Summary
Microglia, the resident macrophages of the central nervous system (CNS), engage in various CNS-specific functions that are critical for development and health. To better study microglia and the properties that distinguish them from other tissue macrophage populations, we have optimized serum-free culture conditions to permit robust survival of highly ramified adult microglia under defined-medium conditions. We find that astrocyte-derived factors prevent microglial death ex vivo and that this activity results from three primary components, CSF-1/IL-34, TGF-β2, and cholesterol. Using microglial cultures that have never been exposed to serum, we demonstrate a dramatic and lasting change in phagocytic capacity after serum exposure. Finally, we find that mature microglia rapidly lose signature gene expression after isolation, and that this loss can be reversed by engrafting cells back into an intact CNS environment. These data indicate that the specialized gene expression profile of mature microglia requires continuous instructive signaling from the intact CNS.
Introduction
Within the CNS, microglia occupy a complex niche. All tissues have resident macrophage populations that contribute to development, homeostasis, and injury responses through canonical macrophage functions such as phagocytosis and chemokine/cytokine signaling (Gordon et al., 2014). Beyond these critical but generic innate immune functions, each tissue’s macrophage populations are shaped by environmental cues that drive tissue-specific gene expression patterns and enable specialized functions (Lavin et al., 2015; Okabe and Medzhitov, 2014). Several independent characterizations of tissue macrophages in general or microglia in particular have identified a number of microglia signature genes, including Tmem119, P2ry12, and Sall1, that are expressed by macrophages of the CNS but not other myeloid cell populations (Bennett et al., 2016; Butovsky et al., 2014; Gautier et al., 2012; Haynes et al., 2006; Hickman et al., 2013; Koso et al., 2016; Lavin et al., 2014).
Microglia oversee an intricate yet plastic network of synaptic connections and glial signaling networks. Microglia contribute to the proper development and homeostasis of the nervous system through synaptic pruning of neural circuits and apoptotic cell clearance in CNS germinal zones (Brown and Neher, 2014; Schafer et al., 2012). As the CNS matures, microglia acquire a ramified morphology, surveying the parenchyma through extension and retraction of dynamic processes (Davalos et al., 2005; Nimmerjahn et al., 2005). Human genetic studies indicate that proper microglial function continues to be important for adult CNS integrity. For instance, mutations in TREM2, a gene exclusively expressed by microglia in the intact CNS, drive or increase the risk of neurodegeneration (Guerreiro et al., 2013; Jonsson et al., 2013; Paloneva et al., 2002).
Specification of the mature microglial fate occurs in multiple waves during CNS development. In early embryonic stages, erythro-myeloid progenitors from the yolk sac migrate to and colonize the CNS rudiment, relying on CSF1R signaling, Pu.1, and Irf8 (Ginhoux et al., 2010; Kierdorf et al., 2013). These progenitors differentiate and expand, eventually comprising most if not all CNS-associated macrophages, and their lineage is preserved in adult animals through continuous self-renewal (Ajami et al., 2007; Gentek et al., 2014; Ginhoux and Guilliams, 2016). Microglia, like other tissue macrophages, rely on sustained CSF1R signaling to survive (Elmore et al., 2014; Ginhoux et al., 2010), which in the CNS is driven by the secreted growth factors CSF-1 and IL-34 (Greter et al., 2012; Wang et al., 2012; Witmer-Pack et al., 1993). Additionally, TGF-β signaling in early embryonic stages is required for complete microglial maturation and has been suggested to be a CNS-specific cue driving microglial specialization (Butovsky et al., 2014; Gosselin et al., 2014). Rodent microglia begin to acquire a mature CNS-specific signature of gene expression just before birth, but microglial maturation is not complete until a second phase of specification during the second postnatal week (Bennett et al., 2016; Matcovitch-Natan et al., 2016).
Efforts to understand microglial function in mechanistic detail have been hindered by the lack of microglial culture models that recapitulate the properties of mature microglia in vivo. Varied methods of isolating intact microglia have been established, but these cells generally assume an amoeboid morphology and are highly proliferative in culture, thereby resembling microglia in injured tissue (Stansley et al., 2012; Witting and Moller, 2011). Similarly, microglia in cultured brain slices rapidly lose their characteristic ramified morphology and mature marker expression (Haynes et al., 2006). Microglia-depleted organotypic hippocampal slices can be repopulated to generate highly ramified microglia in a culture paradigm (Masuch et al., 2016), but the presence of other cells limits some applications. Along a similar vein, microglial morphology and some other properties more closely resemble mature, quiescent microglia if the cells are cultured on an astrocyte feeder layer or in the presence of astrocyte-conditioned medium (ACM) (Sievers et al., 1994; Tanaka and Maeda, 1996). Indeed, several astrocyte-secreted factors, including CSF-1 and TGF-β, have been identified that can partially sustain aspects of mature, quiescent microglia in cultured cells (Butovsky et al., 2014; Liu et al., 1994; Salimi et al., 2003; Schilling et al., 2001). Still, doubts persist as to what extent culture models can be relied upon to faithfully recapitulate the properties and functions of microglia in the intact CNS (Kettenmann et al., 2011).
Motivated by the utility of primary culture models in advancing our understanding of other glial populations (Barres et al., 1993; Foo et al., 2011), we here characterize and improve cultures of pure primary microglia. We find that serum factors strongly perturb microglial morphology and function but can be completely eliminated if survival-promoting cues from astrocyte conditioned medium are supplied. We identify three separated factors, CSF-1, TGF-β, and cholesterol, that are critical to the ACM survival-promoting effects. Each of the three is necessary for ACM survival activity, and the combination of all three is sufficient, when supplied in defined serum-free growth medium, to support survival of microglia from postnatal and adult animals. Finally, we find that even fully mature microglia ex vivo revert to immature levels of key signature genes within hours of isolation, exhibiting rapid downregulaltion of genes that distinguish microglia from other tissue macrophage populations. Loss of mature CNS-specific markers can be reversed by engraftment of isolated cells into brains lacking microglia, demonstrating that additional CNS-specific cues are required to sustain microglial specification. In all, we introduce new methods for microglial culture and demarcate multiple modifiers of their viability, specification, and function.
Results
Astrocytes secrete multiple factors that promote microglial survival
Serum is rich in metabolites and growth factors and thus is ubiquitously used to supplement microglial cultures. However, serum also contains many blood-borne molecules that are actively excluded from the CNS, even before embryonic day 15 (E15) (Daneman et al., 2010). To more closely mimic the intact CNS environment, we tested whether microglia could be maintained ex vivo without serum supplementation. Using CD11b-based immunopanning, we isolated IBA1+, F4/80+ cells to >98% purity from postnatal brains (Fig. S1A). RNA-seq data discussed below suggest limited (<1%) contamination from non-microglial cells. Additionally, cells purified from perfused mouse brains based on CD11b immunoreactivity are >98% TMEM119 positive (Bennett et al., 2016), suggesting limited potential for non-microglial myeloid cell contaminants.
The viability of cultured microglia was assessed based on accumulation of calcein-AM dye in the cytoplasm and exclusion of cell-impermeant ethidium dimers from the nucleus. Without serum supplementation, microglia isolated from postnatal rat brains died within days, even in otherwise rich growth medium (Fig. 1A). Given the impact of astrocytic factors on microglial properties in serum-containing cultures (Salimi et al., 2003; Schilling et al., 2001) and the role of astrocytes as liaisons between the blood and the CNS parenchyma, we tested whether astrocytes could substitute for serum in supporting microglial survival in vitro. Growth medium that had been previously exposed to dense astrocyte cultures for three days (astrocyte-conditioned medium, or ACM) was sufficient to sustain microglial survival in the absence of serum, suggesting that astrocytes secrete factors that promote microglial survival (Fig. 1A, B).
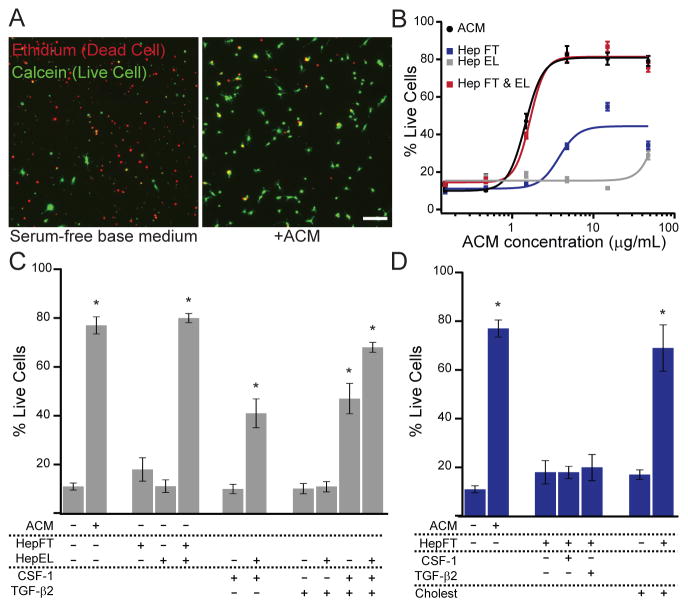
Figure 1. Multiple astrocyte-secreted factors support microglial survival.
(A) Serum-free rat primary microglia cultured in basal medium or astrocyte conditioned medium (ACM, 20 μg/mL) for 5 days. ACM improved survival, as assessed by accumulation of calcein (green) and exclusion of ethidium dimers (red). (B) Dose-response curves of fractionated ACM. Survival activity of unfractionated ACM (black) was not fully recovered in either heparin column flowthrough (Hep FT, blue) or eluate (Hep EL, gray) fractions. Recovered activity with combined fractions (red) indicates multiple active components. Doses represent complete ACM protein concentration or equivalent volume of each fraction. Lines represent Hill equation fits. (C) Survival activity of the Hep EL fraction can be complemented by CSF-1 and TGF-β2. (D) Survival activity of the Hep FT fraction can be complemented by free cholesterol. Some data replotted from (C). Averages are mean ± sem. Scale bars = 80 μm. * P < 0.001 one-way ANOVA across all groups in the panel with Dunnett’s comparison to the leftmost (no additive) condition.
In order to identity factors in ACM responsible for microglial survival, we tracked the survival activity through several biochemical fractionation steps. ACM survival activity could be concentrated in the retentate of centrifugal filter units bearing 30 kilodalton (kDa) molecular-weight cutoff membranes (Fig. S1B), indicating that the activity is associated with relatively large molecules or complexes. We tested whether the ACM survival activity could be enriched based on its affinity for heparin, but ACM survival activity was not entirely recovered in either the flowthrough (FT) fraction or the eluate (EL) fraction (Fig. 1B). However, the full activity of the complete ACM could be recovered by combining both FT and EL fractions (Fig. 1B), indicating that the apparent loss of survival activity is not due to degradation or failed recovery of the active components, but instead occurs due to separation of multiple necessary ACM components.
TGF-β2, CSF-1, and cholesterol reconstitute astrocyte-secreted survival activity
We next endeavored to identify the active components of each heparin affinity fraction individually. We first tested factors previously implicated in microglial survival or maturation. CSF-1 is a well-established trophic cue for microglia (Elmore et al., 2014; Ginhoux et al., 2010; Witmer-Pack et al., 1993), and TGF-β family members have been shown to promote microglial survival in serum-starved cultures (Salimi et al., 2003). We found that activity of the Heparin EL fraction could be partially rescued by the presence of CSF-1 and fully rescued by the presence of both CSF-1 and TGF-β2 (Fig. 1C), factors expressed by astrocytes in vivo (Zhang et al., 2014). Even in the absence of ACM components, the combination of CSF-1 and TGF-β2 promoted a moderate level of microglial survival, although less than complete ACM (Fig. 1C). Thus, CSF-1 and TGF-β2 are sufficient to replace the Heparin FT fraction, but do not fully recapitulate the survival activity of complete ACM.
As no candidate factors we tested complemented the Heparin FT fraction, we turned to further biochemical fractionation of ACM. We found that ACM survival activity was not retained on sugar-binding concanavalin A resin, but was highly enriched in high-salt elution fractions from anion exchange resin (Fig. S1C, D). SDS-PAGE analysis correlated activity with enrichment of a ~36 kDa protein (Fig. S1D). After enrichment over anion exchange resin, the active fraction’s protein content was analyzed by mass spectrometry of tryptic fragments. Astrocytes cultured by two separate methods (MD astrocytes and IP astrocytes, (Foo et al., 2011)) both secreted microglial survival activity and were analyzed in parallel (Fig. S1E). The most abundant proteins found in the active fractions were the apolipoproteins APOE and APOJ, both of which were highly enriched in active fractions over inactive ones (Table S1).
Treatment of microglial cultures with recombinant APOE, APOJ, or both did not promote microglial survival, even in the presence of the Heparin FT fraction (not shown). However, APOE and APOJ are commonly found as components of complexes in both ACM and cerebrospinal fluid that transport cholesterol and other lipids throughout the CNS (Fagan et al., 1999; LaDu et al., 1998). Surprisingly, cholesterol itself was sufficient to rescue survival of microglia in the presence of the Heparin FT fraction, although it conferred no survival advantage when applied alone (Fig. 1D). Therefore, the activity of the Heparin EL fraction is likely attributable to cholesterol transported in APOE and/or APOJ-containing lipoparticles.
