Abstract
This study was undertaken to ascertain whether defined markers of early zebrafish brain development are affected by chronic ethanol exposure or morpholino knockdown of agrin, sonic hedgehog, retinoic acid and fibroblast growth factors, four signaling molecules that are suggested to be ethanol sensitive. Zebrafish embryos were exposed to 2% ethanol from 6–24 hpf or injected with agrin, shha, aldh1a3 or fgf8a morpholinos. In situ hybridization was employed to analyze otx2, pax6a, epha4a, krx20, pax2a, fgf8a, wnt1 and eng2b expression during early brain development. Our results showed that pax6a mRNA expression was decreased in eye, forebrain and hindbrain of both chronic ethanol exposed and select MO treatments. Epha4a expression in rhombomere R1 boundary was decreased in chronic ethanol exposure and aldh1a3 morphants, lost in fgf8a morphants, but largely unaffected in agrin and shha morphants. Ectopic pax6a and epha4a expression in midbrain was only found in fgf8a morphants. These results suggest that while chronic ethanol induces obvious morphological change in brain architecture, many molecular markers of these brain structures are relatively unaffected by ethanol exposure.
Keywords: zebrafish, fgf, sonic hedgehog, ethanol, retinoic acid, FASD
Introduction
Alcohol is a teratogen affecting central nervous system (CNS) development and results in congenital abnormalities of the eyes (Chan et al., 1991; Dangata et al., 1997; Stromland, 1985; Stromland et al., 1994), forebrain (Clarren et al., 1978; Mattson et al., 1996) and hindbrain (Cragg et al., 1985; Goodlett et al., 1990). Understanding the molecular mechanisms that lead to fetal alcohol spectrum disorders (FASD) is a critical emphasis of FASD research. A wealth of evidence indicates that at least some of the effects of ethanol on CNS development may occur via disruption of key extracellular signaling proteins, in particular pathways involving sonic hedgehog (Shh) and fibroblast growth factors (fgfs). Both fgf2 and fgf8 function is affected by ethanol exposure in prenatal mice (Aoto et al., 2008; Hard et al., 2005; Rubert et al., 2006) and in zebrafish fgf signaling is disrupted by ethanol (Zhang et al., 2013). Shh signaling is another critical target of fetal ethanol exposure (Ahlgren et al., 2002; Aoto et al., 2008; Arenzana et al., 2006; Li et al., 2007; Loucks et al., 2009; Zhang et al., 2013), and shh mRNA overexpression in zebrafish rescues many phenotypes induced by ethanol exposure (Loucks et al., 2009; Zhang et al., 2013; 2011).
Agrin is a cell surface and extracellular matrix (ECM) heparan sulfate proteoglycan (HSPG) (Tsen et al., 1995) that regulates zebrafish ocular development via a fgf-dependent mechanism (Liu et al., 2008). We have shown previously that ethanol-mediated disruption of ocular development results from perturbed agrin function (Zhang et al., 2011), and ethanol-mediated disruption of forebrain and cerebellar GABAergic neuron development occurs via a molecular pathway involving Shh and fgfs, which is modulated by agrin (Zhang et al., 2013).
An additional molecular target of prenatal ethanol exposure is retinoic acid (RA) (Duester et al., 1991; Kot-Leibovich et al., 2009; Kumar et al., 2010; Leo et al., 1999; Pillarkat et al., 1991; Sulik et al., 1981; Yelin et al., 2007; 2005; Zachman et al., 1998). Morpholino (MO) knockdown of aldh1a3, an RA biosynthetic enzyme, leads to ocular phenotypes that are similar to ethanol exposure and fgf or shh loss of function (Yahyavi et al., 2013). Our recent work has also shown that aldh1a3 knockdown induces disruption of zebrafish midbrain-hindbrain boundary (MHB) development as observed with ethanol exposure (Zhang et al., 2015).
A complex interplay among multiple extracellular signaling molecules, that are disrupted by prenatal ethanol exposure, likely underlies the hallmark morphological features of FASD (Zhang et al., 2015; 2013; 2011). Our studies have demonstrated synergistic effects of ethanol exposure and agrin, shh, or aldh1a3 MO knockdown. While there are no obvious phenotypes observed in low ethanol exposure or low MO alone, more severe phenotypes occur with combined ethanol and agrin, shh, or aldh1a3 MO knockdown. These data strongly suggest that ethanol exposure may disrupt agrin, shh, or aldh1a3 signaling.
While pronounced alterations in brain architecture result from prenatal ethanol exposure, the molecules responsible for the manifestation of these morphological alterations in brain structure are poorly understood. To further investigate the molecular mechanisms underlying these key features of FASD, we decided to compare the effects of MO treatment to ethanol exposure, as assessed by brain morphology and expression of known markers of zebrafish brain architecture. Employing either chronic ethanol exposure or MOs to agrin, Shha, aldh1a3 or fgf8a (signaling molecules that have been shown previously by numerous laboratories to be ethanol sensitive; Aoto et al, 2008; Loucks and Ahlgren 2009; Zhang et al, 2011; Zhang et al, 2015) we analyzed the expression of well characterized markers of zebrafish eye, forebrain, midbrain, MHB or hindbrain development. Our goal was to ascertain whether ethanol and these targeted MOs would produce common or different molecular changes, thereby providing insight into molecular processes sensitive to ethanol that may lead to the dysmorphogenesis observed in FASD. Our studies showed that fgf8a MO knockdown had the most marked effect on expression of these markers, which ranged from ectopic expression (pax6a, epha4a), to loss of expression in specific brain regions (wnt1, eng2b, pax2a). Chronic ethanol exposure surprisingly did not alter the expression of most of these CNS markers, with only down-regulated pax6a expression in eye, forebrain, hindbrain and decreased epha4a expression in R1 boundary being altered by ethanol. While agrin and shh MOs primarily altered expression of pax6a, aldh1a3 MO produced more marked alterations in the expression of these markers, with decreased MHB pax2a, fgf8a and wnt1, decreased pax6a, and decreased R1 epha4a. Importantly, although both chronic ethanol exposure and specific MOs resulted in morphological disruption of the MHB, the MHB expression of these markers was largely unaffected by many of the MO and ethanol treatments. These data suggest that the morphological development of the MHB is independent of the expression of many known functional markers of this critical brain patterning structure, since ethanol and MO treatments that morphologically disrupt the MHB do not alter expression of well characterized MHB markers.
Materials and methods
Animals
Zebrafish (AB strain) were obtained from Zebrafish International Resource Center and housed in automated fish housing systems (Aquaneering, San Diego, CA) at 28.5° C. All procedures using zebrafish were approved by the NCCU IACUC.
