Summary
Bacterial cell walls are composed of the large cross-linked macromolecule peptidoglycan, which maintains cell shape and is responsible for resisting osmotic stresses. This is a highly conserved structure and the target of numerous antibiotics. Obligate intracellular bacteria are an unusual group of organisms that have evolved to replicate exclusively within the cytoplasm or vacuole of a eukaryotic cell. They tend to have reduced amounts of peptidoglycan, likely due to the fact that their growth and division takes place within an osmotically protected environment, and also due to a drive to reduce activation of the host immune response. Of the two major groups of obligate intracellular bacteria, the cell wall has been much more extensively studied in the Chlamydiales than the Rickettsiales. Here, we present the first detailed analysis of the cell envelope of an important but neglected member of the Rickettsiales, Orientia tsutsugamushi. This bacterium was previously reported to completely lack peptidoglycan, but here we present evidence supporting the existence of a peptidoglycan-like structure in Orientia, as well as an outer membrane containing a network of cross-linked proteins, which together confer cell envelope stability. We find striking similarities to the unrelated Chlamydiales, suggesting convergent adaptation to an obligate intracellular lifestyle.
Introduction
In order to maintain correct cell shape, and the capacity to withstand the high internal turgor pressure, most bacteria synthesise a cell wall comprised largely of the macromolecule peptidoglycan. Peptidoglycan is composed of an alternating N-acetylglucosamine-N-acetylmuramic acid polysaccharide backbone, cross-linked via short peptide side chains. The sequence of the peptides, the degree of cross-linking, and the length of glycan chains vary between species and also stages of growth (Vollmer et al., 2008). However, some form of peptidoglycan is present in almost all known bacterial species with the notable exception of the mycoplasmas. The essentiality and conservation of peptidoglycan has made this an attractive antibiotic target (Schneider and Sahl, 2010), from the early discovery of penicillin to the recent identification of teixobactin (Ling et al., 2015). Peptidoglycan elongation and remodelling are highly complex processes, as they need to occur without compromising the integrity of the cell wall. This is achieved through a coordinated process involving the bacterial cytoskeleton and cell division machinery (Haeusser and Margolin, 2016). A detailed knowledge of the structure and composition of peptidoglycan is thus a prerequisite for understanding the molecular mechanisms of growth and division in bacterial cells.
The vast majority of bacteria grow and divide independently, living in a range of environments including soil, water, and the gut or skin of a mammalian host. Some bacteria, such as salmonella, shigella and listeria, have evolved the means to invade and replicate within mammalian cells during the course of an infection (Pizarro-Cerda and Cossart, 2006). A small group of bacteria, however, have evolved this ability to the extreme that they are no longer able to grow and divide outside their host cells. This unusual and fascinating group of organisms are predominantly comprised of two orders of Gram-negative bacteria: the Chlamydiales and the Rickettsiales. The Chlamydiales include human and animal pathogens, as well as a number of environmental strains that are found in soil-dwelling amoeba (Elwell et al., 2016). The Rickettsiales are a large and diverse group of vector-borne bacteria, that include the promiscuous insect symbiont Wolbachia, a number of human and animal pathogens in the Anaplasmataceae, and a group of typhus-causing pathogens in the Rickettsiaceae. The Rickettsiaceace are also thought to include the closest relatives of the precursor of modern mitochondria (Andersson et al., 1998). Although the Rickettsiales and Chlamydiales are unrelated, they exhibit similarities in their obligate intracellular life cycles. The bacterial cell wall has been extensively studied in the Chlamydiales and the long-standing ‘chlamydial anomaly’ describes the paradox that these bacteria are sensitive to the cell wall targeting drug penicillin, but that peptidoglycan could never be directly detected by chemical analysis (Moulder, 1993; McCoy and Maurelli, 2006; Mohammadi and Breukink, 2014). This was recently resolved using sensitive mass spectrometry techniques (Packiam et al., 2015) (Jacquier et al., 2015; Pilhofer et al., 2013) whilst a combination of novel labelling methods and cryoelectron tomography techniques have shed light on the arrangement of the cell wall of these organisms (Pilhofer et al., 2013; Liechti et al., 2014). In comparison, very little is known about the cell wall biology of the Rickettsiales. Comparative analyses have revealed the presence of peptidoglycan biosynthesis genes in many Rickettsiales (Gillespie et al., 2012), and the presence of the peptidoglycan precursor lipid II has been demonstrated in Wolbachia (Henrichfreise et al., 2009; Vollmer et al., 2013). However, extensive investigations into the composition and structure of peptidoglycan in these organisms have not been performed.
The genus Orientia is a divergent member of the order Rickettsiales, within the family Rickettsiaceae (Tamura et al., 1991; Ohashi et al., 1995). It is the causative agent of the severe mite-borne human disease scrub typhus, which is endemic across large parts of Asia (Phongmany et al., 2006; Mayfong Mayxay et al., 2013; Capeding et al., 2013; Cosson et al., 2015; Dittrich et al., 2015) and which can be life threatening in the absence of effective antibiotic treatment. It is thought to affect at least 1 million people per year (Watt and Parola, 2003). Despite its high incidence and severity, it is less well studied than other rickettsias such as R. prowazekii, R. conorii and R. rickettsii. Orientia is able to infect a range of cell types, including endothelial, fibroblast, monocyte/macrophage and dendritic cells (Paris et al., 2012; Keller et al., 2014; Moron et al., 2001). Similar to the other Rickettsiaceae, but in contrast to the Chlamydia and Anaplasmataceae that live within remodelled vacuoles (Bastidas et al., 2013; Moumène and Meyer, 2016), Orientia escapes from the endo-lysosomal pathway shortly after infection and replicates freely in the host cell cytoplasm (Chu et al., 2006).
Orientia has previously been reported to completely lack both peptidoglycan and LPS (Amano et al., 1987). This conclusion was based on an insensitivity to penicillin (Wisseman et al., 1982), an inability to detect peptidoglycan fragments by chemical analysis (Amano et al., 1987), and an absence of electron-dense material in the periplasmic inter-membrane space by electron microscopy (Silverman and Wisseman, 1978). However, whilst the sequencing of the Orientia genome confirmed an absence of genes required for biosynthesis of LPS, an almost complete complement of peptidoglycan biosynthesis genes were identified (Cho et al., 2007; Min et al., 2008; Nakayama et al., 2008) (Fig. 1). Furthermore, it has been shown that the intracellular host immune receptor Nod1, which recognises fragments of peptidoglycan, is activated in response to infection by Orientia (Cho et al., 2010c). Here, we provide evidence for a peptidoglycan-like structure in the cell envelope of Orientia, and show that cell rigidity is additionally conferred by a highly cross-linked outer membrane protein. This is the first detailed analysis of peptidoglycan in an obligate intracellular bacterium outside the Chlamydiales, and will inform our understanding of rickettsial cell biology as well as enable comparative studies across different bacterial lineages engaging in obligate intracellular life styles.
Figure 1. The peptidoglycan biosynthesis pathway in Orientia tsutsugamushi.