Identification of serum-free conditions for primary microglial culture
Next, we asked whether CSF-1, TGF-β2, and cholesterol are sufficient to promote microglial survival in serum-free, defined-medium cultures. None of the three components individually nor any pairwise combination fully recapitulated the activity of complete ACM, but the combination of all three factors supported robust and reproducible survival of microglial cultures (Fig. 2A). We next tested which components of the base medium were necessary for microglial survival by eliminating basal components. Microglia cultured with CSF-1, TGF-β2, and cholesterol survived in a broad range of base medium formulations, but supplementation with nontoxic levels of sodium selenite, a common cell culture additive (Sato and Kan, 2001) that is incorporated into several redox enzymes, was essential for survival in serum-free medium (Fig. S2A). Additionally, rat, bovine, and human albumin all triggered morphological changes in microglia cultured under our conditions (not shown), and so albumin was eliminated from all cultures.
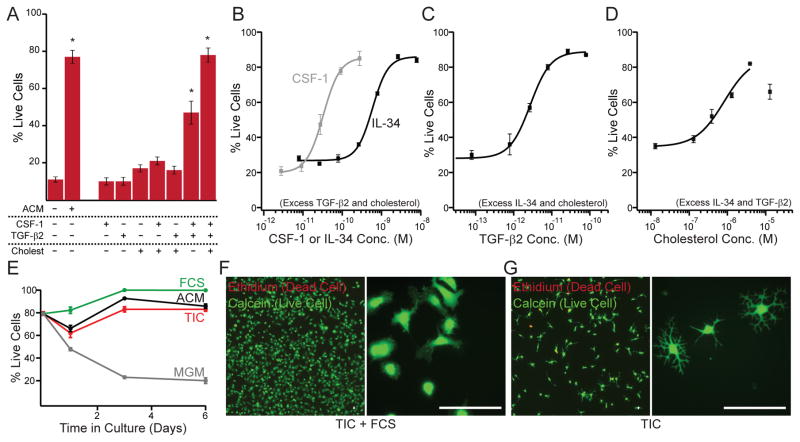
Figure 2. The combination of TGF-β, CSF-1/IL-34, and cholesterol supports survival of ramified microglia in defined medium.
(A) The combination of CSF-1 (10 ng/mL), TGF-β2 (2 ng/mL), and cholesterol (1.5 μg/mL) facilitates survival of primary rat microglia as well as ACM (20 μg/mL). Individual components or pairwise combinations were not sufficient for robust survival (5 div). Some bars are replotted from Fig. 1C for clarity. (B) Dose-response analysis of CSF-1 and IL-34 when supplemented with TGF-β2 (2 ng/mL) and cholesterol (1.5 μg/mL). (C) Dose-response analysis of TGF-β2 when supplemented with IL-34 (100 ng/mL) and cholesterol (1.5 μg/mL). (D) Dose-response analysis of cholesterol when supplemented with IL-34 (100 ng/mL) and TGF-β2 (2 ng/mL). Lines in (B–D) describe Hill equation fits. (E) Time course of microglial survival in base medium (MGM, gray), ACM (black), TIC (red) or TIC+10%FCS (green). (F, G) Calcein/ethidium-labeled microglia grown in TIC supplemented with 10% FCS (F) or TIC alone (G) illustrate serum-induced proliferation (low magnification, left) and altered morphology (high magnification, right). Averages are mean ± sem. Scale bars = 40 μm. * P < 0.001 one-way ANOVA across all groups with Dunnett’s comparison to no additive condition.
We next measured dose-response relations for each factor (CSF-1, TGF-β2, and cholesterol) with excess amounts of the other two factors. In the presence of TGF-β2 and cholesterol, CSF-1 supported microglial survival at concentrations comparable to those described for CSF1R-dependent activity; a second CSF1R ligand, IL-34, promoted equivalent survival at concentrations relevant for CSF1R activation (Wei et al., 2010) (Fig. 2B). In consideration of the poor species cross-reactivity of CSF-1, we elected to perform subsequent experiments using only murine IL-34, which facilitated survival of rat, mouse, and human cells alike in preliminary analyses. The complete cocktail of TGF-β2, IL-34, and cholesterol is hereafter referred to as TIC (applied at 2 ng/mL, 100 ng/mL, and 1.5 μg/mL respectively).
TGF-β2 concentrations relevant for survival tracked with TGF-β2 affinity for the TGF-β receptor (Hoosein et al., 1988) (Fig. 2C). The TGF-β family contains many members that are categorized into several subgroups (Mueller and Nickel, 2012). A sparse sampling of TGF-β family subgroups found that some, but not all, family members could replace TGF-β2 in promoting microglial survival in the presence of IL-34 and cholesterol, including TGF-β1, Activin A, and Bmp10 (Fig S2B). TGF-β has pleiotropic effects on immune activation (Li et al., 2006), and so we considered that TGF-β might promote microglial survival in culture through an anti-inflammatory mechanism. However, other anti-inflammatory factors failed to replace TGF-β family members in promoting survival (Fig. S2B).
A relatively high concentration (1.5 μg/mL) of cholesterol is required to sustain microglial survival (Fig. 2D). Higher concentrations of free cholesterol led to precipitation and cell death, and cholesterol-saturated methyl-β-cyclodextrin did not improve microglial survival beyond that attained at maximal concentrations of free cholesterol. Cholesterol supplementation facilitates survival of various cell types in serum-free media (Sato and Kan, 2001) and is an integral component of mammalian cell membranes that likely participates in multiple general functions.
To further investigate the mechanisms of death, we tested whether microglial death in the absence of TGF-β2, IL-34, or cholesterol could be prevented with known inhibitors. Cells were cultured in the presence of the broad-spectrum caspase inhibitor z-VAD-fmk, the RIPK1 inhibitor necrostatin-1s, or both, and survival was assessed after 3 days. Neither inhibitor nor their combination prevented cell death in the absence of any individual survival cue (Fig. S2C). Because initially plated cells experience a complex signaling milieu, we next tested the effects of removing each survival factor from cells pre-established in serum-free TIC medium for 7 days. Three days after withdrawal, the requirement for continuous CSF1R signaling and cholesterol supplementation for survival was evident, although continued TGF-β2 supplementation was not essential (Fig. S2D). Still, cell death inhibitors were ineffective at preventing microglial death after IL-34 withdrawal from established cultures (Fig. S2E). These data suggest that microglial death in culture does not strictly conform to canonical mechanisms of apoptosis or necroptosis.
TIC medium supported microglial survival over time comparably to ACM (Fig. 2E), and microglia grown in without serum consistently bore closer resemblance to resting microglia in vivo than their serum-exposed counterparts (Fig. 2F, G). Indeed, dramatic differences in process and somatic motility were evident, with serum-free cultures mimicking the dynamic extension and retraction of processes observed in vivo (Movies S1 and S2, (Davalos et al., 2005; Nimmerjahn et al., 2005)). Importantly, the highly ramified morphology of serum-free cultures depends on the continued presence of all three TIC factors, as withdrawal of any one resulted in reduced morphological complexity within three days (Fig. S2F).
CSF-1, TGF-β2, and cholesterol are necessary components of astrocyte-secreted survival activity
CSF-1/IL-34, TGF-β2, and cholesterol are sufficient to promote microglial survival in culture, but are they necessary components of the ACM survival activity? Treatment with the CSF1R inhibitor GW2580 reduced survival of microglia cultured in either TIC or ACM (Fig. 3A). Similarly, the survival activity of both TIC and ACM could be partially eliminated by pretreatment with a TGF-β-neutralizing antibody (Fig. 3B). To determine whether cholesterol was a required component of ACM, we included high concentrations of the cholesterol-chelating agent, methyl-β-cyclodextrin (MβC), which reduced microglial viability (Fig. 3C). The MβC was not inherently toxic, however, as pre-saturating the MβC with free cholesterol eliminated its effects on microglial survival in ACM (Fig. 3C).
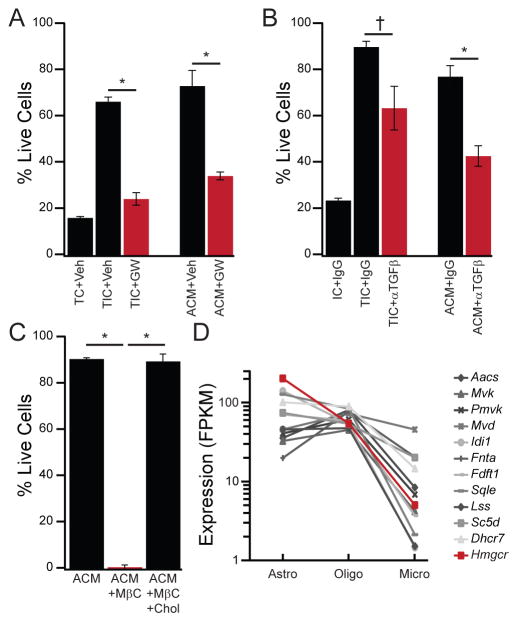
Figure 3. CSF-1/IL-34, TGF-β, and cholesterol are necessary for ACM survival activity.
(A) The CSF1R antagonist GW2580 (10 μM, red), but not vehicle (Veh, black), reduced survival of primary rat microglia in both TIC and ACM. (B) The pan-TGF-β neutralizing antibody 1D11 (5 μg/mL), but not IgG control, partially reversed survival in TIC or ACM (20 μg/mL). (C) The cholesterol-chelating agent methyl-β-cyclodextrin (MβC, 5 mM) eliminated ACM survival activity, but not if it was pre-saturated with cholesterol (MβC+ chol). (D) Abundance of cholesterol biosynthesis machinery mRNA transcripts from published RNA-seq datasets (Zhang et al., 2014) expressed as fragments per kilobase per million reads mapped (FPKM) illustrates lower levels in microglia (Micro) as compared to astrocytes (Astro) and mature oligodendrocytes (Oligo). The rate-limiting enzyme in cholesterol biosynthesis (Hmgcr) is highlighted in red. Averages are mean ± sem. * P < 0.005, † P < 0.05 Student’s t-test.
In consideration of the effects of cholesterol, we consulted published transcriptomic profiles (Zhang et al., 2014) to determine the relative expression of cholesterol biosynthetic genes in glial populations. We found microglia express lower levels of cholesterol biosynthesis genes than either astrocytes or mature oligodendrocytes (Fig. 3D). This pattern is particularly pronounced for the rate-limiting enzyme, Hmgcr, which is expressed ~50-fold more highly in astrocytes than microglia (Fig. 3D). Thus, we propose that microglia are deficient at producing cholesterol autonomously, but instead rely on other CNS cells in vivo or exogenous cholesterol supplementation in vitro.
Serum exposure alters microglial proliferation and phagocytosis
We noticed that addition of serum led to a stable increase of the apparent percentage of surviving microglia in culture to almost 100%, even higher than initial time points (Fig. 2E). This apparent increase in survival over time arose from two confounding factors. First, serum exposure induced rapid proliferation of cultured cells, while cells grown in ACM or TIC showed relatively little change in total cell number (Fig. S2G). Second, dead cells rapidly disappeared from cultures supplemented with serum (Fig. S2G), whereas a small number of cell corpses persist in serum-free cultures. Early after culture, individual serum-exposed cells sometimes associated with multiple ethidium-positive puncta, suggesting that dead cells were being cleared by phagocytosis.
To test whether phagocytosis was altered by serum, we conjugated a pH-sensitive dye, pHrodo, to multiple types of model ‘prey’ particles that have been used to study phagocytosis in vitro (Chung et al., 2013; Freeman and Grinstein, 2014; Mosser and Zhang, 2011). Labeling densities were optimized such that pHrodo fluorescence was only detected at acidic pH, which labeled particles experience only in late-endosomes or lysosomes after phagocytosis. Microglia cultured in TIC and 10% FCS robustly phagocytosed many types of pHrodo-labeled particles, including myelin debris, zymosan, IgG-opsonized erythrocytes, and amine-coated polystyrene beads (Fig. 4A, B, S3A). In stark contrast, microglia cultured in the absence of serum ingested almost no particles of any type over the same period (Fig. 4A, B, S3A). We confirmed that the observed fluorescence arises due to actin-dependent uptake using the actin polymerization inhibitor cytochalasin D, which abolished the pHrodo signal (Fig. 4C). Even with continuous monitoring over 48 hours, little ingested myelin was detected in serum-free cultures, whereas serum-exposed cells phagocytosed all of the provided material (Fig. 4B). Additionally, rat or adult human serum promoted microglial phagocytosis equivalently to fetal calf serum (Fig. 4C, S3F). Serum-free cells were still highly motile and repeatedly contacted myelin, but failed to clear it and transported almost none to acidified compartments (Movie S3), in contrast to serum-exposed cells (Movie S4).
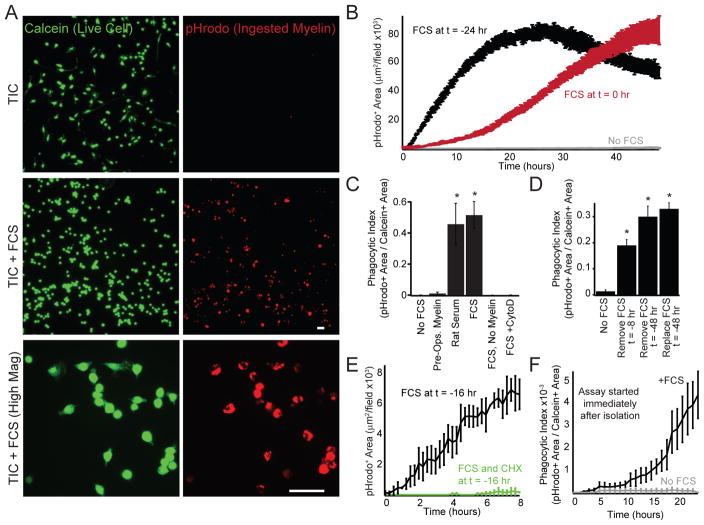
Figure 4. Serum exposure unlocks latent phagocytic potential in serum-free microglial cultures.