Ethanol treatment of zebrafish embryos
Zebrafish embryos in fish water containing a 1:500 dilution of 0.1% methylene blue (to prevent fungal infection) were exposed to 2% ethanol in fish water from 6 to 24 hour post-fertilization (hpf). This duration and concentration of ethanol is not lethal to embryos with a majority of embryos surviving past the larval stage (Zhang et al., 2013), and even 48 h exposure to 2% ethanol in zebrafish embryos has been reported by other laboratories (Zamora et al., 2013). We have previously shown that 2% ethanol results in approximately 65 mM ethanol in zebrafish embryos (Zhang et al, 2013), with this chronic ethanol concentration required to induce FAS phenotypes in a majority of zebrafish embryos. Embryos were incubated in 100 mm plates with the freshly diluted 2% ethanol from 6 hpf, with 10 to 30 embryos per plate. At 24 hpf ethanol was removed and embryos were washed once with fresh fish water. Embryos were then collected and fixed in 4% paraformaldehyde in PBS.
Antisense morpholino oligonucleotides (MOs) injection
Antisense MOs (Gene Tools, Philomath, OR) were designed against exon/intron splice sites for agrin (LG2:5′-CCTCTCCTTTAC GCTGTGAAGACAA-3′) (Kim et al., 2007; Zhang et al., 2011), shha (5′-CAGCACTCTCGTCAAAAGCCGCATT-3′) (Nasevicius et al., 2000; Zhang et al., 2013) and fgf8a (fgf8E2I2:5′-TAGGATGCTCTTACCATGAACGTCG-3′) (Draper et al., 2001). MO to aldh1a3 was designed to target the start codon (TATAGTCCCGTTCTGTGCCATAGC) (Bill et al., 2009; Yahyavi et al., 2013). MOs were solubilized in water at a concentration of 0.5–1.5 mM. 1 nl of agrin-LG2 MO, shha MO, aldh1a3 MO or fgf8a MO was injected into one to two-cell stage embryos. Control MO at the same concentration and volume as that of agrin MO produced no detectable effects (Kim et al., 2007). Shh morphants display phenotypes characteristic of a shh mutation (Nasevicius et al., 2000). Control MO for aldh1a3 does not produce any obvious defects and only wild-type human mRNA for ALDH1A3, but not mutant human mRNA for ALDH1A3/p.Lys190*, rescue the morphant phenotype (Yahyavi et al., 2013). Fgf8a MO injected embryos phenocopy fgf8 mutants (Draper et al., 2001). All of the MOs used have been well characterized and validated in previous publications with regard to specificity and absence of off-target effects (Nasevicius et al., 2000; Draper et al., 2001; Kim et al., 2007; Yahyavi et al., 2013).
Midbrain hindbrain boundary (MHB)
Malformation of the MHB was assessed visually based on absence of the defined border between the midbrain and hindbrain at 24 hpf. The presence of the MHB was defined as the presence of 3 or 4 ridges (tectal and cerebellar boundaries) perpendicular to the anterior-posterior axis of the CNS at the midbrain–hindbrain junction. Absence of this defined border was scored as a disruption of MHB development.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed as previously described, with probe hybridization at 65°C (Kim et al., 2007; Liu et al., 2008). Plasmids to synthesize anti-sense digoxygenin-labeled riboprobes of otx2, epha4a, pax2a, fgf8a, wnt1, eng2b, and krx20 were generous gifts from Yun-Jin Jiang. A plasmid that synthesizes pax6a anti-sense RNA probe was a generous gift from Ju-ahng Lee. Embryos were cleared in 50% glycerol and then viewed using an Olympus MVX-10 stereomicroscope.
Statistical analysis
Each treatment has been repeated at least three times, and combined number of embryos from the three biological replicates was used to calculate ratios of embryos showing effects in response to ethanol exposure and MO injection. Differences in percent of embryos displaying changed marker expression as a result of treatments were analyzed for statistical significance using one-way Anova.
Results
Effects of ethanol and perturbation of agrin, Shh, RA and fgf signaling on forebrain, midbrain and eye development
Otx2 was used as a forebrain and midbrain marker for analysis of mRNA expression. It marks the caudal limit of the developing mesencephalon (Millet et al., 1996) with expression ending at the caudal end of the mesencephalon (Figure 1A–B). We detected no obvious change in otx2 expression in all MO treated embryos, as well as ethanol exposed embryos (Figure 1C–J), an exception being caudally expanded expression in 39/48 of fgf8a morphants (Figure 1K–L; Table 1).
Figure 1. Caudally expanded expression of otx2 mRNA only occurs in fgf8a morphants.
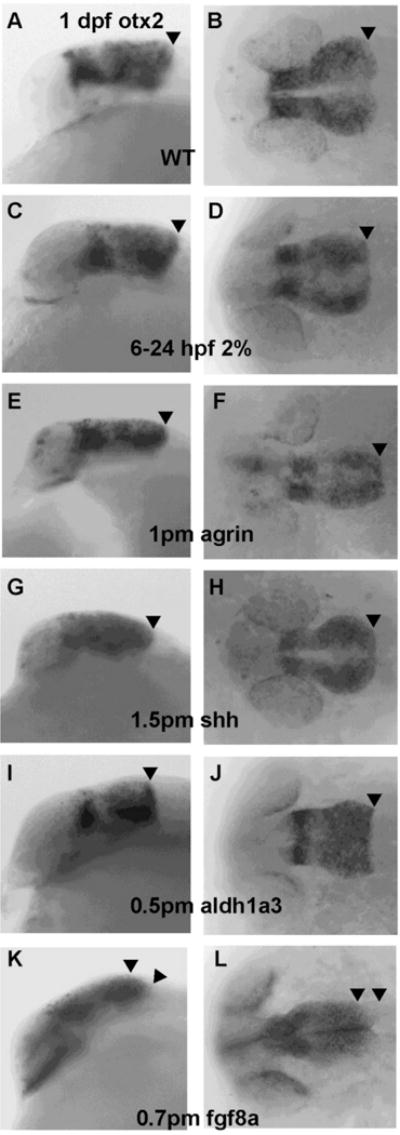
A–B, wild-type embryos (WT); C–D, embryos exposed to 2% ethanol from 6–24 hpf; E–F, embryos injected at the one to two- cell stage with 1 pmol agrin MO; G–H, embryos injected with 1.5 pmol shh MO; I–J, embryos injected with 0.5 pmol aldh1a3 MO; K–L, embryos injected with 0.7 pmol fgf8a MO. A,C,E,G,I, and K embryos are lateral views; B,D,F,H,J, and L embryos are dorsal views. 39/48 of fgf8a morphants have caudally expanded otx2 expression which was not observed with the other treatments. The arrowhead (A–J) points to the caudal expression limit of otx2 at the MHB. The two arrowheads (K–L) mark the caudally expanded territory expression of otx2 in fgf8a morphants.
Table 1.