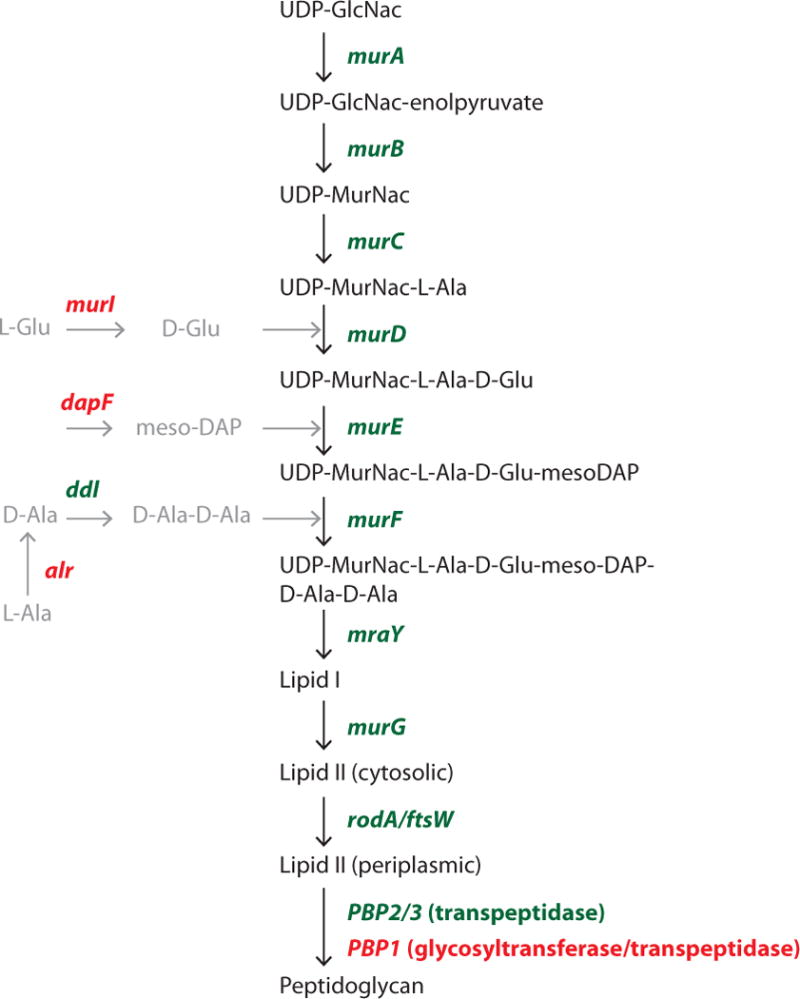
Genes shown in green are found in the genome of O. tsutsugamushi (Boryong and Ikeda strains), genes shown in red have no identifiable homolog.
Results and Discussion
The genome of Orientia tsutsugamushi contains a nearly complete set of genes required for peptidoglycan biosynthesis (Fig. 1) (Cho et al., 2007; Min et al., 2008; Nakayama et al., 2008). However, homologues of three important groups of enzymes are notably absent: (i) amino acid racemases (ii) glycosyltransferases and (iii) some genes in the meso-diaminopimelic acid (meso-DAP) biosynthesis pathway. Intriguingly, these are the same three pathways that are absent or incomplete in the chlamydial genomes (McCoy and Maurelli, 2006). The peptide side-chains of peptidoglycan are characterised by the presence of unusual amino acids, including D-isomer amino acids (usually D-Glu and D-Ala in Gram-negative bacteria) and the diamino acid meso-diaminopimelic acid (meso-DAP) (Vollmer and Bertsche, 2008; Vollmer et al., 2008; Turner et al., 2014). D-amino acids are not usually present in proteins, and are synthesised by the specific activity of amino acid racemases. Although O. tsutsugamushi does not encode the major L-alanine racemase, Alr, it does possess the gene ddl, which encodes a protein catalysing the ligation of the two D-Ala residues found in peptidoglycan side-chains. One possible alternative pathway for generation of D-Ala is through the GlyA enzyme, which was demonstrated to be a plausible candidate for D-Ala biosynthesis in C. pneumonia (De Benedetti et al., 2014; Shostak and Schirch, 1988) and for which a homolog is present in the genome of Orientia (OTBS_0253).
We used a highly sensitive mass spectrometric method (GC/EIMS) to search for the peptidoglycan-specific amino acid DAP in purified Orientia biomass. This method is based on GC/EIMS of amino acids after their transformation into volatile N-heptafluorobutyryl-isobutylester derivatives (Schumann P, 2011), and has previously been used to detect low amounts of peptidoglycan in planctomycetes (Jeske et al., 2015) and Verrucomicrobia (Spring S et al., 2016). Here, we were able to detect DAP in acid hydrolysates of O. tsutsugamushi cells (Fig. 2). The identification was based on a gas chromatographic peak matching that of the DAP standard derivative, with a retention time of 22.2 minutes and a set of fragment ions at 380, 324, 306 and 278 m/z (Jeske et al., 2015). The predicted structures corresponding to these fragment ions are shown in Fig. S1. The detection of DAP is highly suggestive of a peptidoglycan-like structure in O. tsutsugamushi and suggests that an alternative pathway may be used for synthesis of this amino acid. It is important to note, however, that the DAP was detected from isolated bacterial biomass rather than a purified sacculus, and that final proof for DAP-containing peptidoglycan in Orientia will require isolation and chemical analysis of this structure.
Figure 2. Identification of meso-DAP by mass spectrometry analysis.

Total Ion Chromatogram (TIC) and Extracted Ion Chromatograms (EIC) of the DAP derivative (N-heptafluorobutyryl 2,6-diaminopimelic acid isobutylester) from acid hydrolyzed O. tsutsugamushi cells showing a peak with retention time of 22.2 minutes and a set of fragment-ions at 380, 324, 306 and 278 m/z.
In order to determine whether the peptidoglycan biosynthesis pathway is active in O. tsutsugamushi we measured the gene expression levels of several peptidoglycan biosynthesis genes throughout the 7-day bacterial infection cycle using qRT-PCR analysis (Fig. 3). We found that genes murA, murD, murF, ddl and pbp2 were all expressed to a detectable level at some or all time points, when normalised to the housekeeping gene mipZ (Cho et al., 2010a). The gene pbp2, and to a lesser extent murD and murF, showed increased expression levels at early stages of the infection cycle in which bacteria are not yet actively dividing (days 1–3) compared with later stages of the infection cycle (days 5 and 7) potentially reflecting a higher requirement for peptidoglycan precursors during early stages of bacterial cell growth and preparation for division.
Figure 3. Expression of genes in the peptidoglycan biosynthesis pathway.
