-TRF2 and either WT Flag-Rap1 or Flag-Rap1 carrying the I312R mutation (referred to as Rap1ΔTRF2) in 293T cells confirmed that the I312R mutation blocked the interaction between the two shelterin subunits (Figure 4B). We then introduced Myc-Rap1 and Myc-Rap1 ΔTRF2 independently into Rap1-KO MEFs using stable retroviral transduction. Viral titers were diluted to ensure that the expression levels of Rap1-WT and Rap1ΔTRF2 were similar and comparable to endogenous levels of Rap1 in control MEFs (Figure S6). Indirect immunofluorescence indicated that WT Rap1 exhibited a nuclear punctate pattern that completely colocalized with telomeric DNA. On the other hand, Rap1ΔTRF2 failed to localize to telomere repeats, instead displaying a diffused nucleoplasmic labeling pattern (Figure 4C). The expression of mutant Rap1 did not induce a TIF (telomere-dysfunction-induced foci) response (Figure S6) and did not complement the telomere phenotype associated with Rap1 deficiency, which is the repression of aberrant HDR at telomeres (Chen et al., 2011 and data not shown).
Figure 4. Rap1ΔTRF2 Complements Transcriptional Defects of Rap1 Null Cells.
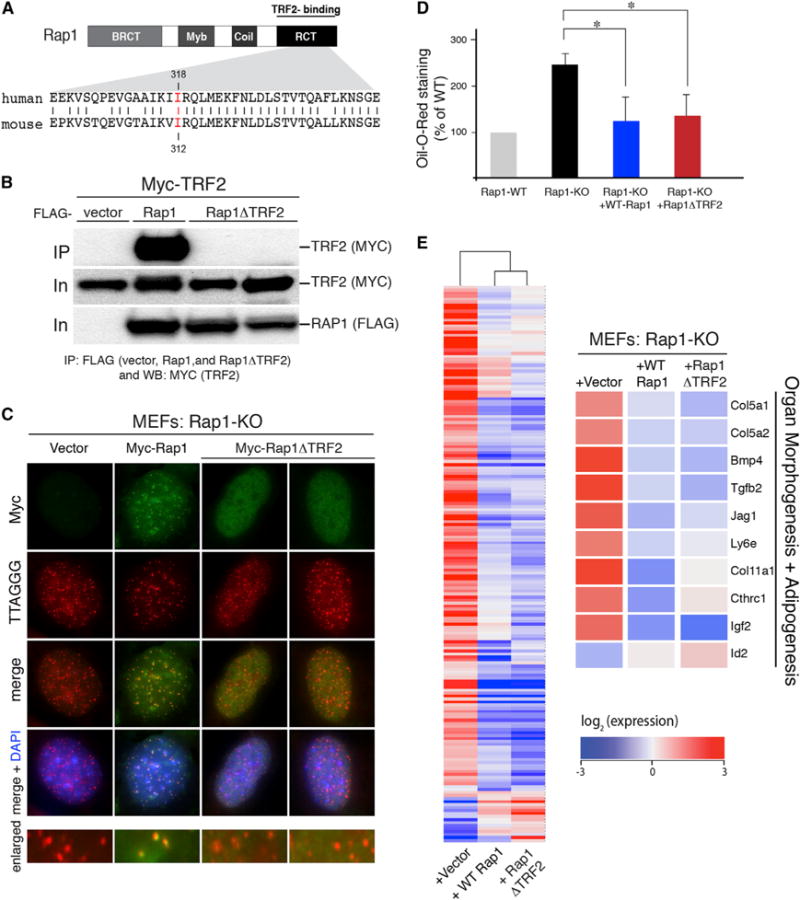
(A) Schematic of mouse Rap1 protein with its different domains: BRCT, Myb, and RCT. Highlighted in red is the amino acid residue that is important for TRF2 interaction and that was mutated in the Rap1ΔTRF2 allele.
(B) Coimmunoprecipitation of TRF2 from 293T cells cotransfected with wild-type Rap1 and mutant Rap1ΔTRF2.