Ratio of embryos that displayed altered marker expression in different treatments.
| Treatments | Ratio of embryos showing altered marker expression | |||||||
|---|---|---|---|---|---|---|---|---|
| otx2 | pax6a | epha4a | krx20 | pax2a | fgf8a | wnt1 | eng2b | |
| WT | 0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
| 6–24 hpf 2% | 0/45 0±0% |
37/45* 82±14% |
57/71* 80±4% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
| 1 pm agrin | 0/45 0±0% |
42/53* 79±1% |
4/51 7±6% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
| 1.5 pm shh | 0/45 0±0% |
32/48* 67±10% |
6/42* 14±7% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
0/45 0±0% |
| 0.5 pm aldh1a3 | 0/45 0±0% |
41/46* 89±10% |
27/45* 60±12% |
0/45 0±0% |
20/57* 36±9% |
19/68* 26±12% |
10/53* 18±5% |
0/45 0±0% |
| 0.7 pm fgf8a | 39/48* 82±11% |
51/56* 91±7% |
36/42* 86±7% |
0/45 0±0% |
45/45* 100±0% |
N.A | 45/45* 100±0% |
45/45* 100±0% |
Each treatment was repeated three times, and the combined number of embryos was used to calculate ratios of embryos showing effects in response to ethanol exposure and MO injection.
indicates P<0.05 comparing the treated embryos to WT embryos.
We used pax6a expression as a marker of eye and forebrain development, and observed decreased expression in 37/45 of embryos exposed to chronic 6–24 hpf 2% ethanol (Figure 2B; Table 1, Table S1), 42/53 of agrin MO-injected embryos (Figure 2C; Table 1, Table S1), 32/48 of shh MO-injected embryos (Figure 2D; Table 1, Table S1), 41/46 of aldh1a3 MO-injected embryos (Figure 2E; Table 1, Table S1) and 51/56 of fgf8a MO-injected embryos (Figure 2F; Table 1, Table S1). Interestingly, ectopic pax6a expression in midbrain was found only in fgf8a morphants (51/56 embryos) (Figure 2F; Table 1, Table S1).
Figure 2. Decreased pax6a mRNA expression in 1 dpf eye, forebrain and hindbrain following chronic ethanol exposure and different MOs treatments.
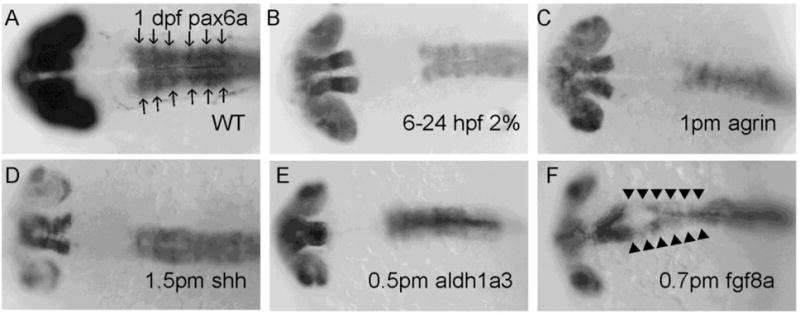
A, WT; B, embryo exposed to 2% ethanol from 6–24 hpf; C, embryo injected with 1 pmol agrin MO; D, embryo injected with 1.5 pmol shh MO; E, embryo injected with 0.5 pmol aldh1a3 MO; F, embryo injected with 0.7 pmol fgf8a MO. All embryos are dorsal views. Noted that decreased pax6a expression was observed in B–F and ectopic expression in midbrain (indicated to by arrowheads) was only observed in fgf8a morphants (F). Arrows in (A) denote rhombomeres R1–R6.
Epha4a was another forebrain and midbrain marker employed, with no obvious change in forebrain and anterior midbrain expression observed in all treatments (Figure 3C–L). Expanded epha4a expression in posterior ventral midbrain was only detected in 36/42 fgf8a morphants (Table 1, Table S1), with this expanded expression extending to the R1 boundary region of hindbrain (Figure 3K–L).
Figure 3. EphA4a mRNA expression in R1 boundary is selectively decreased in embryos exposed to chronic ethanol treatment or aldh1a3 MO.
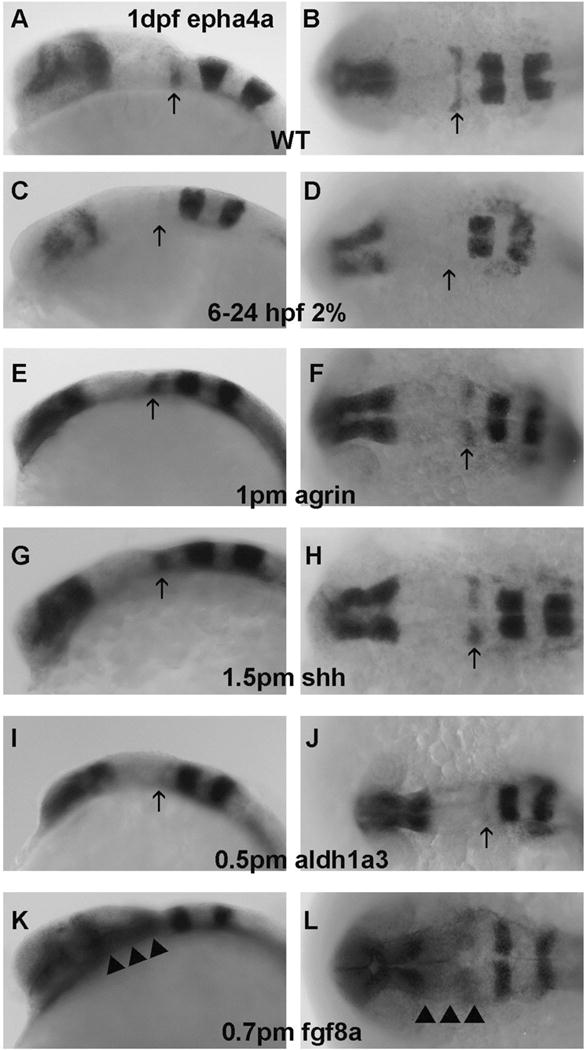
A–B, WT; C–D, embryos exposed to 2% ethanol from 6–24 hpf; E–F, embryos injected with 1 pmol agrin MO; G–H, embryos injected with 1.5 pmol shh MO; I–J, embryos injected with 0.5 pmol aldh1a3 MO; K–L, embryos injected with 0.7 pmol fgf8a MO. A,C,E,G,I, and K embryos are lateral views; B,D,F,H,J, and L embryos are dorsal views. Arrows indicate R1 boundary expression. Note that ephA4a expression in R1 decreased with ethanol exposure (C–D), aldh1a3 MO(I–J) or fgf8a MO (K,L). Ectopic epha4a expression in posterior midbrain (indicated by arrowheads) was only found in fgf8a morphants (K–L).