(A) Phagocytosis (3 hr) of pHrodo-labeled myelin debris (red) by primary rat microglia (5 div in TIC) with (middle, bottom) or without (top) exposure to serum (10% FCS, 24 hr). (B) pHrodo signal monitored over two days. Microglia (5 div in TIC) exposed to 10% FCS for 24 hours before addition of myelin (black) cleared all of the provided myelin. Cells only exposed to 10% FCS when myelin was added (red) were initially less phagocytic than cells pre-exposed to serum, but gradually acquired robust phagocytic capacity. Cells unexposed to serum (gray) showed minimal myelin uptake. (C) Serum-free cultured microglia (5 div in TIC) do not phagocytose myelin pre-opsonized with serum (Pre-Ops. Myelin, 4 hr). Pre-exposure of microglia (24 hr) to adolescent rat serum or fetal calf serum facilitates phagocytosis. Minimal pHrodo signal was observed in the absence of myelin or in the presence of the actin-polymerization inhibitor cytochalasin D (CytoD, 10 μM). Phagocytic index was calculated as the ratio of pHrodo+ area to calcein+ area. (D) 8 or 48 hr after removal of FCS (10%, 24 hr exposure), microglia phagocytose myelin (4 hr) as efficiently as cells continuously maintained with serum. (E) Microglia (5 div in TIC) supplemented with serum before adding myelin (10% FCS at t = −16 hr) or with serum and the translational inhibitor cycloheximide (40 μM, FCS and CHX at t = −16 hr) show that serum-evoked changes in phagocytosis requires new protein synthesis. (F) Microglia (0 div in TIC) do not phagocytose myelin immediately after plating, although phagocytosis gradually increases in cells exposed to 10% FCS. Averages are mean ± sem. Scale bars = 10 μm. * P < 0.001 one-way ANOVA with Dunnett’s comparison to no FCS.
Serum exposure transforms the intrinsic phagocytic capacity of microglia
We hypothesized that the phagocytosis-boosting effects of serum exposure might be attributable to the abundance of serum opsonins, which impact microglial phagocytosis of apoptotic cells (Fourgeaud et al., 2016). Myelin that was pre-incubated with serum before application to serum-free microglial cultures was not phagocytosed (Fig. 4C), suggesting that prey opsonization is insufficient for the serum effect. To explore this further, we exposed microglia to serum only upon addition of myelin particles for phagocytosis. Under these conditions, microglia were initially poor phagocytes, despite the presence of serum opsonins during the assay (Fig. 4B, red trace). Instead of acquiring phagocytic potential immediately upon serum exposure, serum-free microglia slowly acquired maximal phagocytic capacity over the course of 12–24 hours of serum exposure (Fig. 4B, red trace). To confirm that this transformation was intrinsic to the microglia themselves, we tested whether the translational inhibitor cycloheximide (CHX) could prevent the change. Microglia exposed to CHX at the same time as their first exposure to serum never acquired phagocytic potential like cells treated with serum alone (Fig. 4E). Despite the drug’s eventual toxicity, microglia that had previously been exposed to serum were still phagocytic even 16 hours after CHX exposure (Fig. S3B).
We next asked if changes in phagocytosis were reversed by removal of serum. Microglia grown in serum and exchanged into serum-free medium before the addition of myelin still demonstrated increased phagocytosis relative to serum naïve cultures, although serum removal substantially decreased the initial rate of uptake (Fig. S3C). In such cultures, maximal phagocytic capacity could be immediately recovered by re-supplementation with serum (Fig. S3D), or gradually recovered automatically over 8–48 hours (Fig. 4D). These results are consistent with serum-exposed cells requiring additional opsonins, which can be replenished immediately by serum supplementation or gradually by microglial secretion. This differs from serum naïve cells, which are phagocytically inept even in the presence of serum opsonins or pre-opsonized prey. In all, we observe a dramatic and lasting change in the intrinsic phagocytic capacity of microglia in vitro after exposure to serum.
To determine whether serum-induced changes reflect an artificial loss of phagocytic potential induced in culture, we monitored phagocytosis of freshly isolated cells. Microglia fed myelin immediately after isolation did not show appreciable uptake by default (Fig. 4F). Serum-exposed cells gradually acquired robust phagocytic potential, whereas serum-free cells showed minimal myelin uptake at any time point (Fig. 4F, S3E). Additionally, serum activity was retained on a 50 kDa cutoff filter, indicating that the active factor(s) are large and unlikely to be transported across the blood-brain barrier in the intact CNS (Fig. S3F).
Defined-medium cultures for mature microglia
Murine microglia do not acquire their mature transcriptomic signature until after the second postnatal week (Bennett et al., 2016; Matcovitch-Natan et al., 2016), and maturation markers are likewise upregulated in rat microglia between postnatal day 7 (P7) and P21 (Fig. S4B). With these changes in mind, we endeavored to culture fully mature microglia under conditions optimized for younger animals. P21 microglia behaved similarly to cells from younger animals, although survival rates were more variable. To optimize survival of mature cells, we tested an array of substrate coatings and found that collagen IV and heparin sulfate facilitated initial adhesion and overall survival (Fig. S4C). Additionally, transcriptomic databases (Bennett et al., 2016; Lavin et al., 2014) suggest that fatty acid biosynthetic genes Elovl6 and Acaca are downregulated during microglial maturation and are expressed at lower levels in microglia than other macrophage populations (Fig. S4D, E). Inclusion of monounsaturated fatty acids did not reduce survival (Fig. S4F), and was therefore included in cultures of mature microglia. Mature microglia cultured in fully optimized conditions mimicked the morphology and dynamics of resting microglia (Movie S5), and so subsequent experiments were conducted using mature microglia.
Serum exposure alters microglial gene expression
Amoeboid morphology, rapid proliferation, and heightened phagocytic activity are hallmarks of microglia in injured tissue, but also properties of immature microglia in the perinatal CNS (Kettenmann et al., 2011). To determine whether microglia cultured in the presence or absence of serum are “classically” activated (see the Discussion section below for a more nuanced consideration of microglial activation) we measured basal and bacterial lipopolysaccharide (LPS)-induced levels of several canonical activation markers by QPCR. All markers were expressed at levels comparable to or less than freshly isolated cells, and robust induction after LPS stimulation was evident, indicating that cultured microglia do not show hallmarks of overt inflammation (Fig. S4A).
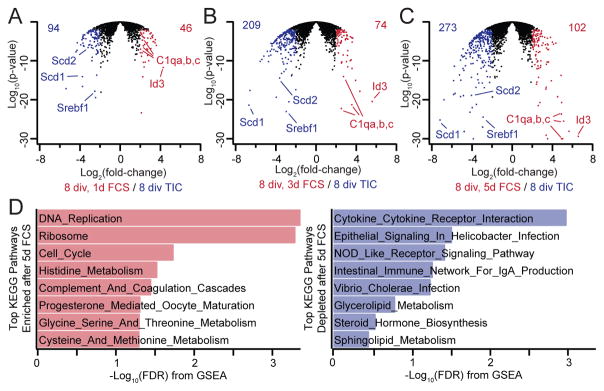
To perform an unbiased assessment of the serum-evoked changes in gene expression, we performed RNA-seq profiling of serum-exposed microglia. We initially cultured P21 rat microglia in serum-free medium to avoid serum-induced survival/expansion of non-microglial cells, then added 10% serum for 1 to 5 days before harvesting RNA. Individual replicates clustered with the appropriate treatment group (Fig. S5A, B), and high-abundance transcripts specific to other major CNS cell types were essentially absent, confirming high microglial purity (Fig. S5C). Gene changes after serum exposure occurred gradually with 140, 283, and 379 genes showing major expression changes (p<0.01 and 4-fold cutoffs) at 1, 3, and 5 days after serum exposure, respectively (Fig. 5A–C, S5B). Several KEGG pathways were found to be altered after serum exposure, with upregulation of cell-cycle and amino acid metabolism pathways and downregulation of cytokine and lipid metabolism pathways (Fig. 5D).
Figure 5. Effects of serum exposure on microglial gene expression.
(A–C) Volcano plots summarizing changes in microglial gene expression (P21, 8 div in TIC) after 1, 3, or 5 days of serum exposure as measured by RNA-seq. Values in the upper corners denote the number of genes upregulated (red) or downregulated (blue) after serum exposure at each time point passing a 4-fold cutoff at P<0.01. Individual genes showing pronounced changes are labeled. (D) KEGG pathways enriched after serum exposure among upregulated (red, left) or downregulated (blue, right) genes. Enrichment was measured using GSEA, and the false-discovery-rate q-value (FDR) is plotted, with larger bars indicating higher confidence of enrichment. Pathways involving proliferation, amino acid metabolism, and complement are upregulated after serum exposure, whereas cytokine pathways and lipid-modifying pathways are downregulated.
Pathway analysis revealed substantial upregulation of a number of complement proteins, including C1qa, C1qb, C1qc, C3, Cfd, and Cfp (Dataset S1), suggesting a possible mechanism underlying serum-induced phagocytic changes. It has previously been shown that macrophage expression of C1q can be augmented by hydrocortisone (Trinder et al., 1995), and we found that hydrocortisone supplementation of serum-free cultures triggered upregulation of complement proteins to a similar degree as serum exposure (Fig. S5D). Despite increasing expression of C1qa-c, hydrocortisone exposure did not facilitate phagocytosis (Fig. S5E). As such, the molecular events underlying serum-evoked phagocytosis changes remain to be determined. Nonetheless, our RNA-seq analysis indicates that serum exposure substantially impacts transcriptional networks in cultured microglia.
Microglia change gene expression profiles in culture
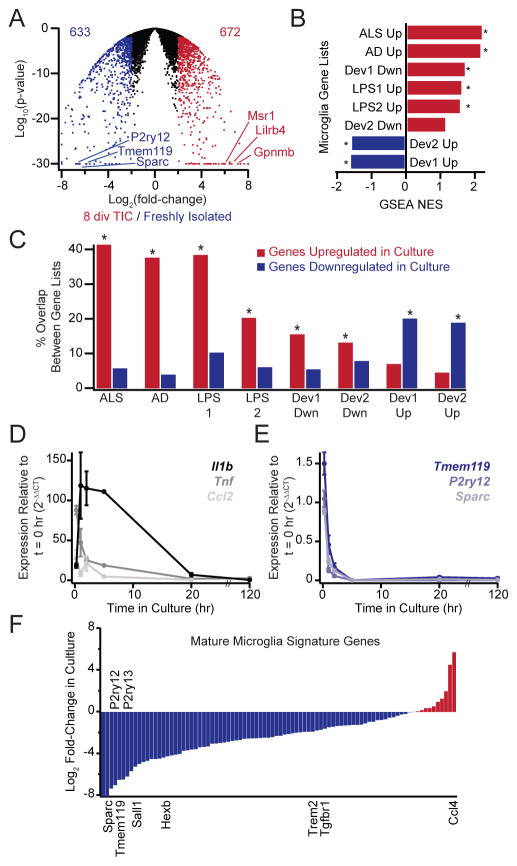
In order to facilitate interpretation of experiments using cultured microglia, we next compared global mRNA expression profiles of serum-free TIC cultures versus freshly isolated microglia. Extensive differences were apparent, involving 1,305 differentially-expressed genes (P < 0.01 and > 4-fold cutoffs, Fig. 6A) out of 10,816 genes detected (FPKM > 1, Dataset S1). To classify these changes, we procured lists of genes that change during neurodegeneration (end stage SOD1G93A mice or 8.5 month old 5XFAD mice (Chiu et al., 2013; Wang et al., 2015)), during systemic inflammation (1 day or 2 days after intraperitoneal LPS injection (Bennett et al., 2016; Chiu et al., 2013)), or over development (E10.5–12.5 or E17 to adulthood (Bennett et al., 2016; Matcovitch-Natan et al., 2016)) using published RNA-seq datasets from purified microglia. Inflammatory genes from each list were greatly enriched in cultured microglia over freshly isolated cells as determined by GSEA (Fig. 6B, Fig. S6A). Likewise, developmentally downregulated genes were strongly enriched in cultured cells, while maturation markers were strongly enriched in freshly isolated cells (Fig. 6B, Fig. S6A). Despite differences in rodent species and methods of cell isolation, each inflammation-associated gene list shared 35–40% overlap with culture-induced genes, whereas overlap with culture-downregulated gene list was only 3–8%, near the level expected by chance (3%, Fig. 6C). The extent of overlap with developmental datasets was less extensive (15–20% overlap), but still highly significant (Fig. 6C).
Figure 6. Transcriptional profiling indicates microglial de-differentiation in culture.
