Abstract
Dengue virus is the leading cause of vector-borne viral disease with four serotypes in circulation. Vaccine development has been complicated by the potential for both protection and disease enhancement during heterologous infection. Secondary infection triggers cross-reactive immune memory responses which have varying functional and epitope specificities that determine protection or risk. Strongly neutralizing antibodies to quaternary epitopes may be especially important for virus neutralization. Cell-mediated immunity dominated by Th1 functions may also play an important role. Determining an immune correlate of protection or risk would be highly beneficial for vaccine development but is hampered by mechanistic uncertainties and assay limitations. Clinical efficacy trials and human infection models along with a systems approach may provide future opportunities to elucidate such correlates.
Keywords: Dengue, Dengue Vaccine, Correlate of Protection, Correlate of Risk, Antibody-dependent Enhancement, Humoral Immunity, Cell-mediated Immunity, Human Infection Model, Systems Immunology, Systems Vaccinology
Introduction
Dengue virus (DENV) is the leading cause of vector-borne viral disease globally with an estimated 390 million infections and 96 million symptomatic cases occurring annually [1]. Four antigenically distinct DENV serotypes (DENV-1 to -4) cause dengue. Infection with one serotype confers long lasting protection against the same serotype, but only short term protection against a different serotype [2]. DENV infection triggers an immune response that can result in protection or enhancement of subsequent infection, thus complicating the effort to develop dengue vaccines [3]. A variety of factors including viral characteristics, host immunity and genetics, and epidemiological context, along with the relative timing of these factors, play a role in ultimately protecting against or enhancing infection [4]. In order to address this problem, vaccine developers have sought to induce simultaneous tetravalent immunity against all four DENV serotypes. However, these efforts have been hampered by an incomplete understanding of the relevant immune responses that contribute to protection or enhancement. A better understanding of these immune parameters is critically needed to inform ongoing dengue vaccine development efforts. The pursuit of dengue immune correlates is further complicated by the limitations of the assays themselves as highlighted by the lack of a standardized neutralization assay making inter-study comparisons difficult [5, 6]. This article will review current knowledge about humoral and cell-mediated immune responses in protection and enhancement of dengue, and prospects to define immune correlates of protection or risk for vaccine development.
Evidence of Immune-mediated Protection or Risk in Natural Infection
Studies of natural DENV infection have shown primary infection typically results in less severe disease than secondary infection [7, 8]. Primary infection confers long lasting protection against the infecting serotype. Paradoxically, the immune response to the primary infection can result in enhancement of subsequent heterologous infection. This potential for enhancement can vary over time. Epidemiological studies have indicated temporary protection occurs against a heterologous serotype with the duration of that protection estimated to vary from two months to three years [2, 9–12]. After 2–3 years, there appears to be a higher likelihood of symptomatic disease, supporting the notion of immune enhancement [9, 10]. This situation is mirrored in infants who appear to have temporary protection by passively acquired maternal antibody during the first few months of life followed by several months of increased risk of disease as maternal antibodies decline to presumably non-protective levels [13, 14]. This disease pattern in infants implies that IgG plays an important role in disease enhancement supporting the theory of antibody-dependent enhancement (ADE) in which non-neutralizing antibodies complexed with DENV facilitate viral entry into host mononuclear cells via Fc receptors [15, 16].
Relatively few field studies have been done in which immune status has been measured prior to natural infection to indicate protection or enhancement. In a cohort study in Thailand, pre-existing neutralizing antibody levels were not consistently associated with lower risk of subsequent homotypic infection, in particular with DENV-2 infection [17]. However, in cluster studies in Thailand, lower neutralizing antibody levels to DENV-1, -2 and -4 did appear to be associated with higher risk of homotypic infection when the neutralizing antibodies were measured within two weeks before infection [18]. Interestingly, the titer thresholds that were associated with increased risk seemed to be much higher for DENV-2 than DENV-1 or -4, although differences in subject ages among serotypes may have affected these thresholds [18]. A cohort study in Sri Lanka demonstrated that the baseline serotype-specific neutralizing antibody profile but not necessarily titer levels was associated with the risk of symptomatic secondary infection suggesting the importance of prior infection history rather than neutralizing titer levels in disease risk [19]. Cell-mediated immunity studies from a Thai cohort found that pre-existing IFN-gamma secreting memory T cells were associated with lower severity of secondary infection [20, 21] suggesting a protective role for specific functional subsets of T cells.
Some epidemiological studies have indicated post-secondary infections (i.e., third or greater sequential infections) are rarely severe [22] and are more likely to be subclinical [23], suggesting repeated exposures to DENV result in cross-protective immunity. The protective immunity induced by sequential exposures and the process by which this immunity is elicited are not well understood. However, heterologous infection occurring sequentially over time is likely to elicit a different immune response from tetravalent vaccination. Sequential heterologous immunization has, in fact, been suggested as an alternative to simultaneous tetravalent administration [24]. Understanding the molecular targets and the functional characteristics of the immune effectors would be invaluable in the identification of immune correlates and development of effective vaccines.
Evidence of Immune-mediated Protection or Risk in Vaccine Studies
Although a great deal of progress has been made in the pursuit of dengue vaccines over the past several years, CYD-TDV, sponsored by Sanofi Pasteur, remains the only vaccine candidate that has undergone clinical efficacy trials [25–29]. Two additional live attenuated tetravalent dengue vaccine candidates, TDV [30], sponsored by Takeda, and TV005 [31, 32], developed by the U.S. National Institutes of Health, are now close to entering clinical efficacy trials while other vaccine candidates are in various stages of early clinical and preclinical development using a variety of strategies including inactivated whole virus, recombinant constructs, DNA, virus vectors, and virus-like particles (VLPs) [33–36].