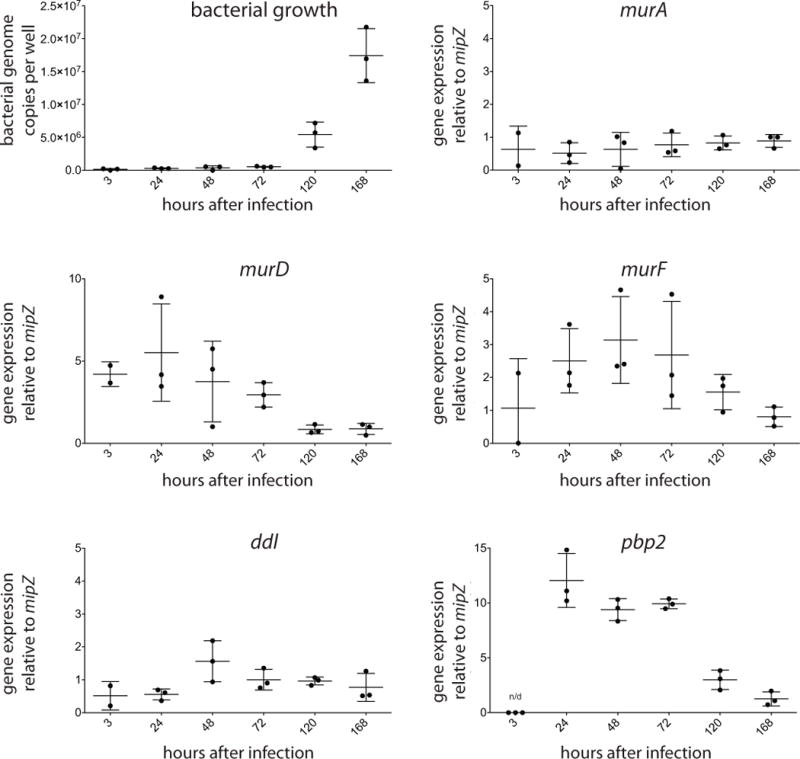
Bacterial genome copy number or relative gene expression of O. tsutsugamushi grown in mouse fibroblast L929 cells over 7 days. Relative expression level was determined by qRT-PCR and normalised to the housekeeping gene mipZ. Graph shows each individual data point, as well as the mean and standard deviation.
Next, we tested whether O. tsutsugamushi was sensitive to drugs that target the cell wall, in order to determine whether intact peptidoglycan was required for bacterial viability. O. tsutsugamushi is known to be insensitive to penicillin (Wisseman et al., 1982), and this is one of the reasons why it was previously reported to lack peptidoglycan. First, we measured the growth of O. tsutsugamushi in the presence of penicillin and also chloramphenicol, the latter of which is used clinically against this organism (Watt et al., 1996). We compared this with bacterial growth in the presence of D-cycloserine and phosphomycin, that target the cell wall biosynthesis proteins Alr/Ddl and MurA respectively (Fig. 4B and F, and Fig. S2 and S3). We found that chloramphenicol inhibited growth and penicillin did not, as expected, but that both D-cycloserine and phosphomycin inhibited the growth of O. tsutsugamushi to the same extent as chloramphenicol. Immunofluorescence microscopy images showed that Orientia cells became large and round in the presence of phosphomycin, and displayed reduced structural integrity in the presence of D-cycloserine (Fig. 4F and Fig. S3). This experiment showed, for the first time, a requirement for peptidoglycan precursor biosynthesis for the replication of O. tsutsugamushi. Next, we attempted to rescue the growth of O. tsutsugamushi treated with D-cycloserine by the addition of exogenous D-Ala. We found that the addition of D-Ala could not completely restore growth, even when added at 10× the molar ratio of D-cycloserine (Fig. 4C). This was in contrast to results from equivalent experiments performed in Chlamydia (Moulder et al., 1963) and may reflect a lack of uptake of D-Ala into O. tsutsugamushi cells or a complete inhibition of Ddl by D-cycloserine.
Figure 4. O. tsutsugamushi treated with cell wall-targeting drugs.
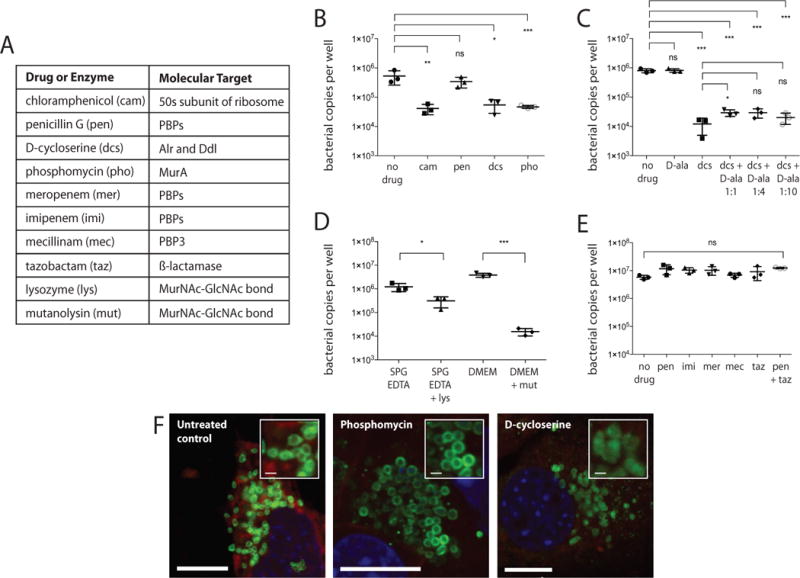
A. Table showing the molecular target of drugs and enzymes used in this study. B-E. Bacterial copy number per well of a 24-well plate after 7 days growth in the presence of different drugs. Three replicate wells were used in each experiment. Graph shows each individual data point, as well as the mean and standard deviation. Statistical significance was determined using an unpaired t test analysis. P values are illustrated as follows: ns (P > 0.05); * (P ≤ 0.05); ** (P ≤ 0.01); *** (P ≤ 0.001). F. Immunofluorescence microscopy of intracellular O. tsutsugamushi cells treated with 40 μg/ml phosphomycin or 250 μg/ml D-cycloserine for 24 hours. Bacteria are labelled in green (using anti-TSA56 antibody); host cytoplasm in red (Evans blue) and nuclei in blue (DAPI). Scale bar = 10 μm (large images) or 1 μm (insets).
The absence of Class A bifunctional PBPs as well as monofunctional glycosyltransferases raises the question of whether Orientia has the enzymatic ability to generate peptidoglycan with polymerised glycan chains. In the case of Wolbachia, which also lacks these key glycosyltransferase genes, it has been postulated that an unpolymerised lipid II precursor may play an essential role in localisation and coordination of the cell division machinery, but that typical a classical peptidoglycan sacculus is absent (Henrichfreise et al., 2009; Vollmer et al., 2013). Recent reports have shown that the SEDS family protein RodA is able to perform previously undescribed glycosyltransferase activity in Bacillus subtilis, and they predict that this may be a general activity of this protein family (Emami et al., 2017; Meeske et al., 2016). Both Orientia and Chlamydia possess homologs of SEDS proteins RodA and FtsW, and it is possible that these could account for the missing glycosyltransferase activity in these organisms. In order to test for the presence of a polymerised glycan chain in peptidoglycan of Orientia we measured the sensitivity to the enzymes lysozyme and mutanolysin. These enzymes cleave the glycan backbone of peptidoglycan at the ß-1,4-linkage between N-acetylmuramic acid (MurNAc) and N-acetyl-glucosamine (GlcNAc). We found that both enzymes caused bacterial cell lysis and resulted in a reduction in bacterial copy number after 7 days (Fig. 4D, Movie 1 and Fig. S2). These results suggest that the peptidoglycan of O. tsutsugamushi may contain at least some degree of MurNAc-GlcNAc disaccharide polymerisation. This could be achieved through the activity of FtsW/RodA SEDS proteins or other unidentified glycosyltransferase genes.