(C) Immunofluorescence showing the localization of WT Rap1 and Rap1ΔTRF2 allele (detected with MYC antibody in green) in Rap1-deficient MEFs. Telomeres are detected with a PNA probe in red and DNA is counterstained with DAPI.
(D) Quantification of lipid accumulation in adipocytes derived from Rap1-KO MEFs expressing vector control, WTRap1, and Rap1ΔTRF2. Cells were stained with oil red O 8 days after hormonal induction and levels of lipid were expressed as percentage relative to adipocytes originating from cells expressing Rap1-WT. Values are mean ±SEM. A Student’s t test was performed for significance (*p < 0.05).
(E) Global transcriptional analysis of Rap1-KO MEFs complemented with vector control, WT Rap1, and Rap1ΔTRF2. Shown on the left is a row-centered heat map with hierarchical clustering carried out on the 187 differentially expressed genes. Represented on the right are examples of differentially expressed genes rescued by the expression of Rap1ΔTRF2.
Strikingly, Rap1ΔTRF2 was capable of complementing adipocyte differentiation of Rap1-KO MEFs to similar extent as the WT allele (Figure 4D). To assess the effect of the Rap1 mutation at the transcriptional level, we generated and analyzed transcript profiles of MEFs expressing Rap1ΔTRF2 and compared them to gene expression profiles of MEFs expressing either WT Rap1 or vector control. Hierarchical clustering with Pearson’s correlation of the differentially expressed genes revealed that cells expressing Rap1ΔTRF2 cosegregate closely with cells expressing WT Rap1 but are very distinct from Rap1 null cells (Figure 4E). A total of 87% of Rap1-deregulated genes were rescued by Rap1ΔTRF2, including genes involved in adipogenesis, organ morphogenesis, and metabolism (Figure 4E; Figure S5; Tables S7, S8, and S9). In conclusion, our results suggest that transcriptional regulation by Rap1 is independent of its TRF2-mediated recruitment to TTAGGG-binding elements found at extratelomeric sites. Therefore, Rap1 most likely relies on alternative transcription factors and/or complexes to control gene expression.
DISCUSSION
Overall, our study sheds light on a yet-unanticipated role for mammalian Rap1 in normal body weight control. Our data establish that Rap1 acts as a molecular determinant of gene expression in various metabolic and signaling pathways. Rap1 deficiency leads to a multisystem metabolic derangement reflected by glucose intolerance, dyslipidemia, liver steatosis, and excess fat accumulation. Ultimately, this manifests as late-onset obesity in the Rap1 knockout mice.
Rap1 has been shown to fulfill telomeric as well as nontelomeric functions. The latter is consistent with the protein acting as a regulator of the NF-κB signaling pathway in the cytoplasm (Teo et al., 2010), in addition to being a general transcriptional activator/repressor in the nucleus (Martinez et al., 2010). This raises the question as to what particular Rap1 function is responsible for the metabolic defect we observe herein. The ability of nontelomeric Rap1ΔTRF2 to rescue metabolic gene expression in MEFs to similar extent as WT Rap1 (Figure 4D) provides sufficient evidence for a telomere-independent role for Rap1 in metabolism. Furthermore, the obesity phenotypes exhibited in Rap1-KO mice are most likely not attributed to its role in the NF-κB pathway, where it was reported that Rap1 activates IKK and therefore enhances NF-κB signaling (Teo et al., 2010). In fact, opposite to what would be predicted based on the positive effect of Rap1 in NF-κB signaling, our microarray data identify a number of inflammation genes that were significantly upregulated when Rap1 was absent (Figures 2D, 3C, and 3D). Furthermore, IKK and Rap1 knockout mice display completely opposite phenotypes, with IKK deficiency counteracting obesity induced by a high-fat diet (Arkan et al., 2005). In conclusion, the metabolic and obesity defects observed in the Rap1-KO mouse are most consistent with mammalian Rap1 being a transcriptional regulator controlling a wide range of metabolic genes. This is reminiscent of the transcriptional activator/repressor function that has long been recognized for budding yeast Rap1 in regulating ribosomal proteins, glycolytic enzymes, and mating-type factors (Shore and Nasmyth, 1987; Kurtz and Shore, 1991).