CYD-TDV is a live attenuated tetravalent chimeric vaccine consisting of the yellow fever virus 17D backbone with DENV pre-membrane (prM) and envelope (E) proteins from each of the four DENV serotypes [25]. CYD-TDV administered in three doses over 12 months was evaluated in a phase IIb clinical trial in Thailand and phase III trials in Asia and Latin America demonstrating good efficacy against DENV-3 and -4, marginal efficacy against DENV-1, poor efficacy against DENV-2, and generally lower efficacy among flavivirus-naïve than primed subjects [26–28]. In addition, vaccinated children 2–5 years of age had an almost eight-fold increased relative risk of dengue hospitalization during the third year of the Asian phase III trial [29]. The vaccine, nevertheless, appeared to have higher overall efficacy against severe disease compared to all symptomatic infections. The poor efficacy shown against DENV-2 was unexpected since immunogenicity as measured by plaque reduction neutralization tests was robust to all four serotypes. Clearly, homotypic neutralizing titer levels after vaccination were insufficient to predict risk of homologous infection. The lower efficacy in flavivirus-naïve subjects was also notable given the possibility that CYD-TDV’s protective effect in primed subjects may have been driven by boosting of pre-existing immune memory. Unfortunately, direct measurement of pre-vaccination immune status was only possible in a subset of subjects for which baseline blood samples were available. A multivariate model applied to five different CYD-TDV phase II trials, however, showed neutralizing antibody response to vaccination was principally predicted by baseline neutralizing antibody titers [37]. A similar model applied to the phase III trials may be able to infer baseline primed status for more subjects and the impact of priming on protection or risk. In terms of cell-mediated immune parameters, peripheral blood mononuclear cells were not obtained in the CYD-TDV clinical efficacy trials preventing direct assessment of the role of cell-mediated immunity in vaccine induced protection.
Correlates of Humoral Immunity in Dengue
DENV genome encodes 10 viral products including three structural proteins: envelope (E), pre-membrane/membrane (prM/M), and capsid (C); and seven non-structural proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. All structural proteins have been shown to be targets of antibodies in natural infection with most of the antibodies being specific to prM and E proteins [38]. Antibodies to non-structural proteins include those specific to NS1, NS3 and NS5 [39, 40].
The surface of the viral particle is made up of prM and E proteins. The E protein binds cellular receptor(s) and mediates viral entry. The crystal structure of the E protein has been solved and is composed of three domains: envelope domains I, II and II (EDI, EDII, and EDIII, respectively). EDI forms the central part of the molecule and is connected to EDII and EDIII by flexible linkers [41]. EDII contains 13 highly conserved amino acids which form a fusion loop that mediates fusion of viral envelope to host cell membrane, a step necessary for the release of viral genome into the cytosol [42]. EDIII serves a critical function in binding host cell receptor(s). Differences in amino acid sequences in EDIII provide the basis for the antigenically distinct serotypes of DENV [43, 44]. These domains are connected to the transmembrane anchor at the C-terminus via a short stem region that is involved in the late stage of the fusion process [45, 46]. PrM/M protein functions as a chaperone for E protein by preventing exposure of the fusion loop that could lead to premature fusion [47]. PrM and E proteins on the viral surface undergo changes in association and arrangement during the maturation process. The surface of immature viral particles consists of 60 prM-E trimeric complexes arranged in a spike pattern with the prM protein interacting with the tip of the spike formed by the fusion loop [48]. In the trans-Golgi network, the surface E proteins rearrange to form 90 units of antiparallel dimers and the prM is cleaved by host protease furin [49]. The pr moiety of prM is released when the viral particle is discharged into the neutral pH environment outside the cell. The extent of prM cleavage varies among different flaviviruses. In DENV, this process is incomplete [50]. Electron microscopy studies have demonstrated that DENV consists of viral particles at various stages of maturity with varying areas of prM on the surface [51]. The presence of surface prM has important implications for the neutralizing and enhancing activity of immune sera [52, 53].
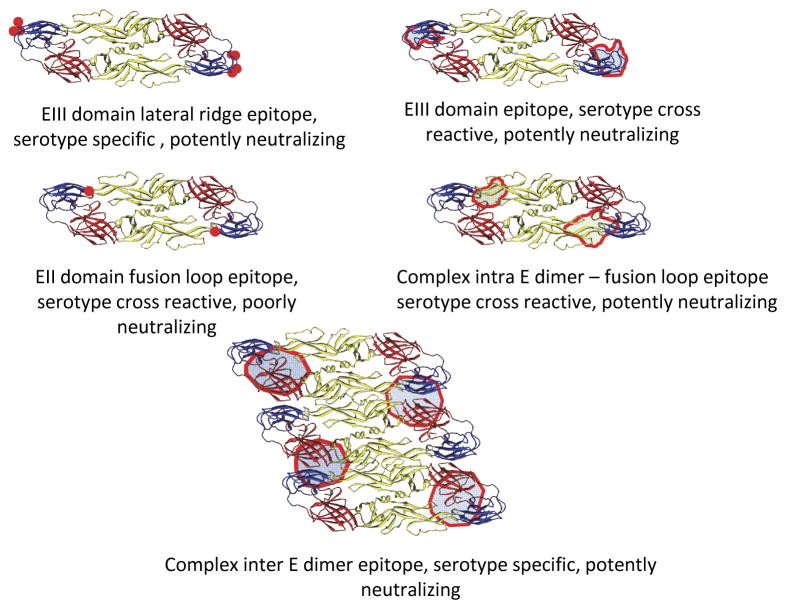
Early evaluations of antibody response focused on an important functional aspect of antibody, namely neutralizing activity. The finding that immune sera after primary infection contained serotype-specific neutralizing activity and that protective immunity to the infecting serotype persisted while cross-protective immunity was short-lived suggested that the envelope protein was the primary target for antibody neutralization [2]. Subsequent studies employing molecular techniques including western blots and recombinant protein antigens demonstrated that during and shortly after acute primary or acute secondary infection, the majority of antibody response to the E protein was actually serotype cross-reactive [38, 54]. The target of these antibodies was mapped to the fusion loop of EDII which is highly conserved among serotypes. The neutralizing activity of immune sera from acute primary infections was low and cross-reactive indicating that the serotype cross-reactive, anti-fusion loop antibodies predominating during early response were non-neutralizing [38]. In contrast, serotype cross-reactive, anti-fusion loop antibodies induced during a secondary infection have been shown to contribute significantly to neutralizing activity of immune sera [24]. Serotype-specific antibodies against EDIII constituted only a small fraction of antibody response [24]. Studies employing mouse monoclonal antibodies and mutated EDIII demonstrated potent neutralizing activity of antibodies targeting EDIII [54]. The binding sites of neutralizing murine and human antibodies specific to EDIII have been mapped by yeast display techniques to two regions: (1) amino acids forming the lateral ridge of EDIII [55], and (2) a closely related site in the A strand of EDIII (figure 1) [56].