In order to understand why O. tsutsugamushi is insensitive to penicillin, we measured bacterial growth in the presence of alternative ß-lactam antibiotics, imipenem and meropenem, which also target penicillin binding proteins (PBPs) but which have improved bacterial permeability properties (Yang et al., 1995), as well as the ß-lactam antibiotic mecillinam that specifically targets PBP2. These were also ineffective against O. tsutsugamushi (Fig. 4E). We then tested growth in the presence of penicillin together with a ß-lactamase inhibitor, tazobactam, but this was unable to modulate the activity of penicillin (Fig. 4E). One possible explanation is that both PBPs in Orientia, PBP2 and PBP3, are intrinsically resistant to ß-lactam antibiotics. In order to test this hypothesis we performed an extensive protein alignment of PBPs 2 and 3 from a diverse set of bacterial species. We analysed known active site amino acid residues (Sauvage et al., 2014) and found that one, K310 (E. coli PBP3 numbering), was strictly conserved across all PBP2/3s tested, but mutated to histidine (PBP3) and glutamine (PBP2) in Orientia (Fig. S4). This mutation could potentially confer the observed ß-lactam insensitivity, but this has not been experimentally tested.
A number of recent studies have developed fluorescent D-amino acid derivatives that can be incorporated into bacterial peptidoglycan and directly visualised by fluorescence microscopy (Kuru et al., 2012; Liechti et al., 2014; Kuru et al., 2015; Siegrist et al., 2013; Shieh et al., 2014; Siegrist et al., 2015). We used one of these, the ‘clickable’ alkyne modified D-ala-D-ala dipeptide EDA-DA (ethynyl-D-alanyl-D-alanine) to visualise peptidoglycan in O. tsutsugamushi cells (Liechti et al., 2014). We found that this labelled the entire bacterial surface, suggesting the existence of a large structure that extends throughout the periplasm of O. tsutsugamushi (Fig. 5A and more examples in Fig. S5). This labelling was abolished in the presence of the cell wall-targeting drugs D-cycloserine and phosphomycin, suggesting that this probe is being incorporated specifically into a peptidoglycan-related structure. We observed a similar pattern of labelling using an alternative peptidoglycan probe, 7-hydroxycoumarin-3-carboxylic acid (HCC)-amino-D-alanine (HADA) (Fig. 5B). To our surprise, we found that the L-isomer of this probe, HCC-amino-L-alanine (HALA), produced similar labelling to that observed with HADA (Fig. 5B) although this labelling was weaker and was not observed every time. In order to study this further we also labelled the model Gram-negative bacteria E. coli with both probes and found that while HADA labelled all cells, as expected, a subpopulation of cells were also clearly labelled with the HALA probe (Fig. 5B). This was not shown in previous reports and may reflect differences in bacterial strains or exact experimental conditions. This result is unlikely to be due to contamination of the HALA probe with D-amino acids, as this would presumably lead to all cells being weakly labelled, rather than a small number being strongly labelled as observed. The exact mechanism of HADA incorporation into peptidoglycan has not been determined, and may vary between different bacterial species and growth stages. Possible routes may involve substitution into existing peptidoglycan by L,D-transpeptidase activity (we could identify no L,D-transpeptidase homolog in O. tsutsugamushi), or uptake into the cytoplasm and incorporation into peptidoglycan precursors by Ddl and MurF (Cava et al., 2011). Our observation of HALA labelling in both E. coli and O. tsutsugamushi may indicate a pathway involving entry into the cytoplasm and racemisation prior to incorporation into peptidoglycan precursors. Alternatively, it is possible that periplasmic peptidoglycan cross-linking or remodelling enzymes mediate the incorporation of D-amino acid probes, and that these are not strictly isomer-specific, especially at the high concentrations of fluorescent amino acids used in these studies (1 mM). In the case of both EDA-DA and HADA we found that the labelling was not positive every time, and was sensitive to parameters such as host cell confluence, multiplicity of infection, and time after initial infection. This suggests that the peptidoglycan biosynthesis machinery may be regulated in response to unknown signals, and this warrants further investigation.
Figure 5. O. tsutsugamushi labelled with peptidoglycan-reactive probes.

A. Fluorescence microscopy of O. tsutsugamushi labelled with EDA-DA probe reacted with an azide modified Alex Fluor 488 via click chemistry (green), antibody against the bacterial surface protein TSA56 (red) and the DNA stain DAPI (blue). Bacteria were grown in the presence or absence of drugs for 24 hours. Drug concentrations were as follows: 40 μg/ml phosphomycin; 250 μg/ml D-cycloserine. B. Fluorescence microscopy of O. tsutsugamushi and E. coli in the presence of the fluorescent D-alanine derivative HADA (blue), the fluorescent L-alanine derivative HALA (blue), antibody against the bacterial surface protein TSA56 (red, O. tsutsugamushi) or the bacterial cytoplasm (Evans blue, red, E. coli). Scale bar = 10 μm.
The EDA-DA and HADA labelling patterns in O. tsutsugamushi are suggestive of an extended sacculus-like structure encasing the bacterial cell, although definitive evidence for this will require isolation of this material and subsequent mass spectrometry and microscopy analysis. Our attempts at biochemical isolation of the peptidoglycan-like structure of Orientia were unsuccessful, and this may reflect low levels of this material as well as unknown regulation of its biosynthesis at different stages of the infection and replication cycle. Our HADA/EDA-DA labelling pattern is in contrast to that observed in pathogenic chlamydiae, in which peptidoglycan labelling localises only to the division septum (Liechti et al., 2016). We never observed septum labelling of Orientia in the experiments reported here. It is worth noting that O. tsutsugamushi possesses both MreB and FtsZ, unlike the Chlamydiales and Planctomycetes that have lost the bacterial cell division cytoskeletal filament FtsZ and have evolved a non-canonical cell division mechanism thought to be driven by the interplay between the bacterial actin homolog MreB and a septal peptidoglycan ring (Liechti et al., 2016; Liechti et al., 2014).
A major role of peptidoglycan is in protecting bacterial cells from osmotic stresses. One hypothesis to explain the lack or reduction of peptidoglycan in obligate intracellular bacteria is that they predominantly occupy an osmotically protected cellular environment and therefore have a reduced need for this structure. We performed fluorescence microscopy on O. tsutsugamushi bacteria that had been isolated from host cells and resuspended in sucrose-phosphate-glutamate buffer (SPG), PBS or water, and found that bacteria did not lyse in the presence of PBS or pure water, indicating the presence of a mechanical stress-bearing structure such as would be provided by a peptidoglycan sacculus (Fig. 6A). We performed quantitative image analysis using both a supervised machine learning approach and a principal component analysis approach (PCA), and this showed that bacteria resuspended in SPG were more ‘smooth’ than those resuspended in water or PBS (Fig. 6B and Fig. S7). This morphology may reflect a disruption in the cell envelope structure, which would be consistent with our previous report that the viability of isolated O. tsutsugamushi was higher when stored in SPG compared with PBS or water (Giengkam et al., 2015).
Figure 6. Structural rigidity of O. tsutsugamushi cells.