Effects of ethanol and perturbation of agrin, Shh, RA and fgf signaling on hindbrain development
We were interested in determining whether ethanol exposure or perturbation of agrin, Shh, RA and fgf signaling would affect expression of hindbrain markers. Pax6a is normally expressed in the entire hindbrain including R1-6 (Figure 2A),and pax6a expression levels were decreased hindbrain of all treatments (Figure 2B–F; Table 1). Epha4a was expressed in rhombomere boundaries R1, R3 and R5 in WT embryos (Figure 3A–B). However, decreased expression in R1 boundary was detected in 57/71 of chronic ethanol exposed embryos (Figure 3C–D; Table 1, Table S1) and 27/45 of aldh1a3 morphants (Figure 3I–J; Table 1, Table S1). Most agrin and shh morphants displayed relatively normal R1 expression (Figure 3E–H; Table 1), with only 4/51 and 6/42 of embryos, respectively, showing noticeably decreased R1 boundary expression. The R1 boundary expression was also lost in 36/42 fgf8a morphants (Figure 3K–L; Table 1, Table S1). Krx20 serves as a hindbrain marker, being normally expressed in R3 and R5 (Figure 4A). We observed no obvious change of the boundary expression of krx20 in R3 and R5 in all treatment groups (Figure 4B–4F; Table 1). However, it can be seen that the pattern of krx20 R3 and R5 expression in all treatment groups was altered from control embryos, likely a result of delayed development resulting from the treatments, that produced a change in R3 and R5 morphology.
Figure 4. Krx20 mRNA expression is unchanged in R3 and R5 boundaries following ethanol or MO treatments.
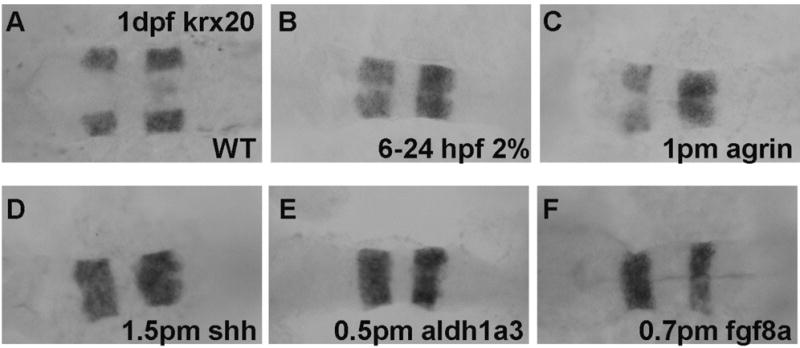
A, WT; B, embryo exposed to 2% ethanol from 6–24 hpf; C, embryo injected with 1 pmol agrin MO; D, embryo injected with 1.5 pmol shh MO; E, embryo injected with 0.5 pmol aldh1a3 MO F, embryo injected with 0.7 pmol fgf8a MO. All embryos are dorsal views.
Analysis of MHB morphological development
We have shown previously that chronic 6–24 hpf exposure of embryos to 2% ethanol (Zhang et al., 2015) and agrin MO (Kim et al., 2007; Liu et al., 2008) disrupted MHB formation. As shown here, Shh MO (Figure 5B) and fgf8a MO treatment perturbed MHB formation, both at the ratio of 30/30 (Figure 5C). To determine if molecular markers of the MHB were also affected by ethanol or MO treatments, we selected several MHB markers including pax2a (anterior MHB), fgf8a, wnt1 (dorsal median midbrain, MHB and dorsal lateral hindbrain), and eng2b (MHB and posterior midbrain) (Thisse et al., 2001; 2004; 2005). Interestingly we observed that although these treated embryos had a similar high ratio of a morphologically disrupted MHB, the molecular changes associated with this altered MHB were different. We detected no obvious change in MHB pax2a expression in chronic ethanol exposed (Figure 5E), agrin (Figure 5F) or shh morphants (Figure 5G). We identified two groups of aldh1a3 morphants based on pax2a expression, Group I showed relatively normal MHB expression (Figure 5H) and Group II (20/57; Table 1) had almost complete loss of MHB pax2a expression (Figure 5I). All fgf8a morphants showed almost complete loss of MHB pax2a expression (Figure 5J; Table 1).
Figure 5. Analysis of pax2a gene expression in ethanol and MO treated embryos.
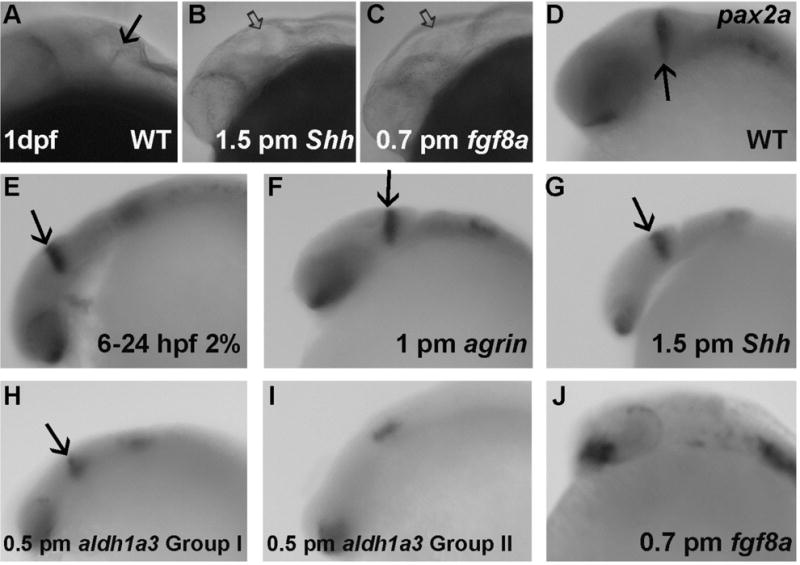
A–C, MHB morphology following shh and fgf8a MO treatment; D–J, WISH of pax2a expression. D, WT; E, embryo exposed to 2% ethanol from 6–24 hpf; F, embryo exposed to 1 pmol agrin MO; G, embryo injected with 1.5 pmol shh MO; H–I, embryos injected with 0.5 pmol aldh1a3 MO, either showing normal pax2a expression (H) or loss of MHB pax2a expression (I); J, embryo injected with 0.7 pmol fgf8a MO; All embryos are lateral views. Solid arrows indicate the morphologically normal MHB or pax2a expression in MHB. The open arrows indicate a morphologically abnormal MHB. Note that MHB morphology was disrupted in B–C and pax2a expression in MHB was lost in I and J.
Fgf8 expression in MHB was relatively normal in ethanol exposed embryos (Figure 6B), agrin morphants (Figure 6C) and shh morphants (Figure 6D). Aldh1a3 morphants could again be divided into two groups based in this case on variable fgf8a expression: Group I had relatively normal MHB expression of fgf8a (Figure 6E) and Group II (19/68 embryos) had markedly decreased MHB fgf8a expression (Figure 6F; Table 1). We observed a similar pattern of marker expression when using wnt1 as a MHB marker with normal dorsal median midbrain, MHB and dorsal lateral hindbrain expression in ethanol exposed embryos (Figure 7C–D), agrin morphants (Figure 7E–F) and shh morphants (Figure 7G–H). While Wnt1 expression in dorsal median midbrain and dorsal lateral hindbrain was similar in aldh1a3, fgf8a morphants and WT embryos (Figure 7I–N), its MHB expression was almost completely lost in Group II aldh1a3 morphants (Figure 7K–L; Table 1; 10/53) and lost in all fgf8a morphants (Figure 7M–N; Table 1). Using eng2b as marker for MHB and posterior midbrain, only fgf8a morphants (45/45 embryos) showed loss of expression in both MHB and posterior midbrain (Figure 8K–L; Table 1).