(A) Volcano plots summarizing changes in microglial gene expression (P21, 8 div in TIC) compared to freshly isolated cells. Values in the upper corners denote the number of genes upregulated (red) or downregulated (blue) in cultured cells (4-fold cutoff at P < 0.01). (B) GSEA comparison of culture-induced gene changes with developmental or inflammatory gene changes. The nominal enrichment score (NES) for each comparison is shown, with positive values (red) indicating enrichment among genes upregulated in culture and negative values (blue) indicating enrichment among genes downregulated in culture. Cultured microglia upregulate genes associated with microglia in neurodegeneration (ALS or AD mouse models) peripheral inflammation (LPS) and early development (developmentally downregulated genes, Dev Dwn), but they downregulate genes associated with maturation (developmentally upregulated genes, Dev Up). *Both FDR q-value and nominal P-value < 0.001. (C) Overlap between genes changing in inflammation/development and genes upregulated (red) or downregulated (blue) in culture. Percent overlap denotes the fraction of genes in each developmental or inflammation gene list that are also up- or down-regulated in culture. Culture-induced gene expression changes partially recapitulate inflammatory changes and demonstrate an inverse relationship to developmental changes. *P < 0.001 hypergeometric distribution. (D, E) QPCR shows rapid but transient upregulation of canonical activation markers accompanied by rapid and sustained downregulation of microglial signature genes over the first 5 days in culture (P19–P21, in TIC). (F) Log2 fold-changes in RNA-seq FPKM values measured for cultured microglia relative to freshly isolated cells for 88 genes that are both upregulated developmentally and preferentially expressed by microglia over other tissue macrophages. The majority of microglial signature genes are downregulated (blue), not upregulated (red) in culture. Averages are mean ± sem.
These analyses reveal two faces to microglial transcriptomic changes in vitro: a direct relation to changes occurring during neuroinflammation and an inverse relation to changes that occur developmentally. In all, cultured microglia gene expression profiles do not perfectly mimic either chronic neuroinflammatory or early developmental profiles, but assume characteristics of each state.
Microglia in culture show dynamic inflammatory responses
To further dissect the timing of gene expression changes, we performed QPCR analysis of individual genes representative of larger cassettes. Immediately after isolation, pronounced upregulation of classical activation markers Il1b, Tnf, and Ccl2 was observed in cultured cells (Fig. 6D). Abundance of these mRNAs increases within 15 minutes of culture, then largely resolves over the first day in vitro (Fig. 6D). We reasoned that one or more of the components of the culture medium might trigger activation. However, microglia cultured for two hours in dPBS, HBSS, or DMEM/F12 without additional factors also showed rapid Il1b upregulation (Fig. S6B), and LPS stimulation after 5 days in culture induced activation markers to levels comparable to those achieved after isolation, suggesting that the initiating signal does not persist in culture (Fig. S4A). We considered that microglial changes might be specific to our methods of isolation, but canonical activation markers were also upregulated in cells enriched using Percoll gradients or by magnetic cell sorting (not shown). Indeed, rapid upregulation of microglial cytokine mRNA is evident even in excised brain tissue held at 37°C in multiple endotoxin-free media formulations (Fig. S6E), suggesting that rapid changes in microglial character are inextricably linked to their isolation. Expression of classical activation markers resolves quickly in culture, and so these genes do not contribute to the aforementioned relationship between microglia in culture and microglia from neuroinflammatory contexts. Instead, the relationship is dictated by genes such as Spp1 (a gene upregulated in ALS, AD, and peripheral LPS models), which demonstrates rapid and sustained upregulation in culture (Fig. S6H).
Microglia rapidly de-differentiate in culture
Among genes showing dramatically altered expression levels in culture are microglia signature genes such as Tmem119, P2ry12, and Sall1, which are more highly expressed by >10-fold in freshly isolated cells (Fig. 6A). To assess whether microglia signature genes are universally downregulated in culture, we assembled a list of highly expressed genes that are enriched in adult microglia over other tissue macrophages and embryonic progenitors (Lavin et al., 2014; Matcovitch-Natan et al., 2016), covering most established microglial markers (Dataset S1). Most signature genes were downregulated by cells in culture (Fig. 6F), consistent with prior reports (Butovsky et al., 2014; Gosselin et al., 2014). However, it was previously reported that TGF-β1 can induce expression of microglial signature markers in FACS-sorted microglia or peritoneal macrophages (Butovsky et al., 2014; Gosselin et al., 2014), and so we considered that the reduction in microglia signature gene expression in our TGF-β containing cultures might be caused by contaminating cell types or progenitors expanding at the expense of microglia. To provide insight into this possibility, we monitored the loss of signature gene expression over time by QPCR. Surprisingly, the expression of mature microglial markers rapidly decreased with a half-life of less than one hour (Fig. 6E), even in the presence of TGF-β2 and despite negligible cell death or proliferation as monitored by live imaging. Tmem119 and P2ry12 downregulation consistently occurred in various independent medium formulations on either collagen-coated or uncoated tissue culture plastic (Fig. S6C, D). Loss of the mature signature coincides with re-expression of a subset of the genes downregulated over development (Fig. 6B, C), suggesting that microglia rapidly de-differentiate in purified cultures even in the presence of functional TGF-β signaling.
Microglial specification requires sustained CNS instruction
Tissue macrophage specialization is dictated largely by tissue-specific environmental cues (Bruttger et al., 2015; Lavin et al., 2015; Okabe and Medzhitov, 2014). As such, we next considered that the apparent loss of mature CNS-specific patterns of gene expression might result from the absence of additional CNS-specific cues. To mimic primary culture manipulations without separating microglia from the CNS environment, we incubated excised brain tissue in isolation buffer or culture medium. QPCR analysis of this tissue did not show downregulation of microglial signature genes Tmem119 or P2ry12 relative to Csf1r, although Il1b upregulation was still apparent, decoupling transient canonical activation from signature gene downregulation (Fig. S6E–G). Thus, preservation of the CNS environment can delay loss of microglial signature gene expression ex vivo although microglial identity does eventually decay in cultured brain slices (Haynes et al., 2006).
To test whether culture-induced changes could be reversed by the native tissue environment, we engrafted cultured microglia back into the CNS parenchyma. While various myeloid cell populations are capable of colonizing the CNS, it is first necessary to open a niche by damaging or eliminating resident microglia (Ajami et al., 2007; Bruttger et al., 2015). To provide this niche, we used CSF1R knockout animals, which lack microglia (Ginhoux et al., 2010) (schematic in Fig. 7A). We first tested whether mouse, like rat, microglia signature gene expression changes after isolation. Within 4 hours of isolation, Tmem119, P2ry12, and Olfml3 mRNA levels were reduced to less than 5% of initial levels, a drop sustained over six days in culture (Fig. 7B). Loss of TMEM119 immunoreactivity followed at a slower rate, but was evident after 4 hours, when staining intensity had been reduced by 50%, and was near-complete by 16 hours in culture (Fig. 7C, E). We conclude that TMEM119 protein production is essentially halted in isolated cells and that the extant surface protein decays with a half-life of approximately 4 hours.
Figure 7. Re-introduction into the CSF1R−/− CNS rescues microglial character.
(A) Schematic showing experimental paradigm, left to right: isolation of microglia by magnetic bead separation, culture for 4 hours in TIC + 5% FCS, intracranial injection into CSF1R−/−pups (P0 to P2), 2 weeks incubation in vivo, analysis of engrafted cells. (B) Cultured mouse microglia demonstrate loss of signature gene expression by 4 hours that is sustained at 6 days. (C) Cultured microglia (blue) show reduced immunoreactivity to Tmem119 after 4 hours (left) or 16 hours (right) in culture compared to acutely purified cells (black) by flow cytometry. (D) FACS histograms of CD11b+CD45Lo gated cells from microglia-injected CSF1R−/− brains or WT controls showing that engrafted microglia show near-WT levels of Tmem119 immunoreactivity two weeks after injection. (E) The geometric mean florescence intensity for Tmem119 immunostaining rapidly decreases in culture, but returns to WT levels in cells cultured for 16 hr then re-engrafted. (F) Engrafted brains have Tmem119 immunopositive ramified microglia, whereas non-engrafted CSF1R−/− brains lack these cells. Averages are mean ± sem. Scale bar = 50 μm. * P < 0.001 one-way ANOVA with Dunnett’s comparison to 0 hr.
We next performed intracranial injections into CSF1R−/− brains using strain-matched adult microglia that were allowed to lose microglia signature gene expression over 4 to 16 hours in culture. Both freshly isolated cells and cells used for engraftment preserved CD45 and CD11b expression, although the signal increased after culture (Fig. S7A). Two weeks after injection, CSF1R knockout brains had been partially repopulated with CD11b+ CD45+ cells (Fig. S7B), which are absent from non-engrafted brains (Ginhoux et al., 2010). Virtually all CD11b+ CD45Lo cells are TMEM119+ (Fig. 7D), whereas CD11b− cells from these brains lack TMEM119 immunoreactivity (Fig. S7C). Among CD11b+ CD45Lo cells, TMEM119 mean fluorescence intensity is indistinguishable between WT littermate controls and animals engrafted with cells cultured for 16 hours (Fig. 7E), although a small population of CD11b+ CD45Hi Tmem119− cells was also detected in engrafted brains (Fig. S7B, C). We next checked engrafted brain slices for microglia-like cells and found repopulation of IBA1+, TMEM119+ cells that assume morphologies similar to mature microglia, a population completely absent from saline-injected CSF1R−/− animals (Fig. 7F). Thus, we find that mature microglia can successfully engraft into CSF1R−/− brain parenchyma and that these cells obtain mature marker expression, even if expression was previously lost after isolation. In all, these data indicate that cues within the intact CNS are both necessary to prevent rapid loss of signature gene expression and sufficient to reverse loss of microglial character.
Discussion
Here we describe a novel approach for culturing adult microglia under defined-medium conditions, thereby identifying new modes of microglial regulation and opening up new avenues of study. First, defined-medium conditions have allowed us to identify microglial survival-promoting activity for TGF-β and cholesterol (Fig. 2, 3). Second, we have revealed roles for as-yet unidentified serum factors in transforming microglial morphology and phagocytic capacity (Fig. 2, 4). Third, the radically different morphology and motility in the absence of serum (Movies S1, S2, S5) allows for the study of microglial dynamics in isolation and suggests that surveillance behavior relies, at least in part, on microglia-intrinsic mechanisms. Fourth, elimination of serum from microglial growth medium will permit co-culture experiments with other glial populations whose behavior is also radically altered by serum exposure (Foo et al., 2011; Raff et al., 1983). Fifth, our culture methods support survival of adult microglia, revealing a rapid and sustained decrease in microglial signature gene expression in fully differentiated cells after removal from the CNS environment (Fig. 6, 7). Lastly, we utilize a novel microglial transplantation system to show that signals from the intact CNS environment continuously instruct microglial fate (Fig. 7). The implications of these findings are discussed below.
TGF-β and exogenous lipids as microglial survival cues
In this study, we show that TGF-β2 (or other TGF-β family members) and cholesterol support survival of isolated microglia. Mice lacking TGF-β1 in the CNS fail to develop cells expressing microglia-specific markers (Butovsky et al., 2014), and Cre-ER dependent deletion of TGFBR2 (a necessary receptor for TGF-β1 and TGF-β2 signaling) from adult microglia leads to loss of mature microglial character in the CNS (Buttgereit et al., 2016). The latter study concluded that microglial loss after TGFBR2 deletion arises from de-differentiation, not death, of mature cells based on recombination-dependent Rosa26-YFP expression. YFP signal was detected in cells lacking microglial character after TGFBR2 excision (Buttgereit et al., 2016), suggesting de-differentiation of mature microglia. However, the proportion of YFP-positive cells dropped from ~95% to ~55% among CNS CD45+ Ly6C− Ly6G− F4/80Hi cells, introducing the possibility of expansion of cells from a non-microglial origin. The survival activity of TGF-β we observe ex vivo suggests that loss of constitutive TGF-β signaling may also contribute to eventual microglial death after rapid decrement of mature microglial marker expression. A complete understanding of the specific contributions of TGF-β signaling in microglial survival and maturation will require further study.
In addition to protein factors, we find that microglia in culture require exogenous cholesterol for prolonged survival. Macrophages in peripheral tissues have ready access to circulating lipoparticles from the blood, which transport low-solubility lipid species throughout the body. However, circulating lipids are not in exchange with cerebrospinal fluid (Pfrieger and Ungerer, 2011). Instead, cholesterol synthesized de novo within the CNS is transported via APOE- and/or APOJ-containing lipid nanodiscs (LaDu et al., 1998; Pfrieger and Ungerer, 2011). Recently, TREM2 was found to be important for binding and microglial uptake of lipidated APOE and APOJ, with disease-associated TREM2 variants showing deficiencies in lipoparticle recognition and engulfment (Yeh et al., 2016). Indeed, variants of APOE, APOJ, and TREM2 are among the strongest alleles linked to Alzheimer’s disease identified to date (Wes et al., 2016), and APOE variants alter A-beta clearance and microglial cholesterol content (Lee et al., 2012). Our data delineating the importance of exogenous lipid species in microglial survival suggest that deficiencies from these alleles could result, in part, due to generalized microglial dysfunction related to insufficient delivery of cholesterol or other lipid species.
Serum exposure transforms microglial properties
Microglial phagocytosis of synapses, protein aggregates, and myelin is highly regulated in vivo. Dysregulation of microglial phagocytosis of synapses has been implicated in the pathology of diverse neurological diseases (Hong et al., 2016; Lui et al., 2016; Vasek et al., 2016), failure to clear myelin debris contributes to CNS regenerative failure (Vargas and Barres, 2007), and problems with microglial uptake and clearance may contribute to aging-associated cognitive decline (Safaiyan et al., 2016). Our data suggest that serum-induced alteration of microglial phagocytosis in vitro may result from phagocytosis-augmenting cues normally excluded from the CNS environment. Indeed, neuroinflammatory models in which the BBB is maintained, such as peripheral LPS injection, typically lack microglial proliferation and evidence of phagocytosis (Chen et al., 2012), whereas proliferation and debris clearance are readily observed in microglia after stroke or contusion injuries that compromise BBB integrity (Greenhalgh and David, 2014; Schilling et al., 2005). As these functions are thought to be critical for neuroprotection and recovery, serum factors that regulate microglial phagocytosis and their signaling pathways are likely to have profound consequences secondary to CNS trauma and may represent valuable therapeutic targets for manipulating microglial function across diverse disease states.