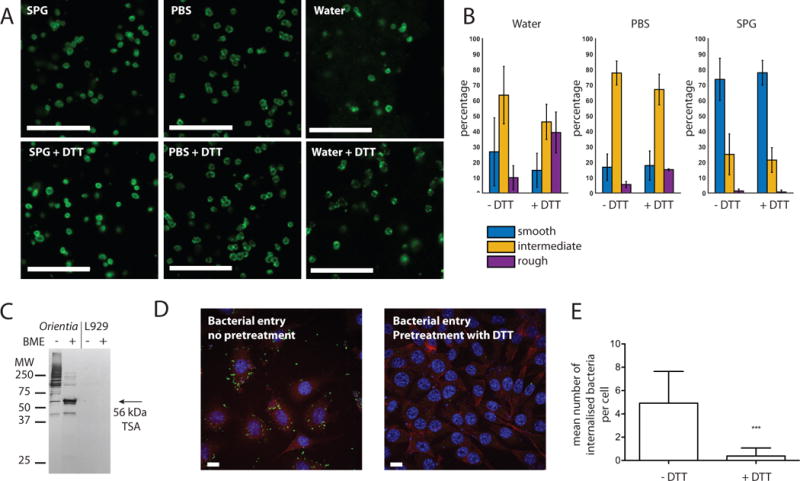
A. Immunofluorescence microscopy showing purified O. tsutsugamushi resuspended in sucrose-phosphate glutamate buffer (SPG), PBS or water, with and without 1 mM DTT. Bacteria are labelled using an antibody against the TSA56 protein. Scale bar = 10 μm. B. Quantitative analysis of images of O. tsutsugamushi treated as described in A. Graph shows morphological differences of O.tsutsugamushi in different solutions as described in A, using Random Forest classification using 18 features. Training dataset includes more than 40 cells for each condition. Bar graph shows fractions of bacteria that exhibit ‘Smooth’, ‘Intermediate’ and ‘Rough’ morphology. See also Fig. S8 for example images from each morphological class. Graph shows mean and SD. C. Western blot of O. tsutsugamushi or uninfected L929 host cells in the presence or absence of ß-mercaptoethanol (BME), using a monoclonal antibody against the TSA56 protein. D. Immunofluorescence microscopy images of L929 cells infected with O. tsutsugamushi that had been mock-treated (left) or pre-treated with 1 mM DTT then washed prior to infection (right). Cells were fixed 24 hours after infection. Bacteria are shown in green (anti-TSA56 antibody), host actin in red (phalloidin) and nuclei in blue (DAPI). Scale bar = 10 μm. E. Average number of bacteria inside host cells after mock treatment or pre-treatment with DTT. 100 host cells were randomly selected, imaged, and manually counted. The mean and standard deviation is shown, with statistical significance determined using an unpaired t test analysis. P values are illustrated as follows: ns (P > 0.05); * (P ≤ 0.05); ** (P ≤ 0.01); *** (P ≤ 0.001).
The most abundant protein in the outer membrane of O. tsutsugamushi is an integral membrane protein called the 56kDa type-specific antigen (TSA56). This highly immunostimulatory protein has no known homolog in any other species, and contains regions of high diversity between Orientia strains. This variability has enabled the use of this gene to classify Orientia strains, and corresponds closely to serotype classification of different strains. In spite of the high sequence variability, we observed that this protein has three conserved cysteine residues (C33, C190 and C416 in strain UT76) and therefore wondered whether this highly abundant protein might form a cross-linked network across the surface of the outer membrane, similar to that formed by the major outer-membrane proteins (MOMP) of the Chlamydiales (Bavoil et al., 1984; Hatch et al., 1986) and in place of LPS which usually covers the surface of other Gram negative bacteria. The TSA56 protein has previously been shown to form higher order intermolecular aggregates in the absence of reducing agent, using western blot analysis, (Urakami et al., 1986) and we were able to reproduce this result using the karp-like UT76 strain of O. tsutsugamushi used in the current report (Fig. 6C). We used fluorescence microscopy to monitor the effect of exposing purified bacteria to the reducing agent DTT and found that this had little additional structural effect when cells were resuspended in SPG or PBS buffer (Fig. 6A and B). However, when isolated bacteria were incubated with water, the addition of DTT resulted in more patchy membrane labelling and a higher background, which may indicate greater bacterial fragility (Fig. 6A). Quantitative image analysis confirmed a greater difference in cell morphology upon DTT treatment in the presence of water compared with PBS or SPG (Fig. 6B and Fig. S7). Control experiments showed that E. coli did not undergo morphological changes under the same conditions (Fig. S6). This result supports the hypothesis that a highly cross-linked outer membrane confers some degree of osmotic protection to bacterial cells.
We then tested the importance of this cross-linked surface for bacterial infectivity and found that bacteria exposed to DTT had a dramatically reduced ability to invade host cells compared with untreated bacteria (Fig. 6D and E). The TSA56 protein is known to be required for bacterial entry into host cells through interaction with extracellular fibronectin, and it is possible that the addition of DTT resulted in the depletion of higher-order structures of TSA56 required for efficient interaction with fibronectin (Lee et al., 2008; Cho et al., 2010b; Lee et al., 2008; Cho et al., 2010b). Taken together, these results support the hypothesis that the O. tsutsugamushi outer membrane is stabilised by a cross-linked network of proteins including the TSA56 protein and potentially other partners, and that this structure is important for bacterial cell integrity and infectivity.
Conclusion
In this study we have analysed the structure of the cell envelope of the obligate intracellular human pathogen O. tsutsugamushi. We present evidence for a peptidoglycan-like molecule as well as disulphide cross-linked outer membrane proteins, and demonstrate that these components are required for bacterial growth, cell integrity and host cell invasion. Important outstanding questions include the identity of the hypothesised glycosyl transferase, amino acid racemase and meso-DAP biosynthesis enzymes and the exact chemical composition and structure of the O. tsutsugamushi peptidoglycan-like network. This first study paves the way for a detailed analysis of the molecular mechanisms of growth and division in this important human pathogen and model intracellular organism. We also show that both the D-isomer and L-isomer of the fluorescent alanine amino acid probe are able to label at least some O. tsutsugamushi and E. coli cells, and this raises further questions about the mechanism of incorporation of these probes. The reduced level of peptidoglycan found in this organism, the absence of key peptidoglycan biosynthesis enzymes, and the presence of a cross-linked protein network embedded in the outer membrane of these bacteria are all highly reminiscent of the unrelated Chlamydiales and suggest convergent adaptation to an obligate intracellular life style.
Materials and Methods
Bacterial strains, cell lines and growth conditions
All experiments were performed using the mouse fibroblast cell line L929 (ATCC CCL-1) which was a kind gift from Dr. Blacksell at the Mahidol Oxford Tropical Medicine Research Unit, Bangkok, Thailand. The Thai Karp-like clinical isolate Orientia tsutsugamushi strain UT76 (Giengkam et al., 2015), and E. coli strain NEB 5 alpha (New England Biolabs, USA, catalog number C2987I) was used throughout. Cell culture was performed using DMEM media with 10 % FBS at 37 °C (uninfected cells) or 35 °C (infected cells) and 5% CO2. For routine propagation bacteria were grown in 25 or 75 cm2 culture flasks as described previously (Giengkam et al., 2015). All experiments were performed using bacteria harvested from the intracellular fraction of infected cells, 7 days after infection.
Mass spectrometry analysis
Mass spectrometry analysis was performed on biomass extracted from between 15 and 45 ml infected cells, which had been infected at an MOI of 10:1 to 1000:1. Bacteria were grown in L929 cells for 7 days, then purified using mechanical lysis of host cells and filtering with a 2 μm filter (Giengkam et al., 2015). The sample was pelleted and washed in 600μl of 300mM ice-cold sucrose two times. The supernatant was removed, and the pellet was autoclaved prior to extraction. The pellet was hydrolysed using 4M HCl at 100 °C for 16 hours, then amino acids were transformed into volatile N-heptafluorobutyryl-isobutylester derivatives using Protocol 10 of Schumann, 2011 (Schumann P, 2011). The samples were analysed by GC/MS (320 Singlequad, Varian) and diaminopimelic acid was identified as a gas chromatographic peak with a retention time of 22.2 minutes and a set of fragment ions at 380, 324, 306 and 278 m/z(Jeske et al., 2015).