Figure 1.
Binding sites of envelope protein-specific antibodies. Antibody binding sites were determined by binding characteristics to mutated recombinant proteins or virus-like particles with point mutations, or by cryogenic electron microscopy. Antibody binding sites are depicted as red shaded areas on envelope protein molecules.
Certain monoclonal antibodies have been observed to bind to whole virion but not to the purified ectodomain of E protein suggesting that they recognize quaternary structures on the viral particle surface (figure 1) [57]. Cryogenic electron microscopy (Cryo-EM) and binding studies using virus-like particles presenting complex surface antigen structures have identified another type of antibody that binds complex quaternary epitopes made up of more than one molecule of envelope protein. Human monoclonal antibodies with serotype-specific neutralizing activity were shown to bind three adjacent E protein regions [58, 59]. Half of the epitope was on EDIII and the other half on EDI and EDI-EDII hinge of a neighboring E protein. A recent study demonstrated that human monoclonal antibodies with broadly neutralizing activity bind across the surface of adjacent E protein molecules that form a dimer (figure 1) [53, 60]. These E dimer-dependent epitope (EDE) antibodies can be divided into two groups based on the epitopes they recognize: (1) EDE1 antibodies that recognize the fusion loop, b-strand, and ij loop of EDII on one side, and EDI and EDIII on the other side of the dimer; and (2) EDE2 antibodies that recognize similar EDII structures as do EDE1 antibodies but interact with the 150 amino acid loop of EDI and glycan chain at N153 on the other side of the dimer. While EDE antibodies recognize the fusion loop in a conformational-dependent manner, another group of antibodies recognizes the linear fusion loop epitope (FLE) formed by the amino acid Tryp 101 and nearby residues. EDE antibodies exhibited broadly neutralizing activity against all four virus serotypes, and neutralized virus derived from either a mosquito cell line or human dendritic cells. In contrast, linear FLE antibodies showed neutralizing activity against mosquito cell-derived virus with a higher prM content but exhibited poor neutralizing activity against dendritic cell-derived virus with low prM content [53].
A major target of the humoral antibody response is prM protein. Although antibodies to prM are detectable during acute primary infection, the response is more rapid and stronger during acute secondary infection [38]. Antibodies to prM are cross-reactive among serotypes and have been shown to be weakly neutralizing. Importantly, these antibodies exhibited infection enhancing activity in vitro particularly in Fc receptor expressing cells [52], although one recent study showed no difference in anti-prM antibody levels between mild and severe disease [61]. Nevertheless, antibodies to prM have been implicated in ADE during secondary infection.
NS1 is a major viral product that elicits an antibody response particularly during secondary infection [38]. NS1 proteins from different serotypes differ significantly in amino acid sequences, and both cross-reactive and serotype-specific antibodies are elicited [62]. Considering the high amounts of NS1 in circulation during infection, it is likely that anti-NS1 antibodies form antigen-antibody complexes particularly during secondary infection [38]. In addition, NS1 has been shown to both activate and inhibit complement depending on the context [63, 64]. NS1 is produced as a membrane-associated and soluble molecule; the soluble form is able to bind glycoaminoglycans and deposit on the surface of many cell types [65]. It is conceivable that anti-NS1 antibodies bind to NS1 on the host cell surface and promote cell injury by complement activation. Alternatively, a recent study has demonstrated a permeability enhancing effect of NS1 on endothelium [66]. Consistent with this finding, immunization against NS1 has been shown to be protective against severe dengue in mice [66–68].
Mechanisms of antibody-mediated neutralization
Antibodies can neutralize viruses by multiple mechanisms. Antibodies against EDIII may inhibit viral binding to cellular receptors. The stoichiometric requirement for neutralization likely depends on a number of factors including the affinity of the antibodies and the accessibility of target epitopes. Some neutralizing antibodies have been shown to bind epitopes which are predicted to be buried but are accessible during conformational changes with rearrangement of E protein at higher temperatures in vivo [56]. The number of binding sites that need to be occupied to prevent infection has been estimated to be between 10–50% for serotype-specific antibody binding to EDIII of DENV-2 [69]. Another important mechanism of neutralization is prevention of viral membrane fusion by binding of the fusion loop or the internal surface of envelope dimers, thus preventing the rearrangement of E proteins into trimers which is required for viral membrane fusion [60, 70]. Other mechanisms that antibodies use to clear virus include various immune effector mechanisms such as complement-mediated lysis of virus and virus-infected cells, and antibody-dependent cell-mediated cytotoxicity (ADCC) [64, 71]. These mechanisms require expression of viral antigens on the infected cell surface. However, NS1 may be displayed on the cell surface of uninfected cells through attachment of soluble NS1 protein rendering these cells targets for cytolysis by complement or natural killer cells.