Microglial transcriptional changes in culture include inflammatory signatures
Through extensive functional and transcriptomic characterization, we illustrate that in spite of the ramified morphology observed in serum-free cultures, unresolved shortfalls exist in modeling the native, resting state of mature microglia ex vivo. A subset of the microglial gene expression changes in culture are mirrored in microglia from the inflamed CNS (Fig. 6A–C). Are cultured microglia, then, activated? Macrophages have historically been classified based on a limited number of markers into three different states: resting, classically activated, and alternatively activated. The advent of genome-wide sampling of gene expression, however, has expanded the dimensionality of this issue and complicated it considerably (Ransohoff, 2016). It is now appreciated that gene expression patterns of microglia isolated from various inflammatory contexts do not conform to a small number of transcriptional programs and are not typically analogous to profiles induced by canonical inflammatory agents such as LPS (Chiu et al., 2013; Wang et al., 2015). Freshly isolated microglia transiently upregulate classical activation markers, but this canonical response resolves within hours to days. A subset of the more persistent gene expression changes reflect alterations typically only observed in microglia from the inflamed CNS. Ex vivo preparations will be critical for defining co-regulated gene cassettes and connecting transcriptional readouts to microglia-specific signaling pathways and effector functions.
Microglial fate is continuously instructed by CNS-specific cues
Our findings that microglia rapidly de-differentiate in culture reecho and expand upon previous reports (Butovsky et al., 2014; Gosselin et al., 2014). We find that microglia in culture still show a pronounced reduction in signature gene expression relative to freshly isolated cells despite the inclusion of TGF-β and CSF-1. Still, even after >10-fold reductions in transcript levels, mature markers are expressed by cultured microglia at levels beyond what can be detected in other tissue macrophages. By taking advantage of the open microglial niche in the CSF1R knockout, we have shown that downregulation of microglial signature genes in culture is reversed by reintroducing these cells back into the CNS and is therefore not a permanent consequence intrinsic to cell isolation manipulations. Based on these findings, we propose that mature microglial fate requires continuous instruction from additional cues specific to the CNS environment.
Loss of microglial signature gene expression occurs extremely rapidly in culture, with maturity markers decaying with a half-life of less than one hour. As such, acutely isolated microglia do not offer a workaround to improve the fidelity of in vitro assays, as these cells suffer from similar shortcomings as prolonged cultures on top of canonical activation responses engaged immediately after isolation. Additionally, studies of microglia isolated from human brains may be subject to substantial changes in gene expression over ethically-achievable post-mortem intervals. Rapid changes in microglial signature gene expression is consistent with rates reported for global mRNA turnover in macrophage-like dendritic cells stimulated with LPS (Rabani et al., 2014). We conclude that microglia are capable of rapid transformations of gene expression profiles to quickly accommodate different tissue contexts, and that the imminent plasticity of these cells makes them an attractive target for therapeutic intervention. Despite their shortcomings, microglial cultures remain an important resource for uncovering the signaling mechanisms and functions of these specialized CNS immune cells.
STAR Methods
Contact for Reagent and Resource Sharing
Step-by-step protocols, reagents, datasets, and other information will be shared upon request to the Lead Contact, Christopher Bohlen (cjbohlen@gmail.com). RNA-seq data and analyses have been deposited in the GEO database, accession number GSE96995.
Experimental Model and Subject Detail
All procedures involving rodents conformed to Stanford University guidelines, which comply with national and state laws and policies. Sprague-Dawley rats were obtained from Charles River, Hollister, CA. Animals less than fourteen days old were euthanized with a lethal dose of ketamine/xylazine (100 μL of 24 mg/mL ketamine, 2.4 mg/mL xylazine); older animals were euthanized by carbon dioxide asphyxiation. CSF1R knockout animals (Dai et al., 2002) on the FVB background (FVB.129X1-Csf1rtm1Ers) were a generous gift from Dr. Richard Stanley. WT FVB/NJ littermate were bred in house from CSF1R+/− animals.
Rat microglia cell isolation
Animal unresponsiveness to toe pinch were transcardially perfused with ice-cold dPBS containing 50 μg/mL heparin (Sigma). Immediately after perfusion, brains were rapidly dissected and placed into ice-cold dPBS. Cerebella and spinal cords were discarded and the remaining brains were chopped to ~1 mm3 with a scalpel blade and transferred to an ice-cold dounce homogenizer (Wheaton) with ice-cold dPBS containing 200 μL of 0.4% DNaseI (Worthington) per 50 mL of dPBS. Tissue chunks were subjected to three successive rounds of 3 to 10 gentle strokes of the homogenizer piston and centrifuged. For animals less than 12 days old, tissue was centrifuged at 500g for 15 minutes at 4°C. For animals greater than 12 days old, PercollPlus was added during centrifugation to separate myelin from cells. The cell suspension volume was adjusted to 33.4 mL with dPBS and 10 mL of 100% isotonic Percoll (9 mL Percoll PLUS (GE Healthcare), 1 mL 10× dPBS without Ca and Mg, 9 μL 1 M CaCl2, 5 μL 1 M MgCl2) was added and thoroughly mixed (23% isotonic Percoll final). Suspensions were centrifuged (15 min, 500g, 4°C) and the supernatant and top layer of myelin were discarded. Cell pellets were resuspended in 15 mL of dPBS containing 2 mg/mL peptone from milk solids (Sigma), passed through a 70 μm cell strainer (BD Biosciences), and applied directly to positive-selection immunopanning dishes coated with mouse anti-rat CD11b monoclonal antibodies (OX42 clone, Bio-Rad).
Immunopanning dishes were prepared the day before as follows. First, a sterile petri dish was incubated in a solution of 50 mM Tris adjusted to pH 9.5 with HCl that contained 6 μg/mL goat-anti-mouse IgG secondary antibody (Jackson ImmunoResearch) at 37°C for 1–2 hours. Dishes were rinsed three times with dPBS and transferred to a solution of dPBS containing 1 μg/mL OX42 antibody (Bio-Rad) and 2 mg/mL of peptone from milk solids (Sigma) as a blocking agent. Dishes were coated overnight at room temperature and rinsed three times with dPBS immediately before addition of cell suspensions for immunopanning.
Cell suspensions were allowed to interact with the immunopanning dish for 20 min at room temperature. Unbound cells and debris were removed by washing the dish with dPBS ten consecutive times. For freshly isolated cell lysates used in RNA-seq and QPCR experiments, cells were lysed directly from the panning dish in RLT buffer (QIAGEN). Otherwise, immunopanned cells were trypsinized (10 min at 37°C) in 12 mL dPBS without calcium and magnesium containing 200 μL of a 30,000 units/mL stock solution of trypsin (Sigma). Microglia were still strongly adherent, and so the trypsin solution was discarded, panning dish was rinsed twice with dPBS and replaced with cold MGM unless otherwise noted. Panning dishes were placed on ice for one minute to weaken cell interaction with the surface, and the cells were recovered by repeated pipetting. Recovered cells were centrifuged (500g, 15 minutes at 4°C) and all but 1 mL of the supernatant was carefully withdrawn and discarded. Cells were gently resuspended in of the reserved 1 mL of MGM. For experiments testing gene expression changes after two hours, cells were resuspended in alternative medium instead.
Rat microglia culture
Base microglial growth medium (MGM) was sterile-filtered and stored at 4°C for up to one month and was comprised of: DMEM/F12 containing 100 units/mL penicillin, 100 μg/mL streptomycin, 2mM glutamine, 5 μg/ml N-acetyl cysteine, 5 μg/ml insulin, 100 μg/mL apo-transferrin, and 100 ng/mL sodium selenite, all from Gibco or Sigma. ACM, 10% heat-inactivated fetal calf serum (FCS), growth factors, or drugs were added after filtering. For preparation of cholesterol-containing medium, MGM and a 1.5 mg/mL ethanolic stock solution of cholesterol (Avanti Polar Lipids) were warmed to 37°C and an appropriate volume of cholesterol stock was added and rapidly mixed to avoid precipitation. For experiments involving methyl-β-cyclodextrin (MβC), MβC (Sigma) was dissolved to 5 mM in MGM and passed through a 20 μm syringe filter before addition of other factors. For cholesterol-saturated MβC, 1 mg of cholesterol powder was added to 10 mL of 5 mM MβC in MGM and the mixture was incubated at 37°C for one hour before being passed through a 20 μm syringe filter and addition of other factors.
TIC medium was comprised of MGM containing human TGF-β2 (2 ng/mL, Peprotech), murine IL-34 (100 ng/mL, R&D Systems), and ovine wool cholesterol (1.5 μg/mL, Avanti Polar Lipids) unless otherwise noted. Microglia isolated from animals older than 12 days old, heparan sulfate (1 μg/mL, Galen Laboratory Supplies) was included to maximize cell adhesion and process extension. Cells were plated either directly onto tissue culture plastic for animals less than 12 days old, or onto plastic coated for 15–60 min at 37°C with collagen IV (2 μg/mL, Corning) for animals over 12 days old and aspirated off immediately before plating cells. Different coatings (poly-D-lysine, poly-L-ornithine, laminin, matrigel, or collagen VI) had minimal impact on the findings described, although the most extensive ramification of cells was observed with collagen IV or laminin coating and inclusion of heparan sulfate in the culture medium. For microglia cultures from P21 rats, monounsaturated fatty acids (0.1 μg/mL oleic acid, 0.001 μg/mL gondoic acid) were included in the growth medium unless otherwise stated.
Cells were plated at various densities depending on the experiment. For survival or immunohistochemical assays, cells were cultured at low density to minimize the influence of neighboring cells. 3,500 cells were pre-plated in 24-well plates in 15 μL, cells were allowed 10 minutes at room temperature to lightly adhere to the plate, and 500 μL of culture medium was gently added to each well before transfer to the incubator. For phagocytosis assays, 10,000 cells were added per 96-well well or 3,500 cells were added per 384-well well. Cells were briefly centrifuged (30 s, 100g, room temperature) to promote adhesion and plates were transferred to the incubator. For gene expression analysis, 20,000 to 40,000 cells were seeded in 24-well plates and allowed to settle for 10 minutes at room temperature before transfer to the incubator. Cells were grown in a humidified incubator held at 37°C and 10% CO2.
For survival assays, cells were typically not fed in order to minimize disruption of dead cell counts, although for time points > 7 days, one 50% medium change was performed every 5 days. For other assays, 50% medium changes were performed every 2 to 3 days.
Mouse cell isolation, culture, and transplantation
For intracranial transplant experiments and associated mouse microglial cultures, brain cell dissociation was performed as previously described (Bennett et al., 2016). Briefly, mice were euthanized by CO2 asphyxiation and whole brains from 3–5 week-old FVB WT mice were dissected and homogenized in ice cold HBSS supplemented with 15mM HEPES and 0.5% glucose by 5 gentle strokes in a 7 mL glass dounce homogenizer. After resuspension in MACS buffer (PBS with 2mM EDTA and 1% BSA) Dissociated cell suspensions were passed through a 70 micron cell strainer (Falcon), and myelin was depleted using myelin removal beads II (Miltenyi) and the MACS system (Miltenyi). After myelin depletion, cell suspensions were positively selected for CD11b expression by CD11b magnetic bead (Miltenyi) separation using the MACS system. Dissociated brain suspensions were incubated in 20 μL of microbeads in 100 μL of MACS buffer per brain, and 1–3 brains were applied to each LS column (Miltenyi), otherwise according to manufacturer instructions.
CD11b purified mouse microglia were cultured in TIC medium supplemented with 5% heat inactivated FCS for 4 hours to 6 days at 37°C and 10% CO2, and subsequently harvested for RNA isolation, flow cytometry, or intracranial injections. Cells were cultured on tissue culture plastic, and harvested on ice by 3–5 washes with ice cold FACS buffer (PBS, 25 mM HEPES, 2mM EDTA, and 2% FCS) and repeated pipetting.
P0–P2 CSF1R KO pups and WT controls were provided local cryoanesthesia and intracranially injected by hand using a pulled glass microcapillary tube (WPI) in an electrode holder connected by silicon tubing to a syringe. One microliter containing 105 cultured microglia in PBS was slowly injected bilaterally over 3 seconds into cortex, 1–2 mm anterior and 2–3 mm lateral to lambda at a depth of 0.5 mm. Host animals were harvested after 14 days, and single cell suspensions were generated and cleared of myelin as described above. Two to three animals were pooled for each biological replicate in knockout animals, while a single WT control animal was used as an engraftment and staining control.
Method Details
Live/Dead assay
Cell survival was assayed on the fifth day in vitro unless otherwise noted. Calcein AM dye and ethidium homodimer-1 (Invitrogen) were diluted into MGM, then applied to cells at a final concentration of 1.33 μM Calcein AM and 2.5 μM ethidium homodimer-1. Cells were incubated at 37°C for 10 min before imaging on an Axio Observer. Z1 (Zeiss). Identical acquisition and illumination conditions were used within each experiment. Images were exported to ImageJ, where custom macros were used for cell counts. After background subtractions, images were thresholded and particles were counted automatically using particle size cutoffs to prevent counting of subcellular fragments or noncellular debris. Meaningful counting was checked visually for every image, and at least one image per session was counted manually to confirm the accuracy of automated counting values.