Bacterial quantification by qPCR
Bacterial quantification was performed as described previously (Giengkam et al., 2015). Briefly, the supernatant and/or cellular fraction of infected L929 cells were isolated and DNA extraction was performed using the alkaline lysis method. Quantitative PCR was performed using a primer/probe set (Table S1) within the TSA47 gene (OTT_1319) and the bacterial genome copy number was calculated using a standard curve.
RNA extraction and gene expression analysis by qRT-PCR
At each time point, infected cells were placed on ice and rapidly resuspended in RNAProtect Bacteria Reagent (Qiagen, catalog number 76506), then stored at −80 °C until use. Total RNA extraction was performed using the Qiagen RNeasy Plus kit (Qiagen, catalog number 74136) according to manufacturer’s instructions. Purified RNA (10 μg) was treated with DNaseI (Thermo Fisher Scientific, catalog number AM2238) at 37 °C for 30–60 minutes, then DNaseI-treated RNA was converted to cDNA using the iScript reverse transcription supermix (Biorad, catalog number 170-8841) with random primers. Complete removal of genomic DNA was verified using a reverse transcriptase-free control reaction, where the absence of any PCR product demonstrated the absence of contaminating genomic DNA in the RNA sample. The cDNA was stored at −20°C until use. cDNA was used as a template for qPCR using gene-specific primers for mipZ, murA, murD, murF, ddl, and pbp2 (Table S1). qPCR was performed using SYBR green qPCR mix (Biotools, Houston, USA, catalog number 10.609) and the expression levels were normalised relative to the housekeeping gene mipZ.
Drug sensitivity assays
Bacteria were grown in L929 cells in 24 well plates in the presence of different drugs, and the bacterial genomic copy number after 7 days was determined by qPCR. All drugs were added directly to the wells shortly before adding isolated bacteria, with the exception of lysozyme (which cannot enter mammalian cells). In the case of lysozyme, purified bacteria were resuspended in SPG buffer +/− lysozyme and incubated at 37 °C for 15 mins before being added to L929 cells for infection. For microscopy analysis, bacteria were grown in microscopy chamber slides in the presence of drug (phosphomycin, D-cycloserine) or bacteria were incubated in TE buffer (10mM Tris + 1mM EDTA) + 5 mg/ml lysozyme before being dried onto a regular glass slide (lysozyme only). Concentrations of enzymes, drugs and chemicals used were as follows: chloramphenicol (cam) 100 μg/ml; penicillin G (pen) 150 μg/ml; D-cycloserine (dcs) 250 μg/ml; phosphomycin (pho) 40 μg/ml; D-alanine (D-ala) 250 μg/ml (1:1), 1,000 μg/ml (1:4), 2,500 μg/ml (1:10); EDTA 1 mM, lysozyme (lys) 5 mg/ml; mutanolysin (mut) 80 μg/ml; imipenem (imi) 10 μg/ml; meropenem (mer) 10 μg/ml; mecillinam (mec) 2 mg/ml; tazobactam (taz) 100 μg/ml.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software using a parametric, unpaired t test. P values are illustrated as follows: ns (P > 0.05); * (P ≤ 0.05); ** (P ≤ 0.01); *** (P ≤ 0.001).
Immunofluorescence microscopy
Bacteria were grown in host cells directly in glass chamber slides (Ibidi, USA) or dried onto a regular glass slide, then fixed with 4 % paraformaldehyde. Cells were permeabilised with 0.5 % triton X at 4 °C for 5 minutes, and then labelled using primary (Table S2) and fluorescently conjugated secondary antibodies. For labelling with the probes HADA and HALA, bacteria were grown in host cells on glass chamber slides, and the probe was added directly to the growth media to a final concentration of 1 mM (a dilution of 1/200). The cells were incubated at 35 °C with 5% CO2 for 3 hours, then washed with PBS and fixed as above. For labelling with EDA-DA, the probe was added to growing bacteria (3 days post infection) at 1 mM concentration and incubated for 24 hours at 35 °C with 5% CO2. When labelling was performed in the presence of drugs these were added at the same time as EDA-DA and incubated for 24 hours at 35 °C with 5% CO2. Cells were fixed and permeabilised as above, and the probe was reacted with azide-derived Alexa fluor 488 using the Click-iT cell reaction buffer kit from Invitrogen (catalog number C10269) following the manufacturers instructions.
Quantitative image analysis and classification
We applied quantitative imaging techniques to extract morphometric parameters for the individual O.tsutsugamushi using Cell Profiler (Broad Institute). The extracted 155 features (shown in Table S3) describe size, shape, intensity, granularity, texture and radial distribution of each bacterial cell.
To perform phenotypic classification, we used the Cell Profiler Analyst (Broad Institute) to apply Random Forest classification algorithm to classify O.tsutsugamushi into 3 main phenotypes, 1) ‘rough’ phenotype from cells in water with DTT, 2) ‘Intermediate’ phenotype from cells in PBS and 3) ‘Smooth’ phenotype from cells in SPG. These training image sets were taken separately from the images to be used for final prediction. The classification algorithm identified 18 features (Table S4) that can best distinguish training images from the three buffer conditions with 85% accuracy (Figure S7). We then applied these classification rules to determine the morphological phenotypes for the rest of the images (Figure 6B).
Principal component analysis was performed on MATLAB (Mathworks, USA, 2015b), using the 18 morphometric parameters previously identified from the Random Forest algorithm. The first three principal components explain over 95% of the observed variance, representing excellent performance for PCA model (Table S6 and S7). To compare phenotypic differences in different buffer solutions, we performed a pair-wise comparison of PCA loadings using three-dimensional Kolmogorov-Smirnov Test, with the significance level of 0.05.
Western blot analysis
Infected or uninfected L929 cells were mechanically lysed in a Bullet Blender (Next Advance, USA), at power 8 for 1 min and pelleted, washed with PBS 1–2 times, and resuspended in a small volume of PBS. The protein concentration was measured by nanodrop analysis and the sample volume was adjusted to ensure equivalent loading. Samples were mixed with SDS-PAGE buffer with or without 20% ß-mercaptoethanol and samples were then loaded on 12% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-rad, USA, catalog number 4561043). Western blots were probed using a monoclonal antibody against the 56 kDa type-specific antigen (Rat, Table S2), and then probed with a secondary antibody conjugated with alkaline phosphatase (Promega, USA, catalog number S3831) and developed using an alkaline phosphatase detection kit (Promega, USA, catalog number S3841)
Quantification of bacterial entry into host cells
Bacteria were isolated and resuspended in growth media with or without 1 mM DTT. Samples were incubated for 20 minutes at room temperature, washed by pelleting two times, and resuspended in fresh media before being added to L929 cells grown in glass chamber slides. Cells were fixed 24 hours after infection and the number of bacteria inside host cells counted manually for 100 cells.
Supplementary Material
Movie 1. Live imaging of purified Orientia in water with and without 80 μg/ml mutanolysin. Bacteria were purified from host cells and labelled with 1 mM SYTO9 as described in (Atwal et al., 2016). Images are acquired every 10 minutes, and the addition of mutanolysin is indicated in the movie. The large clumps accumulating prior to mutanolysin addition reflect natural aggregation of purified bacteria over time.