Measuring Humoral Immune Correlates
Conceptually, techniques used in assessment of antibody response can be divided into two major categories: (1) binding assays which assess physical interaction between antibodies and antigens, and (2) functional assays which measure biological effects of antibodies including neutralization and enhancement of infection. The advantages of binding assays such as ELISA and western blot include the relative ease of performance compared to functional assays, the potential for high throughput, and the ability to identify molecular structures and epitopes recognized by the antibodies. However, binding ability does not necessarily correlate with biological function which can only be measured with functional assays such as neutralization tests.
Measuring protective antibody response in dengue is complicated by a number of factors at both biological and technical levels. Considering the emerging evidence that most antibodies that recognize quaternary epitopes are potently neutralizing, assays should ideally use antigens with quaternary structures such as whole virus or VLPs. Even with these optimized antigens, the usual format of binding assays in which antigen is typically attached on a solid surface does not mimic in vivo binding dynamics characterized by dynamic rearrangement of E proteins on the viral particle surface, which can provide antibodies access to cryptic epitopes not exposed in a static condition [56, 72].
Functional assays can inform potentially relevant biological roles of antibodies. Neutralization potency is a result of the net effect of a mixture of antibodies targeting different antigens and epitopes with different neutralizing and enhancing capacities. Heterogeneity in DENV preparations in terms of the proportions of virus with varying amounts of viral surface prM poses a technical challenge since this could affect assay performance in detecting neutralization or enhancement.
Standardization of virus preparations to ascertain levels of prM and E protein would be an important step in improving reproducibility of these functional assays. Furthermore, the use of characterized cell lines with known and consistent expression of relevant molecules such as Fc receptors may be needed. The threshold for determining neutralizing activity should also be considered since low neutralizing antibodies have been shown to meet the 50% PRNT cut off used in many laboratories. Measurement of antibody response may include a composite of tests that combine binding assays using strategically designed viral antigens that display conformational and linear epitopes to measure the relative proportions of EDE and FLE antibodies, and an assay for antibodies to prM as a marker of non-neutralizing/potentially enhancing antibodies.
Correlates of Cell-mediated Immunity in Dengue
Enhanced cellular immune activation during secondary infection has been postulated to increase disease severity [73]. Studies comparing activation markers and the expansion of antigen-specific CD8+ T cells in dengue fever (DF) and dengue hemorrhagic fever (DHF) cases have demonstrated enhanced T cell activation and expansion in DHF patients [74, 75]. The expansion of antigen-specific T cells is characterized by activation of T cells that recognize serotype cross-reactive epitopes [75]. However, other studies did not find such an association [76, 77]. Although it has been assumed that the magnitude of T cell expansion will be more pronounced during secondary infection, a study comparing the frequencies and kinetics of antigen-specific T cells in primary and secondary infection demonstrated similar expansion of dengue-specific CD8+ T cells [78]. Functional analysis of these cross-reactive T cells demonstrated various patterns of response with regard to the types of cytokine production and cytolytic activity depending on stimulating variant peptides [78]. These findings suggest that disease severity may be related to the functional phenotypes of these cross-reactive T cells activated during both primary and secondary infection. The reported associations between certain HLA loci with disease severity support the role of T cell immunity in the pathogenesis of dengue [79]. However, some studies have failed to demonstrate a similar association. The interplay with other genetic loci encoding gene products relevant to disease mechanisms such as other HLA molecules, Fc receptors, and cytokines likely modify the influence of certain HLA loci on disease severity [80–82].
Most of the studies of T cells have involved analysis of samples collected from DF and DHF patients during an acute infection. Such studies focused mainly on the markers associated with disease severity rather than protection from infection. Nevertheless, inferences can be made on the functional immune parameters that appear to be comparatively defective in more severe (i.e., DHF) cases. One study demonstrated that T cells from severe cases exhibited low cytolytic activity as determined by CD107 expression suggesting that cytolytic function may correlate with protection against severe disease [83]. Consistent with this notion, severe dengue can result in hemophagocytic syndrome which is found in severe viral infections in patients with genetic defects in cytolytic function [84, 85].
Measuring Cell-mediated Immune Correlates
Most cell-mediated immunity assays are functional assays which require well preserved peripheral blood mononuclear cells. Variation in sample collection, cell preservation, and laboratory techniques can affect test performance and is an important obstacle in comparing results from different studies. Analysis of T cell functional profile in an individual prior to and during an acute infection could provide critical information about the type and magnitude of cell-mediated immune responses relevant to disease severity. Although it is possible that viral clearance and tissue injury may be mediated through different mechanisms, the more likely scenario is that outcome of an infection depends on the kinetics of viral replication relative to the activation of memory T cell response. As such, analysis of serial samples during an acute infection will be informative in defining this kinetics. An ideal assay should measure multiple functional aspects of cell-mediated immunity, particularly those aspects speculated to exert antiviral effects and those that may contribute to plasma leakage. These include markers for cytolytic function such as CD107 or perforin, and key cytokines including IFN-gamma, IL-2, and TNF-alpha. Elevated levels of IL-10 have been reported in severe dengue and the frequencies of T cells expressing this regulatory cytokine may be an important marker of disease severity. Multifunctional analysis of T cell immunity will furnish a large and complex dataset that may require analytical tools of systems biology to facilitate data analysis and interpretation [86, 87].