Figure 6.
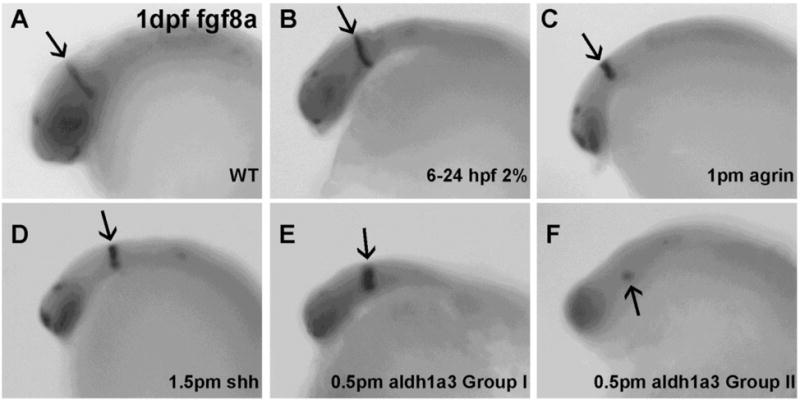
Decreased fgf8a mRNA expression in MHB is observed in aldh1a3 morphant embryos. A, WT; B, embryo exposed to 2% ethanol from 6–24 hpf; C, embryo injected with 1 pmol agrin MO; D, embryo injected with 1.5 pmol shh MO; E–F, embryos injected with 0.5 pmol aldh1a3 MO. All embryos are lateral views. Arrows indicate fgf8a expression in MHB. Noted that fgf8a expression in MHB was noticeably decreased in a subset of embryos injected with aldh1a3 MO (F).
Figure 7. Loss of wnt1 expression in MHB occurs selectively in aldh1a3 and fgf8a morphants.
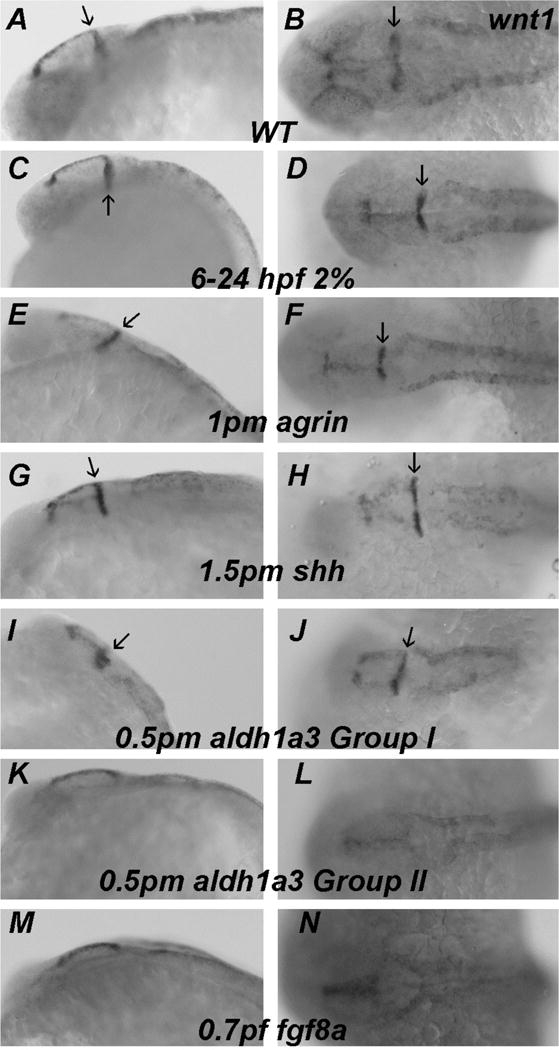
A–B, WT; C–D, embryos exposed to 2% ethanol from 6–24 hpf; E–F, embryos injected with 1 pmol agrin MO; G–H, embryos injected with 1.5 pmol shh MO; I–J, embryos injected with 0.5 pmol aldh1a3 MO that had relatively normal MHB expression; K-L, embryos injected with 0.5 pmol aldh1a3 MO that lost MHB expression; M–N, embryos injected = with 0.7 pmol fgf8a MO. A,C,E,G,I,K and M embryos are lateral views; B,D,F,H,J,L and N embryos are dorsal views. Arrows indicate MHB expression of wnt1. Note that wnt1 expression in MHB is lost in a subset of embryos injected with aldh1a3 MO (K,L) and in all embryos injected with fgf8a MO (M,N).
Figure 8. Eng2b expression is unchanged with ethanol exposure and is selectively lost in MHB and posterior midbrain in fgf8a morphants.
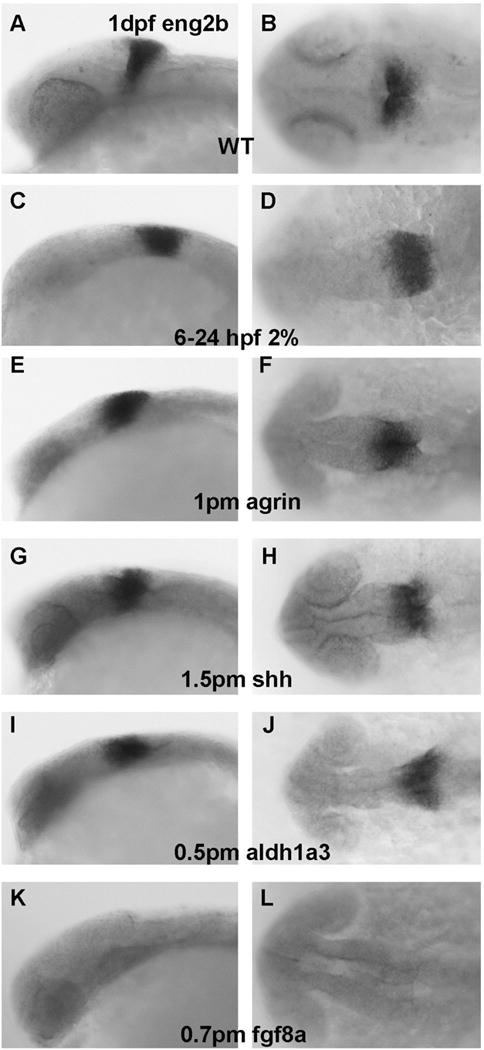
A–B, WT; C–D, embryos exposed to 2% ethanol from 6–24 hpf; E–F, embryos injected e with 1 pmol agrin MO; G–H, embryos injected with 1.5 pmol shh MO; I–J, embryos injected with 0.5 pmol aldh1a3 MO; K–L, embryos injected with 0.7 pmol fgf8a MO. A,C,E,G,I, and K embryos are lateral views; B,D,F,H,J, and L embryos are dorsal views. Note that eng2b expression in MHB and posterior midbrain is only lost with fgf8a MO treatment (K,L).