It will be important to determine the mechanism by which Rap1 regulates transcription. The majority of genes that were deregulated upon Rap1 loss were devoid of the TTAGGG-consensus motif (Martinez et al., 2010; Yang et al., 2011). Furthermore, upon introducing into Rap1 null cells a Rap1ΔTRF2 allele that is unable to bind to the TTAGGG-bearing sequence, gene expression was largely rescued (Figure 4E). These results argue against a possible role for the Rap1-TRF2 complex in transcriptional control. Another possibility is that Rap1 binds directly to DNA, as was recently reported using in vitro approaches (Arat and Griffith, 2012). We find this scenario to be highly unlikely given the strong preference of Rap1 for double-stranded/single-stranded DNA junctions, uncommonly present in the genome. Alternatively, our preferred model is that the effect of Rap1 on gene expression stems from its ability to act as a cofactor for a yet-to-be-identified transcriptional complex. This is in line with recent protein network analyses that highlighted potential Rap1 interactions with histone proteins and other chromatin modifiers (Lee et al., 2011).
Although the mechanism underlying the metabolic phenotype of Rap1 deficiency remains to be delineated, our observation draws an interesting link between telomere function and metabolic regulation. One could speculate that mammalian cells use Rap1 as a general sensor monitoring the stability of chromosome ends. Telomere dysfunction was recently shown to compromise metabolic function in vivo by repressing PGC-1α-dependent processes (Sahin et al., 2011). Although this effect might be due to the activation of p53 at deprotected telomeres (Sahin et al., 2011), it is also possible that Rap1 could be the factor that relays the message from the terminal structure to the rest of the genome. In the latter scenario, significant telomere shortening yields insufficient binding sites for shelterin, leading to a reduction in TRF2 levels (Fujita et al., 2010). In turn, this would alter the overall levels of Rap1 in the nucleus, ultimately influencing genome-wide transcription. This scenario might explain the correlation that was recently described between telomere attrition and diabetes as well as insulin resistance (Gardner et al., 2005; Murillo-Ortiz et al., 2012; Shen et al., 2012; Salpea and Humphries, 2010).
Obesity is a major global public health challenge that has now reached epidemic proportions. Obese individuals are faced with a markedly increased risk of cardiovascular disease, type 2 diabetes, osteoarthritis, and a number of metabolic syndromes. Of the many genes that have been reported to impact obesity in mice, only a handful of heritable genes have been identified in human genetic studies. Whether mutations in Rap1 are associated with human metabolic syndromes remains to be identified. Nevertheless, it is intriguing to note that linkage analysis using a genome-wide association study pertaining to high-adiposity phenotypes among American Samoans has revealed a suggestive linkage with the locus containing Rap1 (16q23.1) (Aberg et al., 2009). 16q23.1 was also linked to childhood-obesity phenotypes in a French population (Meyre et al., 2004) and was found in a recent study to be differentially methylated in carriers of the FTO allele (fat mass and obesity-associated protein) (Almén et al., 2012).
Finally, our study establishes that the loss of a bona fide telomeric protein leads to major metabolic derangement, both at the cellular and organismal level. This underscores how mammalian cells establish crosstalk between telomere homeostasis and metabolism, two seemingly unrelated biological processes.
EXPERIMENTAL PROCEDURES
All animal studies were conducted In accordance with the National Institutes of Health (NIH) guidelines for treatment of animals and were approved by the Institutional Animal Care and Use Committees at NYU Langone Medical Center and at The Rockefeller University. Weight recordings, the DEXA scan, and the cold-endurance test were done on mice that were fed ad lib. GTT, ITT, and blood analysis was performed on mice that were fasted overnight. Microarray analysis was done on 6-week-old mice. RNA was purified from liver, iBAT, WAT, and cells using TRIzol and RNeasy Mini spin columns. Microarray hybridization was performed using Affymetrix GeneChip Mouse Genome 430 2.0 arrays. The array data were analyzed using Bioconductor. For further details, please refer to Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We acknowledge the generous support of Titia de Lange, in whose lab some experiments were initiated. We are grateful to Devon White for his contribution to the mouse work involved in this study. We thank Eros Lazzerini-Denchi for helpful discussions and for commenting on the manuscript. Lastly, we thank members of the Genome Technology Center at NYU Langone Medical Center for their excellent support. The Fernández-Hernando laboratory is supported by grants from the NIH (R01HL107953 and R01HL106063). C.M.R. is supported by a postdoctoral fellowship from the American Heart Association (12POST9780016).