Future Opportunities
Definitive demonstration of vaccine-induced immune correlates of protection or risk will require reproducible measures of pre-infection immunity with documentation of infection or disease in vaccinated and unvaccinated individuals. In the absence of accurate animal models of dengue, this can be done only through large clinical trials with clinical endpoints or through dengue human infection models. Clinical efficacy trials have the advantage of identifying authentic natural infections that can lead to disease. However, the enormous resources required to conduct such studies limit the number that can be done. Even within these large trials, the actual number of infections, the nature, quality and volume of biological samples collected, and the number of collection time points are limited. Thus, the amount of data that can be generated and analyzed is also limited. Currently, only the phase IIb and phase III trials of Sanofi Pasteur’s CYD-TDV vaccine have samples and data available with associated clinical outcomes. However, as other vaccine candidates begin to enter phase III trials, the number and variety of these important reagents will increase, providing expanded opportunities to evaluate immune correlates. Nevertheless, just as immunity from naturally-acquired sequential infections may differ from vaccine-induced immunity, alternative vaccine designs may rely on mechanisms of immunity that vary substantially among vaccine candidates. Investigations of potential immune correlates need to take these differences into account.
Unlike large clinical efficacy trials, human infection models have the advantage of occurring in a controlled setting where the infecting virus strain, host immune status, and timing of infections can be manipulated [88]. Furthermore, a larger number and volume of biological samples can be collected at more frequent time points; and multiple vaccine candidates can be tested with relatively fewer resource requirements. A higher number of immune measures can, therefore, be assessed, with more data generated for analysis. More frequent collection time points also allow for empiric determination of the relevant times for measuring certain immune parameters. However, the experimental infecting virus strain, the timing of sequential infections, and clinical outcomes in human infection models do not reflect natural infections. Thus, immune parameters found to be relevant in human models may not necessarily be applicable to protection or risk in natural infections. This is especially the case for severe disease which cannot be systematically evaluated in human infection models for ethical reasons. Nonetheless, human models could play an important role in suggesting exploratory correlates which could be further validated using samples from clinical efficacy trials.
Ideally, immune correlates should be shown to have a mechanistic role, especially when attempting to elucidate pathophysiology or vaccine design strategies. However, demonstration of an association with clinical endpoints may also be sufficient, especially if the intent is to support regulatory approval [89, 90]. Potential mechanistic correlates such as those involving complex quaternary epitopes are promising. However, given the complexity of the immune response to DENV infection and vaccination, and the myriad interactions among the different serotypes, pre-existing immunity and other contributory variables, determination of a single mechanistic or statistically-associated correlate of protection or risk may not necessarily be feasible. Recent approaches to assessing correlates in diseases such as yellow fever and influenza have incorporated systems analysis of a wide array of parameters [87, 91]. A systems approach has also been used to identify signatures to predict immunity induced by different classes of vaccines [92]. Systems analysis is made possible by new computational tools along with continuing advancement of high throughput techniques such as transcriptomics and proteomics [93]. A systems approach could potentially identify computationally-determined correlates, or lead to unexpected mechanistic markers which could be further explored. The large amount of data that should optimally feed into such analyses could be obtained more readily from human infection models in which samples and data are more accessible. At the same time, clinical efficacy trials with a larger number and variety of subjects could better yield data about certain other parameters such as host genetics. In either case, inclusion of such tests and analyses should be considered in the design of future studies.
Expert Commentary
Efforts and resources applied to dengue vaccine development have accelerated dramatically over the past decade. However, the issues which had concerned dengue researchers for decades have complicated current development efforts. The complex interactions among the four dengue virus serotypes, pre-existing immunity, host genetics, viral characteristics and numerous other factors that can lead to protection or disease enhancement during secondary heterologous infection have become real-world issues in Sanofi Pasteur’s CYD-TDV phase III trials. Key elements of the humoral and cell-mediated immune responses that may be beneficial or detrimental continue to be elucidated. The potentially important role of quaternary envelope dimer-dependent epitope (EDE) is one such example. However, much uncertainty remains, and determining immune correlates of protection or risk will be a challenge. Nevertheless, the existence of vaccine trials with clinical outcomes to validate laboratory findings provides opportunities for progress which had not previously been available. In conjunction with ongoing development of dengue human infection models and other new tools, the dengue research community may be able to accelerate their pursuit of immune correlates in dengue.
Five-year View
With one dengue vaccine candidate already far advanced in phase III trials, two others close to initiating phase III trials, and several other candidates in phase I or II trials, a vast amount of clinically-relevant data will become available in the near future. This may be a mixed blessing as the information is likely to present a confusing picture of the mechanisms of protection and risk. The different vaccine candidates each have unique design strategies which will make the results of any future clinical trials difficult to compare among vaccines. Furthermore, one or more of the vaccine candidates will likely be licensed and introduced in certain endemic countries in the near future. Information from such vaccine introduction(s) will be difficult to interpret compared to data from clinical trials. In the meantime, dengue researchers investigating various immune mechanisms will continue to make further advances as they have already been doing. Within this mix, many opportunities will arise to link research and real-world outcomes greatly adding to our understanding of dengue immunity which will assist both vaccine development and disease management.
Key Issues.
Dengue is caused by four antigenically distinct dengue virus (DENV) serotypes with secondary heterologous infection leading to more severe disease than primary infection.
Six dengue vaccine candidates currently in active development have entered human clinical trials. The most advanced vaccine candidate, Sanofi Pasteur’s CYD-TDV, has undergone phase IIb and phase III trials with good efficacy against two serotypes, marginal or poor efficacy against the other two serotypes, and increased relative risk of dengue hospitalization in vaccinated children 2–5 years of age.
Cross-reactive memory B and T cells from primary infection are activated during secondary DENV infection and may contribute to immune enhancement.
Humoral immune responses to structural proteins are dominated by antibodies to pre-membrane and envelope protein.
Strongly neutralizing antibodies to complex quaternary epitopes may be important in humoral immunity.
Cell-mediated immunity may be important in protection or enhancement depending on functional T cell subsets and epitope specificity.
Data and samples with clinical outcomes from ongoing and upcoming phase III clinical trials and from dengue human infection models can be used to explore and validate potential immune correlates of protection or risk.
A systems approach to data generation and analysis may be useful to determine complex computational correlates or suggest unexpected mechanistic correlates for further exploration.