ACM preparation
Astrocyte cultures were prepared as described (Foo et al., 2011). In brief, MD astrocytes were obtained from P1 rat cortices that were dissociated by 30 minutes of papain (Worthington) digestion and repeated pipetting. Dissociated cells were grown in astrocyte medium then shaken at three days to dislodge non-astrocytic cells. At five days, 10 μM cytosine-arabinoside was added, and cells were passaged at seven days and every subsequent seven days in astrocyte medium [DMEM (high glucose), 10% FCS, 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamine, and 1 mM sodium pyruvate, 5 μg/ml N-acetyl cysteine, 5 μg/ml insulin, 10 μM hydrocortisone, and 10 μM forskolin, all from Gibco or Sigma]. IP astrocytes were prepared from P7 rat cortices dissociated into a single cell suspension as for MD astrocytes. The cell suspension was subjected to negative immunopanning using subsequent petri dishes coated with rat anti-mouse CD45 antibodies (BD Pharmingen), the lectin BSL-1 (Vector Labs), or O4 hybridoma supernatant (mouse IgM, grown in-house) before a positive immunopanning selection step using petri dishes coated with mouse anti-human ITGB5 antibodies (eBioscience). Cells were recovered from ITGB5 dishes by trypsinization followed by centrifugation in a solution containing ovomucoid trypsin inhibitor. Cells were plated at a density of 1 to 3 million cells per 15 cm tissue culture plate in IP astrocyte growth medium [50% DMEM, 50% Neurobasal, 100 units/mL penicillin, 100 μg/mL streptomycin, 1 mM sodium pyruvate, 2mM glutamine, 5 μg/ml insulin, 100 μg/mL transferrin, 100 μg/mL bovine serum albumin, 16 μg/mL putrescine, 60 ng/mL progesterone, 40 ng/mL sodium selenite, 5 μg/ml N-acetyl cysteine, all from Gibco or Sigma] and allowed to expand for one week before being used for conditioning medium.
Conditioned medium was collected from MD and IP astrocytes in the same way. Each dish was washed four times with dPBS, then incubated at 37°C for 10 minutes in dPBS to facilitate complete medium exchange. The medium was then replaced with MGM lacking transferrin and insulin, and the cells were cultured for another three days. The conditioned MGM was collected, centrifuged to remove cellular debris (10 min, 3,400g, 4°C), and concentrated over a 30 kDa MWCO centrif ugal filter unit (Millipore) to approximately 1% of the original volume (15 min intervals interrupted by mixing of the sample, 3,400g, 4°C). ACM protein concentration was determined using a Bradford assay (Bio-Rad), and ACM was stored at 4°C for up to 1 month without loss of survival activity. ACM was applied to cells at a final concentration of 20 μg/mL unless otherwise noted. Neutralizing antibodies against TGF-β (clone 1D11, R&D Systems) or IgG control antibodies were incubated at 100 μg/mL with MGM containing 6.6 ng/mL TGF-β2 or 200 μg/mL ACM for 16 hours at 4°C prior to 10-fold dilution into IL-34 and cholesterol-containing medium and addition to cells.
ACM fractionation and analysis
All fractionation was performed at room temperature using pre-packed 1 mL columns (GE Healthcare) without exceeding the binding capacity. Solutions were applied manually using a syringe inside of a tissue-culture hood to maintain sterility. ACM was diluted five-fold into loading buffer appropriate for each column resin. Each column was washed with an additional 5 mL of loading buffer (included in the FT fraction) after loading, followed by 5 mL of wash or elution buffers. Buffers were applied at approximately 1 mL per minute (FF columns) or 0.2 mL per minute (other columns), and protein was loaded onto the columns at one-fifth the normal flow rate. All buffers contained 20 mM Tris and were adjusted to pH 7.5 with HCl. HiTrap Heparin HP columns were run with 0 mM NaCl (20 mM Tris, pH 7.5 only) as loading buffer and 1 M NaCl as elution buffer. HiTrap Q FF columns were run with 200 mM NaCl load buffer, followed by a 5 mL wash with 500 mM NaCl wash buffer, then elution with 1 M NaCl elution buffer. HiTrap Concanavalin A Sepharose 4B columns were run with 1 mM CaCl2, 1 mM MnCl2 as loading buffer and 500 mM NaCl, 500 mM methyl α-D-mannoside as elution buffer. HiTrap Phenyl FF (High Sub) columns were run with 2 M NaCl load buffer and 0 mM NaCl elution buffer.
For testing activity, each fraction was concentrated to the original ACM volume using a 30 kDa MWCO centrifugal filter unit, diluted ten-fold into DMEM/F12, then re-concentrated before a second consecutive round of ten-fold dilution and re-concentration. Fractions were stored at 4°C for less than one week before application to cells. SDS PAGE analysis was performed according to manufacturer’s instructions using 4–20% gels (Bio-Rad), SYPRO orange (Invitrogen), and a Typhoon imager (GE Healthcare, blue channel 825 V). For protein identification by mass spectrometry, an active fraction (Q column eluate) and a negative fraction (Q column unbound fraction) were analyzed for comparison. Proteins were acetone-precipitated, reduced with DTT, alkylated with iodoacetamide, and fragmented with trypsin. Multiply charged peptides were selected by abundance for sequencing using MS/MS on an Applied Biosystems machine. Peptides were identified and counted using the manufacturers’ software and the NCBI database.
Phagocytosis assay
For myelin debris isolation, perfused rat brains from adult female rats were dissociated with a dounce homogenizer and myelin fragments were isolated by successive rounds of osmotic shock and ultracentrifugation in sucrose gradients as described (Larocca and Norton, 2007). The weight of wet myelin pellets was used to estimate the concentration in suspensions. Zymosan A (Sigma) was suspended in dPBS to 10 mg/mL, pipetted repeatedly, then passed through a 40 μm cell strainer to remove large particles and rinsed with dPBS twice by centrifugation (5,000g, 1 min, room temperature). Sheep red blood cells (MP Biomedicals) were opsonized as described (Mosser and Zhang, 2011). For beads, 1.20 μm diameter amino-coated polystyrene particles (Spherotech) were rinsed with dPBS twice by centrifugation (13,000g, 5 min, room temperature) before further labeling. Each prey particle was labeled with pHrodo Red succinimidyl ester (ThermoFisher Scientific) by incubation at room temperature for 30 min in dPBS at the following ratios: 50 mg/mL myelin with 6 μM pHrodo; 106 RBCs per μL with 250 μM pHrodo; 40 mg/mL zymosan with 60 μM pHrodo; 2.5% (w/v) beads with 10 μM pHrodo. Dye concentrations were optimized for the maximum level of labeling that did not produce fluorescent signal when particles alone were imaged at neutral pH. Prey particles were centrifuged (5,000g, 1 min, room temperature for RBCs or zymosan; 13,000g, 5 min, room temperature for myelin or beads) and washed a total of four times after labeling to remove excess dye before storage at 4°C for up to one month.
Prey particle suspensions were added to cells at the following concentrations, which were titrated to ensure that settled particles covered most of the well’s surface area: myelin, 0.2 mg/mL; zymosan, 0.5 mg/mL; RBCs, 104 RBCs per μL; beads, 0.025% (w/v). Culture medium was gently mixed with a pipette after addition of prey, and smaller quantities of myelin (0.05 mg/mL) were added for the movies shown to enhance cell visibility. For conditions involving removal of serum, wells were gently washed 4 times with dPBS before transfer into serum-free growth medium. For experiments involving cycloheximide (40 μM) or cytochalasin D (10 μM), drugs were added 5 min before addition of prey unless otherwise noted. Rat serum was collected from P21 rat blood that underwent platelet aggregation for 30 min at room temperature before centrifugation (10 min, 2,000g, 4°C) to remove clots and cells. Serum was heat-inactivated (20 min, 55°C), aliquoted, and stored at −20°C.
All phagocytosis assays were performed with P7 to P21 rat microglia at 5 to 7 div unless otherwise noted. Images were acquired with an Incucyte ZOOM (Essen Bioscience) at 37°C and 10% CO2 without perturbing the plates. Custom analysis scripts within the Incucyte ZOOM Software were used to measure phagocytosed particles (pHrodo-positive area), and intensity and particle size cutoffs were used to discriminate signal from noise. Identical image acquisition and analysis parameters were used for all experiments, and analysis script parameters were adjusted for each batch of prey particles to ensure accurate reporting. Cellular localization of quantified pHrodo signal was confirmed visually. Movie image stacks were aligned in ImageJ using the cvMatch_Template plugin (Tseng et al., 2012). Single time point measurements were captured with an Axio Observer.Z1 (Zeiss) an analyzed with custom ImageJ scripts.
Flow cytometry
Single cell suspensions were stained with the dead cell marker LIVE/DEAD for 5 min on ice (1:1000; Life Technologies), and then immunostained using antibodies specific to TMEM119 (Bennett et al., 2016). Each lot is titrated to achieve maximal staining of WT tissue with zero staining of Tmem119 knockout mouse tissue; then stained with Brilliant Violet 421 Donkey anti-rabbit IgG (1:300; Biolegend), PE-Cy7-CD45 (Clone 30-F11; 1:250; eBioscience), and PerCP/Cy5.5-CD11b (1:250; Biolegend). Samples were analyzed immediately or fixed in 1% PFA, using an LSR II or Aria (Becton Dickinson), and data processed using Flowjo software (Treestar). Data was collected on an instrument in the Stanford Shared FACS Facility obtained using NIH S10 Shared instrument Grant (S10RR027431-01). Debris, doublets, and dead cells were excluded using fsc/ssc, fsc-h/fsc-w, and live cell gates, respectively. Tmem119 mean florescence values represent the geometric mean of florescence calculated on CD45/CD11b double positive cells.
Immunohistochemistry
Cells were plated on hand-punched Aclar coverslips. After 5 days in culture, cells were rinsed with dPBS and fixed for 10 min with 4% PFA. After three PBS rinses, cells were blocked and permeabilized in PBS containing 20% goat serum and 0.2% Triton X. After three more PBS rinses, cells were incubated for 16 hours at 4°C overnight in PBS containing 2% goat se rum and primary antibodies rat anti-F4/80 (1:100; Abcam); rabbit anti-Iba1, (1:500; Wako); rabbit anti-GFAP (1:1000 Dako). Fixed cells were subjected to three five minute washes in PBS before addition of PBS containing 2% goat serum and highly cross-absorbed secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 594 (1:500; Life Technologies). Cells were subjected to three five minute washes in PBS before mounting onto SuperFrost Plus glass slides (Fisher) with Dapi Fluoromount-G (Southern Biotech).
For tissue staining, tissue pieces cut from intracranially injected cortices were immersion fixed in 4% PFA overnight at 4°C, sunk in 30% sucrose-PBS, and embedded in OCT (Fisher). 12 micron cryosections on SuperFrost Plus slides (Fisher) were blocked for 1 hour with PBS/10% donkey serum/0.5% Triton x-100 at room temperature, then stained overnight with primary antibodies at 4°C; rabbit anti-Tmem119 custom Ab hybridoma supernatant (Bennett et al., 2016); goat anti-Iba1 (1:500; Abcam) in PBSTx (PBS/1% donkey serum/0.5% triton x-100). After washing, slides were incubated for 2 hours at RT with highly cross absorbed Alexa-conjugated secondary antibodies (1:500, Life Technologies) in PBSTx. Coverslips were mounted with Vectashield with DAPI (Vector labs). Images were acquired on a Zeiss Axioimager M1 with 20x 0.8 NA Plan Apo objective. Images were adjusted for brightness and black values in ICY (http://icy.bioimageanalysis.org).
QPCR
20,000 to 40,0000 cells or ~1 mm3 of brain tissue were quickly rinsed with dPBS then lysed in chilled RLT buffer (Qiagen) and immediately frozen until further processing. RNA was isolated using the RNeasy Micro Kit (Qiagen) with on-column DNase treatment. RNA was reverse-transcribed using a High-Capacity RNA-to-cDNA Kit (ThermoFisher Scientific) and used as template for SYBR Green PCR Master Mix (ThermoFisher Scientific) reactions (20 μL volume), which were assembled in 96-well PCR plates (Fisher Scientific) for cycling in an Eppendorf Realplex 4 QPCR machine. Primers were designed to bridge splice-junctions when possible using Primer3 or previously published sequences. Primers were synthesized at the Stanford PAN facility and used at a final concentration of 100 nM each. Primer efficiency was measured for each primer set and were only used for experiments if efficiency was between 85% and 115% (r2 > 0.98). Rat primer sequences are provided in Supplemental Table 3, and other primer sequences were previously reported (Bennett et al., 2016). CT values were registered using noiseband thresholding, and melting curves were checked to ensure formation of a single product within the expected size range. ΔCT values were calculated relative to the housekeeping gene RPLP0 except for excised tissue samples shown in Fig. S6, for which ΔCT values were calculated relative to Csf1r to account for potential variation in the fractional abundance of myeloid cells across samples. Consistent ΔCT values of a second housekeeping gene, EEF1A1, were measured across samples, arguing against the possibility that RPLP0 expression levels change dramatically across conditions. All CT values were detected before completion of the 38th cycle.
For mouse qPCR experiments, RNA isolation was performed as described above, however Vilo Master Mix (Thermofisher) was used for reverse transcription, Powerup Sybr Green (Thermofisher) for qPCR, and samples were run on a QuantStudio 3 thermocycler (Thermofisher). Mouse primers were used at 800 nM.