Discussion
Zebrafish embryos exposed to ethanol display the hallmark craniofacial defects of FASD (McCarthy et al., 2013; Swartz et al., 2014), including microphthalmia, which are dependent on the dosage and timing of ethanol exposure (Ali et al., 2011; Zhang et al., 2014). Many of the hallmark phenotypes of FASD are also produced with disruption of gene function in zebrafish (Eberhart et al., 2016). Relevant to these studies, our previous work showed agrin morphants displayed similar ocular defects to those induced by ethanol exposure, suggesting agrin as a target of prenatal ethanol exposure (Zhang et al., 2011). Disruption of Shh, fgf and RA signaling also produced FASD phenotypes such as microphthalmia and abnormal MHB (Duester et al., 1991; Kot-Leibovich et al., 2009; Kumar et al., 2010; Leo et al., 1999; Marrs et al., 2010; Pillarkat et al., 1991; Sulik et al., 1981; Yelin et al., 2007; 2005; Zachman et al., 1998; Zhang et al., 2011). The ability to rescue FASD phenotypes with shh mRNA overexpression (Loucks et al., 2009; Zhang et al., 2011; 2013) or RA treatment (Muralidharan et al., 2015; Peng et al., 2004; Zhang et al., 2015) also provided support for these signaling pathways serving as molecular targets of ethanol. There also appears to be crosstalk between multiple molecular signaling pathways that are impacted by ethanol, since Shh, fgf and RA rescued ethanol induced phenotypes in zebrafish, and even overexpression of Shh or fgf8/fgf19 rescued FASD phenotypes induced by combined ethanol and agrin treatment (Zhang et al., 2011; 2013).
To further understand possible molecular targets of ethanol exposure that manifest in FASD, we chose to analyze several markers expressed in brain regions known to be affected by ethanol exposure: eye, forebrain, midbrain, MHB (isthmus in rodent) and hindbrain. The genes selected were well-characterized markers of zebrafish brain development, and the main objective of the present study was to compare the expression of these established markers as a result of ethanol exposure to gene knockdown, using MOs to genes that have been suggested to be ethanol sensitive (agrin, Shha, aldh1a3 and fgf8a) and thus potentially responsible for the brain dysmorphologies observed in FASD. Surprisingly, chronic ethanol exposure that induced FAS phenotypes in 100% of embryos only had modest effects on expression of these CNS markers. Most of the markers analyzed exhibited no change in expression following chronic ethanol exposure, and included well-characterized genes such as fgf8, krox20, wnt1, eng2b, and otx2. Ethanol exposure decreased ocular expression of pax6a as previously reported (Zhang et al., 2011; 2015), but also decreased pax6a expression in forebrain and hindbrain. These data are consistent with studies in Xenopus and mouse, that showed reduced pax6 in these regions (Aronne et al., 2008; Peng et al., 2004). Ethanol also decreased epha4a expression in rhombomere R1 boundary, but not in other brain regions. As R1 gives rise to the cerebellum (Moens and Prince, 2002), which is known to be sensitive to prenatal ethanol exposure (Cragg et al., 1985; Goodlett et al., 1990; Zhang et al., 2013), this finding is of interest as it provides possible new insight into the mechanisms underlying ethanol disruption of cerebellar development. Interestingly, our laboratory has shown that chronic ethanol exposure disrupted zebrafish cerebellar development at early stages by a mechanism involving Shh and fgf signaling (Zhang et al., 2013), yet shh MO did not disrupt R1 boundary expression of epha4a while ethanol had this effect.
Although chronic ethanol exposure disrupted the morphological development of the MHB, well known markers of the MHB, such as pax2a and fgf8a, did not display altered expression levels. With regard to pax2a, our data confirm earlier studies that showed this MHB marker was not affected by early ethanol exposure (Coffey et al, 2013; Joya et al, 2014), although Coffey et al (2013) showed decreased pax2a expression following later exposure to ethanol. Pax2a MHB expression was only lost in a small subset of aldh1a3 morphants and in fgf8a morphants in the present study. Similarly, fgf8a MHB expression was only markedly decreased in some aldh1a3 morphants, but not in chronic ethanol embryos or agrin or shh morphants. These data are in contrast to mouse studies that found ethanol exposure to cause decreased fgf8 expression in telencephalon (Aoto et al., 2008), although our analysis focused on MHB fgf8a expression. Not surprisingly, we found loss of wnt1 MHB expression in all fgf8a morphants. The loss of MHB gene expression in fgf8a morphants was consistent with studies using the fgf8 ace mutant, where the initial molecular identity of the isthmic domain (MHB) was not maintained (Jászai et al., 2003). However, our current results clearly showed that the perturbation of morphological MHB formation did not disrupt expression of genes typically associated with the MHB. These data suggest that even in the absence of a defined MHB, genes that may be critical to midbrain and hindbrain patterning are still expressed, indicating that the cellular and functional integrity of the MHB was intact even in the absence of a well-defined morphological MHB. Adding to the complexity of this picture is our recent work showing that RA treatment rescued a morphologically abnormal MHB induced by combined aldh1a3 MO and ethanol treatment, or Shh MO combined with ethanol treatment, but Shh mRNA overexpression could not rescue MHB phenotypes in aldh1a3 MO combined with ethanol treatment (Zhang et al., 2015). Taking into account the current analysis of expression of molecular markers of the MHB, these data suggest that RA signaling may act upstream of pax2a and fgf8a, and that ethanol may be acting downstream of these genes, thus accounting for an absence of an effect on their expression.
Numerous studies have shown that the MHB, or the isthmus organizer in rodent, is a critical patterning center in the CNS (Joyner et al., 2000; Pera et al., 2002), with isthmic fgf8 required for both tectum and cerebellum development (Chi et al., 2003). Both the strength and duration of fgf8 MHB expression are crucial for patterning different tectal-isthmo-cerebellar structures (Sato et al., 2009). It therefore was not surprising that perturbed MHB fgf8a expression resulted in loss of expression of pax2a/wnt1/eng2 in MHB and ectopic expression of pax6a/epha4a in midbrain. We detected relatively normal fgf8a expression in MHB in all chronic ethanol exposed embryos, agrin and shh morphants and most aldh1a3 morphants, despite the absence of a morphologically normal MHB. This was despite our previous studies that showed either fgf8 or fgf19 mRNA overexpression was capable of rescuing decreased GAD1 and Atonal1a expression phenotypes in combined ethanol and agrin or shh MO treated embryos (Zhang et al., 2013). We also detected decreased fgf19 expression in forebrain following chronic ethanol exposure and in agrin and shh morphants (Zhang et al., 2013). These data therefore suggest redundancy in fgf function, with fgf19 having a more critical role in ethanol pathology, and manifestation of some features of FASD, than fgf8. Our data also suggest that morphological disruption of the MHB occurs while expression of MHB molecular markers is maintained in ethanol exposed embryos and agrin or shh morphants. The observation that the early markers of neural domains and MHB are not affected in ethanol-exposed embryos suggests that the morphologically disrupted MHB is a downstream effect that likely is not due to defects in the patterning of the neural primordium. However, the observed changes in pax6a and epha4a gene expression as a consequence of ethanol exposure suggest these genes may be important mediators of the effects of ethanol in the manifestation of FASD. When considered in the context of our previous studies that have suggested a role for agrin, shh, fgf8/19 and RA in ethanol teratogenicity (Zhang et al, 2013; 2015), and our demonstration that genes such as Gli1 and fgf19 were reduced in expression, this body of work begins to shed more light on possible molecular pathways that are disrupted by ethanol exposure during embryogenesis.