Acknowledgments
Financial Disclosure
This work was supported in part by the National Institutes of Health (grant P01 AI034533) and the Bill and Melinda Gates Foundation (grant OPP1053432). The opinions expressed are those of the authors and do not represent the official position of the National Institutes of Health or the Bill and Melinda Gates Foundation.
Contributor Information
Anon Srikiatkhachorn, Address: Division of Infectious Diseases and Immunology, Department of Medicine, University of Massachusetts Medical School, Worcester, Massachusetts 01655, USA, Tel: +66-2-696-2752, Fax: +66-2-644-4760
In-Kyu Yoon, Address: Dengue Vaccine Initiative, International Vaccine Institute, SNU Research Park, 1 Gwanak-ro, Gwanak-gu, Seoul, 151-742 Korea, Tel: +82-2-872-2801, Fax: +82-2-872-2803
References
- 1.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabin AB. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1:30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- 3.Halstead SB. Observations related to pathogensis of dengue hemorrhagic fever. VI. Hypotheses and discussion. Yale J Biol Med. 1970;42:350–362. [PMC free article] [PubMed] [Google Scholar]
- 4.Guzman MG, Harris E. Dengue. Lancet. 2015;385(9966):453–65. doi: 10.1016/S0140-6736(14)60572-9. [DOI] [PubMed] [Google Scholar]
- 5.Rainwater-Lovett K, et al. Variation in dengue virus plaque reduction neutralization testing: systematic review and pooled analysis. BMC Infect Dis. 2012;12:233. doi: 10.1186/1471-2334-12-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salje H, et al. Variability in dengue titer estimates from plaque reduction neutralization tests poses a challenge to epidemiological studies and vaccine development. PLoS Negl Trop Dis. 2014;8(6):e2952. doi: 10.1371/journal.pntd.0002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halstead SB, Nimmannitya S, Cohen SN. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J Biol Med. 1970;42:311–328. [PMC free article] [PubMed] [Google Scholar]
- 8.Burke DS, et al. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38(1):172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 9.Montoya M, et al. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl Trop Dis. 2013;7(8):e2357. doi: 10.1371/journal.pntd.0002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Anderson KB, et al. A shorter time interval between first and second dengue infections is associated with protection from clinical illness in a school-based cohort in Thailand. J Infect Dis. 2014;209(3):360–8. doi: 10.1093/infdis/jit436. Cohort studies in Thailand showed a shorter time interval between sequential infections was more likely to be associated with subclinical infection in dengue-naïve children supporting the notion of temporary heterologous immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salje H, et al. Revealing the microscale spatial signature of dengue transmission and immunity in an urban population. Proc Natl Acad Sci U S A. 2012;109(24):9535–8. doi: 10.1073/pnas.1120621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich NG, et al. Interactions between serotypes of dengue highlight epidemiological impact of cross-immunity. J R Soc Interface. 2013;10(86):20130414. doi: 10.1098/rsif.2013.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kliks SC, et al. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am J Trop Med Hyg. 1988;38(2):411–419. doi: 10.4269/ajtmh.1988.38.411. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen TH, et al. Dengue hemorrhagic fever in infants: a study of clinical and cytokine profiles. J Infect Dis. 2004;189(2):221–32. doi: 10.1086/380762. [DOI] [PubMed] [Google Scholar]
- 15.Halstead SB. Immune enhancement of viral infection. Prog Allergy. 1982;31:301–64. [PubMed] [Google Scholar]
- 16.Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch Virol. 2013;158(7):1445–59. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- 17.Endy TP, et al. Relationship of Preexisting Dengue Virus (DV) Neutralizing Antibody Levels to Viremia and Severity of Disease in a Prospective Cohort Study of DV Infection in Thailand. J Infect Dis. 2004;189(6):990–1000. doi: 10.1086/382280. [DOI] [PubMed] [Google Scholar]
- *18.Buddhari D, et al. Dengue virus neutralizing antibody levels associated with protection from infection in thai cluster studies. PLoS Negl Trop Dis. 2014;8(10):e3230. doi: 10.1371/journal.pntd.0003230. Cluster studies in Thailand found lower homotypic neutralizing antibody levels had increased risk of DENV infection suggesting some clinical relevance of neutralization assay results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbett KS, et al. Preexisting neutralizing antibody responses distinguish clinically inapparent and apparent dengue virus infections in a Sri Lankan pediatric cohort. J Infect Dis. 2015;211(4):590–9. doi: 10.1093/infdis/jiu481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangada MM, et al. Dengue-Specific T Cell Responses in Peripheral Blood Mononuclear Cells Obtained prior to Secondary Dengue Virus Infections in Thai Schoolchildren. J Infect Dis. 2002;185(12):1697–703. doi: 10.1086/340822. [DOI] [PubMed] [Google Scholar]
- 21.Hatch S, et al. Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J Infect Dis. 2011;203(9):1282–91. doi: 10.1093/infdis/jir012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbons RV, et al. Analysis of Repeat Hospital Admissions for Dengue to Estimate the Frequency of Third or Fourth Dengue Infections Resulting in Admissions and Dengue Hemorrhagic Fever, and Serotype Sequences. Am J Trop Med Hyg. 2007;77(5):910–913. [PubMed] [Google Scholar]
- 23.Olkowski S, et al. Reduced risk of disease during postsecondary dengue virus infections. J Infect Dis. 2013;208(6):1026–33. doi: 10.1093/infdis/jit273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Tsai WY, et al. Complexity of Neutralizing Antibodies against Multiple Dengue Virus Serotypes after Heterotypic Immunization and Secondary Infection Revealed by In-Depth Analysis of Cross-Reactive Antibodies. J Virol. 2015;89(14):7348–62. doi: 10.1128/JVI.00273-15. Sequential heterotypic immunization with live attenuated dengue vaccine induced highly neutralizing serotype cross-reactive antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guy B, et al. From research to phase III: preclinical, industrial and clinical development of the Sanofi Pasteur tetravalent dengue vaccine. Vaccine. 2011;29(42):7229–41. doi: 10.1016/j.vaccine.2011.06.094. [DOI] [PubMed] [Google Scholar]