RNA sequencing
RNA was isolated as above and analyzed for integrity by BioAnalyzer (Eukaryote RNA Pico, Agilent). For each replicate, 10 ng of total RNA (RIN > 9.7) was used to prepare a poly-A enriched cDNA library using Smart-seq2 (Picelli et al., 2014). Libraries were modified for sequencing using the Nextera XT DNA Sample Preparation Kit (Illumina) with 150 pg of cDNA as input material. Libraries were sequenced using an Illumina MiSeq to obtain 75 bp paired-end reads. Data were collected in two batches of library preparation and sequencing, with samples from each experimental condition included in each batch.
Sequencing reads were mapped to the UCSC rat reference genome rn6 using HISAT version 2.0.3 (Kim et al., 2015) via the Galaxy platform (http://usegalaxy.org) (Afgan et al., 2016), resulting in concordant pair alignment rates of >75% for each sample. Transcript FPKM (fragments per kilobase of transcript sequence per million mapped fragments) scores were obtained using Cufflinks version 2.2.1 (Trapnell et al., 2010) with the iGenomes rn6 gtf file as a reference annotation. Sample purity was analyzed using cell type specific marker expression (Zhang et al., 2014), confirming that the sequenced samples were comprised predominantly of microglia mRNA and little to no mRNA from astrocytes, neurons, endothelial cells, or oligodendrocyte lineage cells. Freshly isolated samples contained some neutrophil mRNA, and highly expressed neutrophil genes represent some of the genes listed as downregulated in culture. Identification of differentially expressed genes between conditions was calculated using DESeq2 (Love et al., 2014). For calculating fold-change, a value of 0.1 was added to all FPKM values to allow for meaningful comparison of genes that showed extremely low abundance in at least one conditions.
Gene list generation and comparisons
For comparisons of culture-induced changes with published datasets, we first selected recent RNA-seq datasets characterizing isolated microglia for which related phenomena had been characterized by independent groups. The following groups were compared to obtain the gene lists used: SODG93A endstage versus non-transgenic day 130 for ALS (Chiu et al., 2013); APP+ versus APP- for AD (Wang et al., 2015); intraperitoneal LPS (1 or 2 days post injection) versus saline for LPS 1 or LPS 2 (Bennett et al., 2016; Chiu et al., 2013); Brain E10.5, E11.5, and E12.5 (average) versus Adult cortex, hippocampus, and spinal cord (average) for Dev Down or Dev Up 1 (Matcovitch-Natan et al., 2016); E17 versus P14, P21, and P60 (average) for Dev Down or Dev Up 2 (Bennett et al., 2016). Published tables reporting statistically-significant changes were used, and a 4-fold change cutoff was applied to filter out smaller differences more likely to go undetected across different datasets. We limited our analysis to 13,741 genes sharing identical nomenclature in the most recent mouse (mm10) and rat (rn6) UCSC genome annotations, resulting in lists of 40–489 genes per condition. We did not include genes downregulated in SOD1G93A, 5XFAD, or LPS-injected mice due to the low number of such genes detected in these experiments. Gene list overlap and P-values were calculated using the GeneOverlap package in R (version 3.3.1).
To generate the list of mature microglia signature genes used in Fig. 6F, we started with “Cluster 1” from (Lavin et al., 2014) and identified genes also upregulated in the Dev Up 1 list from above. To facilitate cross-species comparison, we considered only genes sharing identical gene ID annotation in rat and mouse. Because several mouse microglia signature genes were not detected in our freshly isolated rat microglia, we further limited our analysis to the most highly expressed genes, considering only genes expressed at FPKM > 10 in both freshly isolated mouse microglia and freshly isolated rat microglia. This resulted in a list of 88 genes that included most established microglial markers including Tmem119, P2ry12, Trem2, and Sall1.
Quantification and Statistical Analysis
Quantification of live/dead or phagocytosis images was performed in ImageJ using custom macros (available upon request). Micrographs shown were contrast adjusted for display using Adobe Photoshop. Data were analyzed and plotted using Excel (Microsoft) and Igor (WaveMetrics). Averages represent n ≥ 3 independent experiments except for dose-response curves, phagocytosis live-imaging traces, Fig. S4C, and Fig. S4F, which are representative of multiple independent experiments but display data averaged from n ≥ 3 technical replicates within a single experiment of n ≥ 3 pooled biological samples. Micrographs and FACS plots are representative images of n ≥ 3 biological replicates. Unless otherwise stated, error bars are SEM, and p values were calculated using ANOVA followed by Dunnett’s multiple comparison test to the leftmost control group. Gene set enrichment analysis (GSEA (Subramanian et al., 2005)) was run with 10,000 permutations using the log2 Ratio of Classes metric and minimum set size of 25 genes against MSigDB KEGG pathway list (c2.cp.kegg.v5.2.symbols.gmt) or manually uploaded gene lists (Dataset S1). A hypergeometric distribution test was used to verify significance of overlap between gene lists.
KEY RESOURCES TABLE
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-CD11b (clone OX42) | Bio-Rad | Cat# MCA275R |
| Rat monoclonal anti-CD45 | BD Pharmingen | Cat# 550539 |
| Lectin BSL-1 | Vector Labs | Cat# L-1100 |
| Mouse monoclonal anti-ITGB5 | eBioscience | Cat# 14-0497-82 |
| Neutralizing antibodies against TGF-β (clone 1D11) | R&D Systems | Cat# MAB1835 |
| Anti-CD11b microbeads | Miltenyi | Cat# 130-049-601 |
| Rabbit monoclonal anti-TMEM119 | (Bennett et al., 2016) | N/A |
| Rabbit hybridoma supernatant anti-TMEM119 | (Bennett et al., 2016) | N/A |
| Goat anti-mouse IgG (H+L) | Jackson ImmunoResearch | Cat# 115-005-003 |
| Donkey anti-rabbit (H+L), 421 brilliant blue | Biolegend | Cat# 406410 |
| Rat monoclonal anti-mouse CD45 | eBioscience | Cat# 25-0451-82 |
| Rat anti-CD11b, conjugated PerCP/Cy5.5 | Biolegend | Cat# 101228 |
| Rat monoclonal anti-F4/80 | Abcam | Cat# ab6640 |
| Rabbit polyclonal anti-Iba1 | Wako | Cat# 019-19741 |
| Goat polyclonal anti-Iba1 | Abcam | Cat# AB5076 |
| Rabbit polyclonal anti-GFAP | Dako | Cat# Z0334 |
| O4 hybridoma supernatant mouse IgM | (Foo et al., 2011) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Heparan sulfate | Galen Laboratory Supplies | Cat# GAG-HS01 |
| Heparin | Sigma | Cat# M3149 |
| Peptone from milk solids | Sigma | Cat# P6838 |
| Methyl-β-cyclodextrin | Sigma | Cat# C4555 |
| Human TGF-β2 | Peprotech | Cat# 100-35B |
| Murine IL-34 | R&D Systems | Cat# 5195-ML/CF |
| Ovine wool cholesterol | Avanti Polar Lipids | Cat# 700000P |
| Collagen IV | Corning | Cat#354233 |
| Oleic acid | Cayman Chemicals | Cat# 90260 |
| 11(Z)Eicosadienoic (Gondoic) Acid | Cayman Chemicals | Cat# 20606 |
| Calcein AM dye | Invitrogen | Cat# C3100MP |
| Ethidium homodimer-1 | Invitrogen | Cat# E1169 |
| Ovomucoid trypsin inhibitor | Worthington | Cat# LS003086 |
| Cycloheximide | Sigma | Cat# C7698 |
| Cytochalasin D | Sigma | Cat# C8273 |
| Cytosine-arabinoside | Sigma | Cat# C1768 |
| DNaseI | Worthington | Cat# DPRFS |
| Percoll PLUS | GE Healthcare | Cat# 17-5445-02 |
| Trypsin | Sigma | Cat# T9935 |
| DMEM/F12 | Gibco | Cat# 21041-02 |
| Penicillin/ Streptomycin | Gibco | Cat# 15140-122 |
| Glutamine | Gibco | Cat# 25030-081 |
| N-acetyl cysteine | Sigma | Cat# A9165 |
| Insulin | Sigma | Cat# I6634 |
| Apo-transferrin | Sigma | Cat# T1147 |
| Sodium selenite | Sigma | Cat# S-5261 |
| Papain | Worthington | Cat# LS003126 |
| DMEM (high glucose) | Gibco | Cat# 11960-044 |
| Neurobasal | Gibco | Cat# 21103-049 |
| Sodium pyruvate | Sigma | Cat# P5280 |
| Hydrocortisone | Sigma | Cat# H0888 |
| Forskolin | Sigma | Cat# F6886 |
| Putrescine | Sigma | Cat# P-5780 |
| Progesterone | Sigma | Cat# P-8783 |
| Dapi Fluoromount-G | Southern Biotech | Cat# 0100-20 |
| 30 kDa MWCO centrifugal filter unit | Sartorius | Cat# VS2022 |
| HiTrap Heparin HP columns | GE Healthcare | Cat# 17-0406-01 |
| HiTrap Q FF columns | GE Healthcare | Cat# 17-5053-01 |
| HiTrap Concanavalin A Sepharose 4B columns | GE Healthcare | Cat# 28-9520-85 |
| HiTrap Phenyl FF (High Sub) columns | GE Healthcare | Cat# 17-1355-01 |
| 4–20% SDS PAGE gels | Bio-Rad | Cat# 4561096 |
| SYPRO orange | Invitrogen | Cat# S6650 |
| Zymosan A | Sigma | Cat# Z4250 |
| Sheep red blood cells | MP Biomedicals | Cat# 55876 |
| 1.20 μm diameter amino-coated polystyrene particles | Spherotech | Cat# AM-10-10 |
| pHrodo Red succinimidyl ester | ThermoFisher | Cat# P36600 |
| Myelin Removal Beads II | Miltenyi | Cat# 130-096-433 |
| LS columns | Miltenyi | Cat# 130-042-401 |
| QuadroMACS Separator | Miltenyi | Cat# 130-090-976 |
| Microcapillary tube | WPI | Cat# 1B100f-4 |
| OCT | ThermoFisher | Cat# 23-730-571 |
| Vectashield with DAPI | Vector labs | Cat# H-1200 |
| SYBR Green PCR Master Mix | ThermoFisher | Cat# 4385612 |
| Vilo Master Mix | ThermoFisher | Cat# 11755050 |
| Powerup Sybr Green | Thermofisher | Cat# A25741 |
| Human serum | Gemini | Cat# 100-512 |
| GW2580 | Cayman Chemicals | Cat# 15411 |
| Critical Commercial Assays | ||
| LIVE/DEAD | Life Technologies | Cat# L23101 |
| Bradford Assay | Bio-Rad | Cat# 500-0006 |
| RNeasy Micro Kit | Qiagen | Cat# 74004 |
| High-Capacity RNA-to-cDNA Kit | ThermoFisher Scientific | Cat# 4387406 |
| Nextera XT DNA Sample Preparation Kit | Illumina | Cat# FC-131-1024 |
| Nextera XT Index Kit (24 indexes, 96 samples) | Illumina | Cat# FC-131-1001 |
| Deposited Data | ||
| SODG93A endstage versus non-transgenic day 130 for ALS | (Chiu et al., 2013) | |
| Mouse reference genome UCSC annotation mm10 | UCSC Genome Browser | |
| Rat reference genome UCSC annotation rn6 | UCSC Genome Browser | |
| iGenomes reference annotation gtf for rn6 | Illumina | |
| APP+ versus APP-for AD | (Wang et al., 2015) | |
| Intraperitoneal LPS (1 or 2 days post injection) versus saline for LPS 1 or LPS 2 | (Bennett et al., 2016; Chiu et al., 2013) | |
| E17 versus P14, P21, and P60 (average) for Dev Down 2 or Dev Up 2 | (Bennett et al., 2016) | |
| Brain E10.5, E11.5, and E12.5 (average) versus Adult cortex, hippocampus, and spinal cord (average) for Dev Down 1 or Dev Up 1 | (Matcovitch-Natan et al., 2016) | |
| Cluster 1 – microglia versus other tissue macrophages | (Lavin et al., 2014) | |
| Experimental Models: Organisms/Strains | ||
| Rat: Sprague-Dawley | Charles River | Cat# 400 |
| Knockout Mouse: FVB.129X1-Csf1rtm1Ers | (Dai et al., 2002) | N/A |
| Mouse: FVB/NJ | Jackson Laboratory | Cat# 001800 |
| Oligonucleotides | ||
| Primers for QPCR (rat)—See Table S2 | This Paper | |
| Primers for QPCR (mouse) | (Bennett et al., 2016) | |
| Recombinant DNA | ||
| Software and Algorithms | ||
| ImageJ | NIH | |
| Incucyte ZOOM | Essen Bioscience | |
| cvMatch_Template ImageJ plugin | https://sites.google.com/site/qingzongtseng/templatematching-ij-plugin | |
| Primer3 | The Whitehead Institute | |
| Flowjo | Treestar | |
| HISAT version 2.0.3 | (Kim et al., 2015) | |
| Galaxy platform | http://usegalaxy.org | |
| Cufflinks version 2.2.1 | (Trapnell et al., 2010) | |
| DESeq2 | (Love et al., 2014) | |
| R (version 3.3.1) | The R Foundation | |
| GeneOverlap plugin for R | https://www.bioconductor.org/packages/release/bioc/html/GeneOverlap.html | |
| Pvclust plugin for R | https://cran.rproject.org/web/packages/pvclust/index.html | |
| Igor | WaveMetrics | |
| Other | ||
| Dounce homogenizer | Wheaton | Cat# 357424 |
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
-
IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
Please see the sample Table at the end of this document for examples of how reagents should be cited. To see how the typeset table will appear in the PDF and online, please refer to any of the research articles published in Cell in the August 25, 2016 issue and beyond.