Footnotes
ACCESSION NUMBERS
The microarray data reported in this paper have been deposited in the Gene Expression Omnibus under the accession number GSE46209.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and nine tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.05.032.
References
- Aberg K, Dai F, Sun G, Keighley ED, Indugula SR, Roberts ST, Zhang Q, Smelser D, Viali S, Tuitele J, et al. Susceptibility loci for adiposity phenotypes on 8p, 9p, and 16q inAmerican Samoa and Samoa. Obesity (Silver Spring) 2009;17:518–524. doi: 10.1038/oby.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almén MS, Jacobsson JA, Moschonis G, Benedict C, Chrousos GP, Fredriksson R, Schiöth HB. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–137. doi: 10.1016/j.ygeno.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Arat NO, Griffith JD. Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere. J Biol Chem. 2012;287:41583–41594. doi: 10.1074/jbc.M112.415984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, Wynshaw-Boris A, Poli G, Olefsky J, Karin M. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATMmediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol. 2011;18:213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costet P, Legendre C, Moré J, Edgar A, Galtier P, Pineau T. Peroxisome proliferator-activated receptor alpha-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem. 1998;273:29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]
- deLange T. Shelterin:the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Fujita K, Horikawa I, Mondal AM, Jenkins LM, Appella E, Vojtesek B, Bourdon JC, Lane DP, Harris CC. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat Cell Biol. 2010;12:1205–1212. doi: 10.1038/ncb2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JP, Li S, Srinivasan SR, Chen W, Kimura M, Lu X, Berenson GS, Aviv A. Rise in insulin resistance is associated with escalated telomere attrition. Circulation. 2005;111:2171–2177. doi: 10.1161/01.CIR.0000163550.70487.0B. [DOI] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Kabir S, Sfeir A, de Lange T. Taking apart Rap1: an adaptor protein with telomeric and non-telomeric functions. Cell Cycle. 2010;9:4061–4067. doi: 10.4161/cc.9.20.13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991;5:616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7(7A):1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- Lee OH, Kim H, He Q, Baek HJ, Yang D, Chen LY, Liang J, Chae HK, Safari A, Liu D, Songyang Z. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol Cell Proteomics. 2011;10:M110.001628. doi: 10.1074/mcp.M110.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gómez-López G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyre D, Lecoeur C, Delplanque J, Francke S, Vatin V, Durand E, Weill J, Dina C, Froguel P. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31–q23.2. Diabetes. 2004;53:803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- Murillo-Ortiz B, Albarrán-Tamayo F, Arenas-Aranda D, Benítez-Bribiesca L, Malacara-Hernández JM, Martínez-Garza S, Hernández-González M, Solorio S, Garay-Sevilla ME, Mora-Villalpando C. Telomere length and type 2 diabetes in males, a premature aging syndrome. Aging Male. 2012;15:54–58. doi: 10.3109/13685538.2011.593658. [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Kitamura Y, Biggs WH, 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- Pardo B, Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24:3117–3127. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpea KD, Humphries SE. Telomere length in atherosclerosis and diabetes. Atherosclerosis. 2010;209:35–38. doi: 10.1016/j.atherosclerosis.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhao X, Yu L, Zhang Z, Zhou D, Kan M, Zhang D, Cao L, Xing Q, Yang Y, et al. Association of leukocyte telomere length with type 2 diabetes in mainland Chinese populations. J Clin Endocrinol Metab. 2012;97:1371–1374. doi: 10.1210/jc.2011-1562. [DOI] [PubMed] [Google Scholar]
- Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol. 2010;12:758–767. doi: 10.1038/ncb2080. [DOI] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