- 26.Sabchareon A, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. 2012 doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 27.Capeding MR, et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 2014;384(9951):1358–65. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- 28.Villar L, et al. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med. 2015;372(2):113–23. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- *29.Hadinegoro SR, et al. Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N Engl J Med. 2015 doi: 10.1056/NEJMoa1506223. Analysis of data from the phase IIb and phase III trials of Sanofi Pasteur’s CYD-TDV revealed mixed efficacy results and an increased risk of hospitalized dengue in the youngest children during the third year of phase III trial in Asia. [DOI] [PubMed] [Google Scholar]
- 30.Osorio JE, et al. Development of DENVax: a chimeric dengue-2 PDK-53-based tetravalent vaccine for protection against dengue fever. Vaccine. 2011;29(42):7251–60. doi: 10.1016/j.vaccine.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durbin AP, et al. Development and clinical evaluation of multiple investigational monovalent DENV vaccines to identify components for inclusion in a live attenuated tetravalent DENV vaccine. Vaccine. 2011;29(42):7242–50. doi: 10.1016/j.vaccine.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkpatrick BD, et al. Robust and Balanced Immune Responses to All 4 Dengue Virus Serotypes Following Administration of a Single Dose of a Live Attenuated Tetravalent Dengue Vaccine to Healthy, Flavivirus-Naive Adults. J Infect Dis. 2015;212(5):702–10. doi: 10.1093/infdis/jiv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Govindarajan D, et al. Preclinical development of a dengue tetravalent recombinant subunit vaccine: Immunogenicity and protective efficacy in nonhuman primates. Vaccine. 2015;33(33):4105–16. doi: 10.1016/j.vaccine.2015.06.067. [DOI] [PubMed] [Google Scholar]
- 34.Beckett CG, et al. Evaluation of a prototype dengue-1 DNA vaccine in a Phase 1 clinical trial. Vaccine. 2011;29(5):960–8. doi: 10.1016/j.vaccine.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 35.Martinez LJ, et al. Safety and Immunogenicity of a Dengue Virus Serotype-1 Purified-Inactivated Vaccine: Results of a Phase 1 Clinical Trial. Am J Trop Med Hyg. 2015;93(3):454–60. doi: 10.4269/ajtmh.14-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz LM, et al. The dengue vaccine pipeline: Implications for the future of dengue control. Vaccine. 2015;33(29):3293–8. doi: 10.1016/j.vaccine.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Dorigatti I, et al. Modelling the immunological response to a tetravalent dengue vaccine from multiple phase-2 trials in Latin America and South East Asia. Vaccine. 2015;33(31):3746–51. doi: 10.1016/j.vaccine.2015.05.059. Multivariate analysis of five phase II trials of Sanofi Pasteur’s CYD-TDV showed neutralizing antibody response to vaccination was principally predicted by pre-vaccination neutralizing antibody titers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai CY, et al. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol. 2008;82(13):6631–43. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Churdboonchart V, et al. Antibodies against dengue viral proteins in primary and secondary dengue hemorrhagic fever. Am J Trop Med Hyg. 1991;44(5):481–93. doi: 10.4269/ajtmh.1991.44.481. [DOI] [PubMed] [Google Scholar]
- 40.Valdes K, et al. Human Dengue antibodies against structural and nonstructural proteins. Clin Diagn Lab Immunol. 2000;7(5):856–7. doi: 10.1128/cdli.7.5.856-857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn RJ, et al. Structure of dengue virus. Implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allison SL, et al. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75(9):4268–75. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gubler D, Kuno G, Markoff L. In: Flavivirus, Field’s Virology. 5. Knipe M David, Howly P., editors. Lippincott Williams & Wilkins; 2007. pp. 1154–1161. [Google Scholar]
- 44.Hung JJ, et al. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. J Virol. 2004;78(1):378–88. doi: 10.1128/JVI.78.1.378-388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol. 2013;20(1):105–10. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein DE, Choi JL, Harrison SC. Structure of a dengue virus envelope protein late-stage fusion intermediate. J Virol. 2013;87(4):2287–93. doi: 10.1128/JVI.02957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinz FX, Stiasny K, Allison SL. The entry machinery of flaviviruses. Arch Virol Suppl. 2004;(18):133–7. doi: 10.1007/978-3-7091-0572-6_11. [DOI] [PubMed] [Google Scholar]
- 48.Li L, et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319(5871):1830–4. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 49.Yu IM, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–7. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 50.Junjhon J, et al. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J Virol. 2008;82(21):10776–91. doi: 10.1128/JVI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plevka P, et al. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 2011;12(6):602–6. doi: 10.1038/embor.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dejnirattisai W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328(5979):745–8. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dejnirattisai W, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 2015;16(2):170–7. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *54.Beltramello M, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8(3):271–83. doi: 10.1016/j.chom.2010.08.007. Monoclonal antibodies characterized from human immune sera identified a new class of broadly reactive, highly neutralizing antibodies targeting a complex conformational epitope called envelope dimer epitope (EDE), which may be important in virus neutralization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sukupolvi-Petty S, et al. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol. 2007;81(23):12816–26. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *56.Lok SM, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15(3):312–7. doi: 10.1038/nsmb.1382. Monoclonal antibodies from human immune sera demonstrated highly cross-reactive, poorly neutralizing antibodies to pre-membrane (prM) protein which showed antibody-dependent enhancement in vitro. [DOI] [PubMed] [Google Scholar]