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. You do not need to delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell.
TABLE FOR AUTHOR TO COMPLETE
Please upload the completed table as a separate document. Please do not add subheadings to the Key Resources Table. If you wish to make an entry that does not fall into one of the subheadings below, please contact your handling editor.
TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and Virus Strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological Samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 12487-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406P01 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental Models: Cell Lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUU AGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130 415 |
| AAV2/1-hsyn-GCaMP6-WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and Algorithms | ||
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtiebio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
Supplementary Material
Related to Figure 2
Phase images of P20 microglia at 7 div. Each frame is 30 min apart and movies are played at 3 frames per second.
Related to Figure 2
Phase images of P20 microglia at 7 div exposed to 10% FCS for 24 hr before imaging. Each frame is 30 min apart and movies are played at 3 frames per second.
Related to Figure 4
Phase images of P15 microglia at 7 div interacting with pHrodo-labeled myelin debris (red). Each frame is 10 min apart and movies are played at 12 frames per second. Minimal pHrodo signal is observed.
Related to Figure 4
P15 microglia at 7 div exposed to 10% FCS for 24 hr before addition of pHrodo-labeled myelin debris (red). Each frame is 10 min apart and movies are played at 12 frames per second. Robust pHrodo signal is observed even before the added myelin has settled to the bottom of the well.
Related to Figure 5
Phase images of P24 microglia at 14 div plated on collagen-coated Primaria tissue culture plates in TIC with monounsaturated fatty acid and heparan sulfate supplementation. Each frame is 3 min apart and movies are played at 6 frames per second. Timestamp in lower-right corner denotes hours:minutes.
Related to Figure 5
Acknowledgments
We thank members of the Barres laboratory for feedback throughout, especially M Bennett, S Sloan, L Foo, A Lutz, and M Fabian. We thank the Stanford PAN Facility and the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University. This work was supported by the Christopher and Dana Reeve Foundation International Research Consortium on Spinal Cord Injury, the Dr. Miriam and Sheldon G. Adelson Medical research Foundation, the JPB Foundation, the Novartis Institute of Basic Research, a National Institutes of Health (NIH) Grant to B.A.B. (R01 DA015043), and generous contributions from Vincent and Stella Coates. C.J.B. was supported by the Damon Runyon Cancer Research Foundation (DRG-2125-12). F.C.B. was supported by an NIH training grants (5T32MH019938-22, 1K08MH112120).
Footnotes
Author Contributions
C.J.B and B.A.B conceived the project, designed experiments, and wrote the paper. F.C.B conceived, performed, and analyzed experiments related to CNS engraftment with assistance from S.B.M. C.J.B, A.F.T, and H.Y.C conducted all other experiments and analyses.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- Barres BA, Schmid R, Sendnter M, Raff MC. Multiple extracellular signals are required for long-term oligodendrocyte survival. Development. 1993;118:283–295. doi: 10.1242/dev.118.1.283. [DOI] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A. 2016;113:E1738–1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC, Neher JJ. Microglial phagocytosis of live neurons. Nat Rev Neurosci. 2014;15:209–216. doi: 10.1038/nrn3710. [DOI] [PubMed] [Google Scholar]
- Bruttger J, Karram K, Wortge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, et al. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity. 2015;43:92–106. doi: 10.1016/j.immuni.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, Nishinakamura R, Becher B, Greter M. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol. 2016 doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
- Chen Z, Jalabi W, Shpargel KB, Farabaugh KT, Dutta R, Yin X, Kidd GJ, Bergmann CC, Stohlman SA, Trapp BD. Lipopolysaccharide-induced microglial activation and neuroprotection against experimental brain injury is independent of hematogenous TLR4. J Neurosci. 2012;32:11706–11715. doi: 10.1523/JNEUROSCI.0730-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O’Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Holtzman DM, Munson G, Mathur T, Schneider D, Chang LK, Getz GS, Reardon CA, Lukens J, Shah JA, et al. Unique lipoproteins secreted by primary astrocytes from wild type, apoE (−/−), and human apoE transgenic mice. J Biol Chem. 1999;274:30001–30007. doi: 10.1074/jbc.274.42.30001. [DOI] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, Cahoy JD, Daneman R, Zong H, Ellisman MH, et al. Development of a method for the purification and culture of rodent astrocytes. Neuron. 2011;71:799–811. doi: 10.1016/j.neuron.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourgeaud L, Traves PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagorska A, Rothlin CV, Nimmerjahn A, et al. TAM receptors regulate multiple features of microglial physiology. Nature. 2016;532:240–244. doi: 10.1038/nature17630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SA, Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol Rev. 2014;262:193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek R, Molawi K, Sieweke MH. Tissue macrophage identity and self-renewal. Immunol Rev. 2014;262:56–73. doi: 10.1111/imr.12224. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- Gordon S, Pluddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev. 2014;262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh AD, David S. Differences in the phagocytic response of microglia and peripheral macrophages after spinal cord injury and its effects on cell death. J Neurosci. 2014;34:6316–6322. doi: 10.1523/JNEUROSCI.4912-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, Kundig TM, Frei K, Ginhoux F, Merad M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, Julius D. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. 2006;9:1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoosein NM, Brattain DE, McKnight MK, Brattain MG. Comparison of the effects of transforming growth factor beta, N,N-dimethylformamide, and retinoic acid on transformed and nontransformed fibroblasts. Exp Cell Res. 1988;175:125–135. doi: 10.1016/0014-4827(88)90260-1. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koso H, Tsuhako A, Lai CY, Baba Y, Otsu M, Ueno K, Nagasaki M, Suzuki Y, Watanabe S. Conditional rod photoreceptor ablation reveals Sall1 as a microglial marker and regulator of microglial morphology in the retina. Glia. 2016 doi: 10.1002/glia.23038. [DOI] [PubMed] [Google Scholar]
- LaDu MJ, Gilligan SM, Lukens JR, Cabana VG, Reardon CA, Van Eldik LJ, Holtzman DM. Nascent astrocyte particles differ from lipoproteins in CSF. J Neurochem. 1998;70:2070–2081. doi: 10.1046/j.1471-4159.1998.70052070.x. [DOI] [PubMed] [Google Scholar]
- Larocca JN, Norton WT. Isolation of myelin. Curr Protoc Cell Biol. 2007;Chapter 3(Unit 3–25) doi: 10.1002/0471143030.cb0325s33. [DOI] [PubMed] [Google Scholar]
- Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol. 2015;15:731–744. doi: 10.1038/nri3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Tse W, Smith JD, Landreth GE. Apolipoprotein E promotes beta-amyloid trafficking and degradation by modulating microglial cholesterol levels. J Biol Chem. 2012;287:2032–2044. doi: 10.1074/jbc.M111.295451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- Liu W, Brosnan CF, Dickson DW, Lee SC. Macrophage colony-stimulating factor mediates astrocyte-induced microglial ramification in human fetal central nervous system culture. Am J Pathol. 1994;145:48–53. [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, et al. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell. 2016;165:921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuch A, van der Pijl R, Funer L, Wolf Y, Eggen B, Boddeke E, Biber K. Microglia replenished OHSC: A culture system to study in vivo like adult microglia. Glia. 2016;64:1285–1297. doi: 10.1002/glia.23002. [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- Mosser DM, Zhang X. Measuring opsonic phagocytosis via Fcgamma receptors and complement receptors on macrophages. Curr Protoc Immunol. 2011;Chapter 14(Unit 14–27) doi: 10.1002/0471142735.im1427s95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TD, Nickel J. Promiscuity and specificity in BMP receptor activation. FEBS Lett. 2012;586:1846–1859. doi: 10.1016/j.febslet.2012.02.043. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger FW, Ungerer N. Cholesterol metabolism in neurons and astrocytes. Prog Lipid Res. 2011;50:357–371. doi: 10.1016/j.plipres.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Rabani M, Raychowdhury R, Jovanovic M, Rooney M, Stumpo DJ, Pauli A, Hacohen N, Schier AF, Blackshear PJ, Friedman N, et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell. 2014;159:1698–1710. doi: 10.1016/j.cell.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM. A polarizing question: do M1 and M2 microglia exist? Nat Neurosci. 2016;19:987–991. doi: 10.1038/nn.4338. [DOI] [PubMed] [Google Scholar]
- Safaiyan S, Kannaiyan N, Snaidero N, Brioschi S, Biber K, Yona S, Edinger AL, Jung S, Rossner MJ, Simons M. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat Neurosci. 2016;19:995–998. doi: 10.1038/nn.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi K, Moser KV, Marksteiner J, Reindl M, Humpel C. GDNF and TGF-beta1 promote cell survival in serum-free cultures of primary rat microglia. Cell Tissue Res. 2003;312:135–139. doi: 10.1007/s00441-003-0711-7. [DOI] [PubMed] [Google Scholar]
- Sato JD, Kan M. Media for culture of mammalian cells. Curr Protoc Cell Biol. 2001;Chapter 1(Unit 1–2) doi: 10.1002/0471143030.cb0102s00. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling M, Besselmann M, Muller M, Strecker JK, Ringelstein EB, Kiefer R. Predominant phagocytic activity of resident microglia over hematogenous macrophages following transient focal cerebral ischemia: an investigation using green fluorescent protein transgenic bone marrow chimeric mice. Exp Neurol. 2005;196:290–297. doi: 10.1016/j.expneurol.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Schilling T, Nitsch R, Heinemann U, Haas D, Eder C. Astrocyte-released cytokines induce ramification and outward K+ channel expression in microglia via distinct signalling pathways. Eur J Neurosci. 2001;14:463–473. doi: 10.1046/j.0953-816x.2001.01661.x. [DOI] [PubMed] [Google Scholar]
- Sievers J, Parwaresch R, Wottge HU. Blood monocytes and spleen macrophages differentiate into microglia-like cells on monolayers of astrocytes: morphology. Glia. 1994;12:245–258. doi: 10.1002/glia.440120402. [DOI] [PubMed] [Google Scholar]
- Stansley B, Post J, Hensley K. A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J Neuroinflammation. 2012;9:115. doi: 10.1186/1742-2094-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: aknowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J, Maeda N. Microglial ramification requires nondiffusible factors derived from astrocytes. Exp Neurol. 1996;137:367–375. doi: 10.1006/exnr.1996.0038. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinder PK, Faust D, Petry F, Loos M. Modulation of mRNA expression and secretion of C1q in mouse macrophages by anti-inflammatory drugs and cAMP: evidence for the partial involvement of a pathway that includes cyclooxygenase, prostaglandin E2 and adenylate cyclase. Immunology. 1995;84:638–644. [PMC free article] [PubMed] [Google Scholar]
- Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, Thery M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci U S A. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas ME, Barres BA. Why is Wallerian degeneration in the CNS so slow? Annu Rev Neurosci. 2007;30:153–179. doi: 10.1146/annurev.neuro.30.051606.094354. [DOI] [PubMed] [Google Scholar]
- Vasek MJ, Garber C, Dorsey D, Durrant DM, Bollman B, Soung A, Yu J, Perez-Torres C, Frouin A, Wilton DK, et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534:538–543. doi: 10.1038/nature18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, Barrow AD, Diamond MS, Colonna M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Nandi S, Chitu V, Yeung YG, Yu W, Huang M, Williams LT, Lin H, Stanley ER. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J Leukoc Biol. 2010;88:495–505. doi: 10.1189/jlb.1209822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Sayed FA, Bard F, Gan L. Targeting microglia for the treatment of Alzheimer’s Disease. Glia. 2016;64:1710–1732. doi: 10.1002/glia.22988. [DOI] [PubMed] [Google Scholar]
- Witmer-Pack MD, Hughes DA, Schuler G, Lawson L, McWilliam A, Inaba K, Steinman RM, Gordon S. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci. 1993;104(Pt 4):1021–1029. doi: 10.1242/jcs.104.4.1021. [DOI] [PubMed] [Google Scholar]
- Witting A, Moller T. Microglia cell culture: a primer for the novice. Methods Mol Biol. 2011;758:49–66. doi: 10.1007/978-1-61779-170-3_4. [DOI] [PubMed] [Google Scholar]
- Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 2
Phase images of P20 microglia at 7 div. Each frame is 30 min apart and movies are played at 3 frames per second.
Related to Figure 2
Phase images of P20 microglia at 7 div exposed to 10% FCS for 24 hr before imaging. Each frame is 30 min apart and movies are played at 3 frames per second.
Related to Figure 4
Phase images of P15 microglia at 7 div interacting with pHrodo-labeled myelin debris (red). Each frame is 10 min apart and movies are played at 12 frames per second. Minimal pHrodo signal is observed.
Related to Figure 4
P15 microglia at 7 div exposed to 10% FCS for 24 hr before addition of pHrodo-labeled myelin debris (red). Each frame is 10 min apart and movies are played at 12 frames per second. Robust pHrodo signal is observed even before the added myelin has settled to the bottom of the well.
Related to Figure 5
Phase images of P24 microglia at 14 div plated on collagen-coated Primaria tissue culture plates in TIC with monounsaturated fatty acid and heparan sulfate supplementation. Each frame is 3 min apart and movies are played at 6 frames per second. Timestamp in lower-right corner denotes hours:minutes.
Related to Figure 5