Supplementary Material
Acknowledgments
The authors thank Ms. Shanta Mackinnon for zebrafish husbandry. This work was supported by NIH grant U54 AA019765.
Footnotes
Declaration of interest
The authors report no conflicts of interest.
References
- Ahlgren SC, Thakur V, Bronner-Fraser M. Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proc Natl Acad Sci USA. 2002;99:10476–10481. doi: 10.1073/pnas.162356199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Champagne DL, Alia A, Richardson MK. Large-scale analysis of acute ethanol exposure in zebrafish development: a critical time window and resilience. Plos One. 2011;6:e20037. doi: 10.1371/journal.pone.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto K, Shikata Y, Higashiyama D, Shiota K, Motoyama J. Fetal ethanol exposure activates protein kinase A and impairs Shh expression in prechordal mesendoderm cells in the pathogenesis of holoprosencephaly. Birth Defects Res (Part A) 2008;82:224–231. doi: 10.1002/bdra.20447. [DOI] [PubMed] [Google Scholar]
- Arenzana FJ, Carvan MJ, 3rd, Aijon J, Sanchez-Gonzalez R, Arevalo R, Porteros A. Teratogenic effects of ethanol exposure on zebrafish visual system development. Neurotoxicol Teratol. 2006;28:342–348. doi: 10.1016/j.ntt.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Aronne MP, Evrard SG, Mirochnic S, Brusco A. Prenatal ethanol exposure reduces the expression of the transcription factor Pax6 in the developing rat brain. Ann N Y Acad Sci. 2008;1139:478–498. doi: 10.1196/annals.1432.006. [DOI] [PubMed] [Google Scholar]
- Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T, Bowell R, O’Keefe M, Lanigan B. Ocular manifestations in fetal alcohol syndrome. Br J Ophthalmol. 1991;75:524–526. doi: 10.1136/bjo.75.9.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Clarren SK, Alvord EC, Jr, Sumi SM, Streissguth AP, Smith DW. Brain malformations related to prenatal exposure to ethanol. J Pediatrics. 1978;92:64–67. doi: 10.1016/s0022-3476(78)80072-9. [DOI] [PubMed] [Google Scholar]
- Coffey CM, Solleveld PA, Fang J, Roberts AK, Hong SK, Dawid IB, Laverriere CE, Glasgow E. Novel oxytocin gene expression in the hindbrain is induced by alcohol exposure: transgenic zebrafish enable visualization of sensitive neurons. PLoS One. 2013;8:e53991. doi: 10.1371/journal.pone.0053991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg BG, Phillips SC. Natural loss of Purkinje cells during development and increased loss with alcohol. Brain Res. 1985;325:151–160. doi: 10.1016/0006-8993(85)90311-7. [DOI] [PubMed] [Google Scholar]
- Dangata YY, Kaufman MH. Morphometric analysis of the postnatal mouse optic nerve following prenatal exposure to alcohol. J Anat. 1997;19:49–56. doi: 10.1046/j.1469-7580.1997.19110049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: A quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Duester G. A hypothetical mechanism for fetal alcohol syndrome involving ethanol inhibition of retinoic acid synthesis at the alcohol dehydrogenase step. Alcohol Clin Exp Res. 1991;15:568–572. doi: 10.1111/j.1530-0277.1991.tb00562.x. [DOI] [PubMed] [Google Scholar]
- Eberhart JK, Parnell SE. The genetics of fetal alcohol spectrum disorders. Alcoholism Clin Exp Res. 2016;40:1154–1165. doi: 10.1111/acer.13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlett CR, Marcussen BL, West JR. A single day of alcohol exposure during the brain growth spurt induces brain weight restriction and cerebellar Purkinje cell loss. Alcohol. 1990;7:107–114. doi: 10.1016/0741-8329(90)90070-s. [DOI] [PubMed] [Google Scholar]
- Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. J Lab Clin Med. 2005;145:47–54. doi: 10.1016/j.lab.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Jaszai J, Reifers F, Picker A, Langenberg T, Brand M. Isthmus-to-midbrain transformation in the absence of midbrain-hindbrain organizer activity. Development. 2003;130:6611–6623. doi: 10.1242/dev.00899. [DOI] [PubMed] [Google Scholar]
- Joya X, Garcia-Algar O, Vall O, Pujades C. Transient exposure to ethanol during zebrafish embryogenesis results in defects in neuronal differentiation: an alternative model system to study FASD. PLoS One. 2014;10:e112851. doi: 10.1371/journal.pone.0112851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, Liu A, Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr Opin Cell Biol. 2000;12:736–741. doi: 10.1016/s0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Liu IH, Song Y, Lee JA, Halfter W, Balice-Gordon RJ, Linney E, Cole GJ. Agrin is required for posterior development and motor axon outgrowth and branching in embryonic zebrafish. Glycobiology. 2007;17:231–247. doi: 10.1093/glycob/cwl069. [DOI] [PubMed] [Google Scholar]
- Kot-Leibovich H, Fainsod A. Ethanol induces embryonic malformations by competing for retinaldehyde dehydrogenase activity during vertebrate gastrulation. Dis Model Mech. 2009;2:295–305. doi: 10.1242/dmm.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Singh CK, DiPette DD, Singh US. Ethanol impairs activation of retinoic acid receptors in cerebellar granule cells in a rodent model of fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2010;34:928–937. doi: 10.1111/j.1530-0277.2010.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo MA, Lieber CS. Alcohol, vitamin A, and beta-carotene: adverse interactions, including hepatotoxicity and carcinogenicity. Am J Clin Nutr. 1999;69:1071–1085. doi: 10.1093/ajcn/69.6.1071. [DOI] [PubMed] [Google Scholar]
- Li YX, Yang HT, Zdanowicz M, Sicklick JK, Qi Y, Camp TJ, Diehl AM. Fetal alcohol exposure impairs Hedgehog cholesterol modification and signaling. Lab Invest. 2007;87:231–240. doi: 10.1038/labinvest.3700516. [DOI] [PubMed] [Google Scholar]
- Liu IH, Zhang C, Kim MJ, Cole GJ. Retina development in zebrafish requires the heparan sulfate proteoglycan agrin. Dev Neurobiol. 2008;68:877–898. doi: 10.1002/dneu.20625. [DOI] [PubMed] [Google Scholar]
- Loucks EJ, Ahlgren SC. Deciphering the role of Shh signaling in axial defects produced by ethanol exposure. Birth Defects Res (Part A) 2009;85:556–567. doi: 10.1002/bdra.20564. [DOI] [PubMed] [Google Scholar]