- 57.de Alwis R, et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A. 2012;109(19):7439–44. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teoh EP, et al. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med. 2012;4(139):139ra83. doi: 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- 59.Fibriansah G, et al. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun. 2015;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rouvinski A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520(7545):109–13. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 61.Rodenhuis-Zybert IA, et al. Antibodies against immature virions are not a discriminating factor for dengue disease severity. PLoS Negl Trop Dis. 2015;9(3):e0003564. doi: 10.1371/journal.pntd.0003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puttikhunt C, et al. The development of a novel serotyping-NS1-ELISA to identify serotypes of dengue virus. J Clin Virol. 2011;50(4):314–9. doi: 10.1016/j.jcv.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 63.Avirutnan P, et al. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med. 2010;207(4):793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avirutnan P, et al. Vascular Leakage in Severe Dengue Virus Infections: a Potential Role for the Nonstructural Viral Protein NS1 and Complement. J Infect Dis. 2006;193(8):1078–88. doi: 10.1086/500949. [DOI] [PubMed] [Google Scholar]
- 65.Avirutnan P, et al. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 2007;3(11):e183. doi: 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beatty PR, et al. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med. 2015;7(304):304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 67.Costa SM, et al. Protection against dengue type 2 virus induced in mice immunized with a DNA plasmid encoding the non-structural 1 (NS1) gene fused to the tissue plasminogen activator signal sequence. Vaccine. 2006;24(2):195–205. doi: 10.1016/j.vaccine.2005.07.059. [DOI] [PubMed] [Google Scholar]
- *68.Wu SF, et al. Evaluation of protective efficacy and immune mechanisms of using a non-structural protein NS1 in DNA vaccine against dengue 2 virus in mice. Vaccine. 2003;21(25–26):3919–29. doi: 10.1016/s0264-410x(03)00310-4. The frequencies of activated CD8+ T cells were found to be higher in DHF compared to DF cases providing direct evidence of cross-reactive T cell activation during secondary infection. [DOI] [PubMed] [Google Scholar]
- 69.Gromowski GD, Barrett AD. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology. 2007;366(2):349–60. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 70.Fibriansah G, et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. 2015;349(6243):88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sissons JG, Oldstone MB. Antibody-mediated destruction of virus-infected cells. Adv Immunol. 1980;29:209–60. doi: 10.1016/S0065-2776(08)60045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuhn RJ, et al. Shake, rattle, and roll: Impact of the dynamics of flavivirus particles on their interactions with the host. Virology. 2015;479–480:508–17. doi: 10.1016/j.virol.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Halstead SB. Dengue. Lancet. 2007;370(9599):1644–52. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 74.Green S, et al. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Infect Dis. 1999;180(5):1429–1435. doi: 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 75.Mongkolsapaya J, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9(7):921–7. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 76.Dung NT, et al. Timing of CD8+ T cell responses in relation to commencement of capillary leakage in children with dengue. J Immunol. 2010;184(12):7281–7. doi: 10.4049/jimmunol.0903262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friberg H, et al. Cross-reactivity and expansion of dengue-specific T cells during acute primary and secondary infections in humans. Sci Rep. 2011;1:51. doi: 10.1038/srep00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Friberg H, et al. Memory CD8+ T cells from naturally acquired primary dengue virus infection are highly cross-reactive. Immunol Cell Biol. 2011;89(1):122–9. doi: 10.1038/icb.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stephens HA. HLA and other gene associations with dengue disease severity. Curr Top Microbiol Immunol. 2010;338:99–114. doi: 10.1007/978-3-642-02215-9_8. [DOI] [PubMed] [Google Scholar]
- 80.Loke H, et al. Susceptibility to dengue hemorrhagic fever in vietnam: evidence of an association with variation in the vitamin d receptor and Fc gamma receptor IIa genes. Am J Trop Med Hyg. 2002;67(1):102–6. doi: 10.4269/ajtmh.2002.67.102. [DOI] [PubMed] [Google Scholar]
- 81.Fernandez-Mestre MT, et al. TNF-alpha-308A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens. 2004;64(4):469–72. doi: 10.1111/j.1399-0039.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 82.Noecker CA, et al. Contrasting associations of polymorphisms in FcgammaRIIa and DC-SIGN with the clinical presentation of dengue infection in a Mexican population. Acta Trop. 2014;138:15–22. doi: 10.1016/j.actatropica.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 83.Mongkolsapaya J, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. 2006;176(6):3821–9. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 84.Ribeiro E, et al. Primary dengue fever associated with hemophagocytic syndrome: a report of three imported cases, Bordeaux, France. Intern Med. 2014;53(8):899–902. doi: 10.2169/internalmedicine.53.1108. [DOI] [PubMed] [Google Scholar]
- *85.Tan LH, et al. Hemophagocytosis in dengue: comprehensive report of six cases. J Clin Virol. 2012;55(1):79–82. doi: 10.1016/j.jcv.2012.06.005. A systems biology approach revealed transcriptional signatures of antibody responses to different classes of vaccines. [DOI] [PubMed] [Google Scholar]
- 86.Lin L, et al. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat Biotechnol. 2015;33(6):610–6. doi: 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thomas SJ. Dengue human infection model: re-establishing a tool for understanding dengue immunology and advancing vaccine development. Hum Vaccin Immunother. 2013;9(7):1587–90. doi: 10.4161/hv.24188. [DOI] [PubMed] [Google Scholar]
- 89.Plotkin SA, Gilbert PB. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis. 2012;54(11):1615–7. doi: 10.1093/cid/cis238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Connell RJ, Excler JL. HIV vaccine efficacy and immune correlates of risk. Curr HIV Res. 2013;11(6):450–63. doi: 10.2174/1570162x113116660052. [DOI] [PubMed] [Google Scholar]
- 91.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15(2):195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsang JS. Utilizing population variation, vaccination, and systems biology to study human immunology. Trends Immunol. 2015;36(8):479–93. doi: 10.1016/j.it.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]