- Marrs JA, Clendenon SG, Ratcliffe DR, Fielding SM, Liu Q, Bosron WF. Zebrafish fetal alcohol syndrome model: effects of ethanol are rescued by retinoic acid supplement. Alcohol. 2010;44:707–715. doi: 10.1016/j.alcohol.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattso SN, Riley EP. Brain anomalies in fetal alcohol syndrome. In: Abel EL, editor. Fetal Alcohol Syndrome: From Mechanism to Prevention. CRC Press; Boca Raton: 1996. pp. 50–68. [Google Scholar]
- McCarthy N, Wetherill L, Lovely CB, Swartz ME, Foroud TM, Eberhart JK. Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development. 2013;140:3254–3265. doi: 10.1242/dev.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet S, Bloch-Gallego E, Simeone A, Alvarado-Mallart RM. The caudal limit of Otx2 gene expression as a marker of the midbrain/hindbrain boundary: a study using in situ hybridisation and chick/quail homotopic grafts. Development. 1996;122:3785–3797. doi: 10.1242/dev.122.12.3785. [DOI] [PubMed] [Google Scholar]
- Moens CB, Prince VE. Constructing the hindbrain: insights from zebrafish. Dev Dyn. 2002;224:1–17. doi: 10.1002/dvdy.10086. [DOI] [PubMed] [Google Scholar]
- Muralidharan P, Sarmah S, Marrs JA. Zebrafish retinal defects induced by ethanol exposure are rescued by retinoic acid and folic acid supplement. Alcohol. 2015;49:149–163. doi: 10.1016/j.alcohol.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang PH, Ng SS, Wong OG, Liu J, He ML, Kung HF, Lin MC. A critical role of Pax6 in alcohol-induced fetal microcephaly. Neurobiol Dis. 2004;16:370–376. doi: 10.1016/j.nbd.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Pera EM, Kim JI, Martinez SL, Brechner M, Li SY, Wessely O, De Robertis EM. Isthmin is a novel secreted protein expressed as part of the Fgf-8 synexpression group in the Xenopus midbrain-hindbrain organizer. Mech Dev. 2002;16:169–172. doi: 10.1016/s0925-4773(02)00123-5. [DOI] [PubMed] [Google Scholar]
- Pillarkat RK. Hypothesis: prenatal ethanol-induced birth defects and retinoic acid. Alcohol Clin Exp Res. 1991;15:565–567. doi: 10.1111/j.1530-0277.1991.tb00561.x. [DOI] [PubMed] [Google Scholar]
- Rubert G, Minana R, Pascual M, Guerri C. Ethanol exposure during embryogenesis decreases the radial glial progenitor pool and affects the generation of neurons and astrocytes. J Neurosci Res. 2006;84:483–496. doi: 10.1002/jnr.20963. [DOI] [PubMed] [Google Scholar]
- Sato T, Joyner AL. The duration of Fgf8 isthmic organizer expression is key to patterning different tectal-isthmo-cerebellum structures. Development. 2009;136:3617–3626. doi: 10.1242/dev.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromland K. Ocular abnormalities in the fetal alcohol syndrome. Acta Ophthamol. 1985;(Suppl 171):1–50. [PubMed] [Google Scholar]
- Stromland K, Pinazo-Duran MD. Optic nerve hypoplasia: comparative effects in children and rats exposed to alcohol during pregnancy. Teratology. 1994;50:100–111. doi: 10.1002/tera.1420500204. [DOI] [PubMed] [Google Scholar]
- Sulik KK, Johnston MC, Webb MA. Fetal alcohol syndrome: embryogenesis in a mouse model. Science. 1981;214:936–938. doi: 10.1126/science.6795717. [DOI] [PubMed] [Google Scholar]
- Swartz ME, Wells MB, Griffin M, McCarthy N, Lovely CB, McGurk P, Rozacky J, Eberhart JK. A screen of zebrafish mutants identifies ethanol-sensitive genetic loci. Alcohol Clin Exp Res. 2014;38:694–703. doi: 10.1111/acer.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Furthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402) ZFIN Direct Data Submission. 2001 http://zfin.org.
- Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004 http://zfin.org.
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 http://zfin.org.
- Tsen G, Halfter W, Kroger S, Cole GJ. Agrin is a heparan sulfate proteoglycan. J Biol Chem. 1995;270:3392–3399. doi: 10.1074/jbc.270.7.3392. [DOI] [PubMed] [Google Scholar]
- Yahyavi M, Abouzeid H, Gawdat G, de Preux AS, Xiao T, Bardakjian T, Schneider A, Choi A, Jorgenson E, Baier H, El Sada M, Schorderet DF, Slavotinek AM. ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum Mol Genet. 2013;22:3250–3258. doi: 10.1093/hmg/ddt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Kot H, Yelin D, Fainsod A. Early molecular effects of ethanol during vertebrate embryogenesis. Differentiation. 2007;75:393–403. doi: 10.1111/j.1432-0436.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- Yelin R, Schyr RB, Kot H, Zins S, Frumkin A, Pillemer G, Fainsod A. Ethanol exposure affects gene expression in the embryonic organizer and reduces retinoic acid levels. Dev Biol. 2005;279:193–204. doi: 10.1016/j.ydbio.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Zachman RD, Grummer MA. The interaction of ethanol and vitamin A as a potential mechanism for the pathogenesis of fetal alcohol syndrome. Alcohol Clin Exp Res. 1998;22:1544–1556. [PubMed] [Google Scholar]
- Zamora LY, Lu Z. Alcohol-induced morphological deficits in the development of octavolateral organs of the zebrafish (Danio rerio) Zebrafish. 2013;10:52–61. doi: 10.1089/zeb.2012.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Anderson A, Cole GJ. Analysis of crosstalk between retinoic acid and sonic hedgehog pathways following ethanol exposure in embryonic zebrafish. Birth Defects Res A Clin Mol Teratol. 2015;103:1046–1057. doi: 10.1002/bdra.23460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frazier JM, Chen H, Liu Y, Lee JA, Cole GJ. Molecular and morphological changes in zebrafish following transient ethanol exposure during defined developmental stages. Neurotoxicol Teratol. 2014;44:70–80. doi: 10.1016/j.ntt.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ojiaku P, Cole GJ. Forebrain and hindbrain development in zebrafish is sensitive to ethanol exposure involving agrin, Fgf, and sonic hedgehog function. Birth Defects Res A Clin Mol Teratol. 2013;97:8–27. doi: 10.1002/bdra.23099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Turton QM, MacKinnon S, Sulik KK, Cole GJ. Agrin function associated with ocular development is a target of ethanol exposure in embryonic zebrafish. Birth Defects Res (Part A) 2011;91:129–141. doi: 10.1002/bdra.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