Acknowledgments
We are grateful for the support of colleagues at the Microbiology Department of MORU, especially Dr. Blacksell, Prof. Paris, Dr. Limmathurotsakul and Prof. Day. We thank Prof. Waldemar Vollmer of Newcastle University for helpful discussions and comments on this manuscript. This work was funded by a Royal Society Dorothy Hodgkin Fellowship (JS), the Wellcome Trust (JS, SA, SC, SG) and NIH grant AI113172 (MVN).
Footnotes
Author contributions
Conception and design of study – JD, JS, SA, SG. Acquisition, analysis or interpretation of data – JD, JS, MVN, NK, PS, SA, SC, SG, SS. Writing of the manuscript – JD, JS, SS. All in alphabetical order.
References
- Amano K, Tamura A, Ohashi N, Urakami H, Kaya S, Fukushi K. Deficiency of peptidoglycan and lipopolysaccharide components in Rickettsia tsutsugamushi. Infection and Immunity. 1987 doi: 10.1128/iai.55.9.2290-2292.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Atwal S, Giengkam S, VanNieuwenhze M, Salje J. Live imaging of the genetically intractable obligate intracellular bacteria Orientia tsutsugamushi using a panel of fluorescent dyes. J Microbiol Methods. 2016;130:169–176. doi: 10.1016/j.mimet.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. Chlamydial intracellular survival strategies. Cold Spring Harb Perspect Med. 2013;3:a010256. doi: 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavoil P, Ohlin A, Schachter J. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect Immun. 1984;44:479–485. doi: 10.1128/iai.44.2.479-485.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capeding MR, Chua MN, Hadinegoro SR, Hussain II, Nallusamy R, Pitisuttithum P, Rusmil K, Thisyakorn U, Thomas SJ, Huu Tran N, Wirawan DN, Yoon IK, Bouckenooghe A, Hutagalung Y, Laot T, Wartel TA. Dengue and other common causes of acute febrile illness in Asia: an active surveillance study in children. PLoS Negl Trop Dis. 2013;7:e2331. doi: 10.1371/journal.pntd.0002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BA, Cho NH, Min CK, Kim SY, Yang JS, Lee JR, Jung JW, Lee WC, Kim K, Lee MK, Kim S, Kim KP, Seong SY, Choi MS, Kim IS. Global gene expression profile of Orientia tsutsugamushi. Proteomics. 2010a;10:1699–1715. doi: 10.1002/pmic.200900633. [DOI] [PubMed] [Google Scholar]
- Cho BA, Cho NH, Seong SY, Choi MS, Kim IS. Intracellular invasion by Orientia tsutsugamushi is mediated by integrin signaling and actin cytoskeleton rearrangements. Infect Immun. 2010b;78:1915–1923. doi: 10.1128/IAI.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KA, Jun YH, Suh JW, Kang JS, Choi HJ, Woo SY. Orientia tsutsugamushi induced endothelial cell activation via the NOD1-IL-32 pathway. Microb Pathog. 2010c;49:95–104. doi: 10.1016/j.micpath.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Cho NH, Kim HR, Lee JH, Kim IS. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host– cell interaction genes. PNAS. 2007;104 doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Lee JH, Han SH, Kim SY, Cho NH, Kim IS, Choi MS. Exploitation of the endocytic pathway by Orientia tsutsugamushi in nonprofessional phagocytes. Infect Immun. 2006;74:4246–4253. doi: 10.1128/IAI.01620-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson JF, Galan M, Bard E, Razzauti M, Bernard M, Morand S, Brouat C, Dalecky A, Ba K, Charbonnel N, Vayssier-Taussat M. Detection of Orientia sp. DNA in rodents from Asia, West Africa and Europe. Parasit Vectors. 2015;8:172. doi: 10.1186/s13071-015-0784-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti S, Buhl H, Gaballah A, Klockner A, Otten C, Schneider T, Sahl HG, Henrichfreise B. Characterization of serine hydroxymethyltransferase GlyA as a potential source of D-alanine in Chlamydia pneumoniae. Front Cell Infect Microbiol. 2014;4:19. doi: 10.3389/fcimb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich S, Rattanavong S, Lee SJ, Panyanivong P, Craig SB, Tulsiani SM, Blacksell SD, Dance DA, Dubot-Peres A, Sengduangphachanh A, Phoumin P, Paris DH, Newton PN. Orientia, rickettsia, and leptospira pathogens as causes of CNS infections in Laos: a prospective study. Lancet Glob Health. 2015;3:e104–12. doi: 10.1016/S2214-109X(14)70289-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell C, Mirrashidi K, Engel J. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol. 2016 doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, Allenby N, Daniel RA, Errington J. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol. 2017;2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giengkam S, Blakes A, Utsahajit P, Chaemchuen S, Atwal S, Blacksell SD, Paris DH, Day NP, Salje J. Improved Quantification, Propagation, Purification and Storage of the Obligate Intracellular Human Pathogen Orientia tsutsugamushi. PLoS Negl Trop Dis. 2015;9:e0004009. doi: 10.1371/journal.pntd.0004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Nordberg E, Azad A, Sobral BW. Phylogeny and Comparative Genomics: the Shifting Landscape in the Genomics Era. Phylogeny and Comparative Genomics: the Shifting Landscape in the Genomics Era. 2012:84–141. [Google Scholar]
- Haeusser DP, Margolin W. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol. 2016;14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP, Miceli M, Sublett JE. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol. 1986;165:379–385. doi: 10.1128/jb.165.2.379-385.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichfreise B, Schiefer A, Schneider T, Nzukou E, Poellinger C, Hoffmann TJ, Johnston KL, Moelleken K, Wiedemann I, Pfarr K, Hoerauf A, Sahl HG. Functional conservation of the lipid II biosynthesis pathway in the cell wall-less bacteria Chlamydia and Wolbachia: why is lipid II needed? Mol Microbiol. 2009;73:913–923. doi: 10.1111/j.1365-2958.2009.06815.x. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Viollier PH, Greub G. The role of peptidoglycan in chlamydial cell division: towards resolving the chlamydial anomaly. FEMS Microbiol Rev. 2015;39:262–275. doi: 10.1093/femsre/fuv001. [DOI] [PubMed] [Google Scholar]
- Jeske O, Schüler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, Spring S, Jogler C. Planctomycetes do possess a peptidoglycan cell wall. Nat Commun. 2015;6:7116. doi: 10.1038/ncomms8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CA, Hauptmann M, Kolbaum J, Gharaibeh M, Neumann M, Glatzel M, Fleischer B. Dissemination of Orientia tsutsugamushi and inflammatory responses in a murine model of scrub typhus. PLoS Negl Trop Dis. 2014;8:e3064. doi: 10.1371/journal.pntd.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc. 2015;10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cho NH, Kim SY, Bang SY, Chu H, Choi MS, Kim IS. Fibronectin facilitates the invasion of Orientia tsutsugamushi into host cells through interaction with a 56-kDa type-specific antigen. J Infect Dis. 2008;198:250–257. doi: 10.1086/589284. [DOI] [PubMed] [Google Scholar]
- Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. Pathogenic Chlamydia Lack a Classical Sacculus but Synthesize a Narrow, Mid-cell Peptidoglycan Ring, Regulated by MreB, for Cell Division. PLoS Pathog. 2016;12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. A new antibiotic kills pathogens without detectable resistance. Nature. 2015 doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfong Mayxay M, Castonguay-Vanier J, Chansamouth V, Dubot-Peres A, Paris DH, Phetsouvanh R, Tangkhabuanbutra J, Douangdala P, Inthalath S, Souvannasing P, Slesak G, Tongyoo N, Chanthongthip A, Panyanouvong P, Sibounheuang B, Phommasone K, Dohnt M, Phonekeo D, Hongvanthong B, Xayadeth S, Ketmayoon P, Blacksell SD, Moore CE, Craig SB, Burns MA, von Sonnenburg F, Corwin A, de Lamballerie X, Gonzalez IJ, Christophel EM, Cawthorne A, Bell D, Newton PN. Causes of non-malarial fever in Laos: a prospective study. Lancet Glob Health. 2013;1:e46–e54. doi: 10.1016/S2214-109X(13)70008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Maurelli AT. Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 2006;14:70–77. doi: 10.1016/j.tim.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min CK, Yang JS, Kim S, Choi MS, Kim IS, Cho NH. Genome-based construction of the metabolic pathways of Orientia tsutsugamushi and comparative analysis within the Rickettsiales order. Comp Funct Genomics. 2008:623145. doi: 10.1155/2008/623145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Breukink E. The chlamydial anomaly clarified? Chembiochem. 2014;15:1391–1392. doi: 10.1002/cbic.201402143. [DOI] [PubMed] [Google Scholar]
- Moron CG, Popov VL, Feng HM, Wear D, Walker DH. Identification of the target cells of Orientia tsutsugamushi in human cases of scrub typhus. Mod Pathol. 2001;14:752–759. doi: 10.1038/modpathol.3880385. [DOI] [PubMed] [Google Scholar]
- Moulder JW. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis. 1993;2:87–99. [PubMed] [Google Scholar]
- Moulder JW, Novosel DL, Officer JE. Inhibition Of The Growth Of Agents Of The Psittacosis Group By D-Cycloserine And Its Specific Reversal By D-Alanine. J Bacteriol. 1963;85:707–711. doi: 10.1128/jb.85.3.707-711.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumène A, Meyer DF. Ehrlichia’s molecular tricks to manipulate their host cells. Microbes Infect. 2016;18:172–179. doi: 10.1016/j.micinf.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, Urakami H, Ohnishi M, Uchiyama I, Ogura Y, Ooka T, Oshima K, Tamura A, Hattori M, Hayashi T. The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008;15:185–199. doi: 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi N, Fukuhara M, Shimada M, Tamura A. Phylogenetic position of Rickettsia tsutsugamushi and the relationship among its antigenic variants by analyses of 16S rRNA gene sequences. FEMS Microbiol Lett. 1995;125:299–304. doi: 10.1111/j.1574-6968.1995.tb07372.x. [DOI] [PubMed] [Google Scholar]
- Packiam M, Weinrick B, Jacobs WRJ, Maurelli AT. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. Proc Natl Acad Sci U S A. 2015;112:11660–11665. doi: 10.1073/pnas.1514026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris DH, Phetsouvanh R, Tanganuchitcharnchai A, Jones M, Jenjaroen K, Vongsouvath M, Ferguson DP, Blacksell SD, Newton PN, Day NP, Turner GD. Orientia tsutsugamushi in human scrub typhus eschars shows tropism for dendritic cells and monocytes rather than endothelium. PLoS Negl Trop Dis. 2012;6:e1466. doi: 10.1371/journal.pntd.0001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phongmany S, Rolain JM, Phetsouvanh R, Blacksell SD, Soukkhaseum V, Rasachack B, Phiasakha K, Soukkhaseum S, Frichithavong K, Chu V, Keolouangkhot V, Martinez-Aussel B, Chang K, Darasavath C, Rattanavong O, Sisouphone S, Mayxay M, Vidamaly S, Parola P, Thammavong C, Heuangvongsy M, Syhavong B, Raoult D, White NJ, Newton PN. Rickettsial infections and fever, Vientiane, Laos. Emerg Infect Dis. 2006;12:256–262. doi: 10.3201/eid1202.050900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun. 2013;4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Sauvage E, Derouaux A, Fraipont C, Joris M, Herman R, Rocaboy M, Schloesser M, Dumas J, Kerff F, Nguyen-Distèche M, Charlier P. Crystal structure of penicillin-binding protein 3 (PBP3) from Escherichia coli. PLoS One. 2014;9:e98042. doi: 10.1371/journal.pone.0098042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Sahl HG. An oldie but a goodie - cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol. 2010;300:161–169. doi: 10.1016/j.ijmm.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Schumann P. Peptidoglycan structure. Methods Micobiology 2011 [Google Scholar]
- Shieh P, Siegrist MS, Cullen AJ, Bertozzi CR. Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc Natl Acad Sci U S A. 2014;111:5456–5461. doi: 10.1073/pnas.1322727111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shostak K, Schirch V. Serine hydroxymethyltransferase: mechanism of the racemization and transamination of D- and L-alanine. Biochemistry. 1988;27:8007–8014. doi: 10.1021/bi00421a006. [DOI] [PubMed] [Google Scholar]
- Siegrist MS, Swarts BM, Fox DM, Lim SA, Bertozzi CR. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol Rev. 2015;39:184–202. doi: 10.1093/femsre/fuu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8:500–505. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman DJ, Wisseman CL. Comparative Ultrastructural Study on the Cell Envelopes of Rickettsia prowazekii, Rickettsia rickettsii, and Rickettsia tsutsugamushi. Infection and Immunity. 1978:1020–1023. doi: 10.1128/iai.21.3.1020-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring S, Bunk B, Spröer C, Schumann P, Rohde M, Tindall BJ, HP K. Description and complete genome sequence of Kiritimatiella glycovorans gen. nov., sp. nov., the first cultured representative of a novel phylum originally designated Verrucomicrobia subdivision 5. ISME Journal. 2016 doi: 10.1038/ismej.2016.84. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura A, Urakami H, Ohashi N. A comparative view of Rickettsia tsutsugamushi and the other groups of rickettsiae. Eur J Epidemiol. 1991;7:259–269. doi: 10.1007/BF00145675. [DOI] [PubMed] [Google Scholar]
- Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol. 2014;91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakami H, Ohashi N, Tsuruhara T, Tamura A. Characterization of polypeptides in Rickettsia tsutsugamushi: effect of preparative conditions on migration of polypeptides in polyacrylamide gel electrophoresis. Infect Immun. 1986;51:948–952. doi: 10.1128/iai.51.3.948-952.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer J, Schiefer A, Schneider T, Julicher K, Johnston KL, Taylor MJ, Sahl HG, Hoerauf A, Pfarr K. Requirement of lipid II biosynthesis for cell division in cell wall-less Wolbachia, endobacteria of arthropods and filarial nematodes. Int J Med Microbiol. 2013;303:140–149. doi: 10.1016/j.ijmm.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Watt G, Chouriyagune C, Ruangweerayud R, Watcharapichat P, Phulsuksombati D, Jongsakul K, Teja-Isavadharm P, Bhodhidatta D, Corcoran KD, Dasch GA, Strickman D. Scrub typhus infections poorly responsive to antibiotics in northern Thailand. Lancet. 1996;348:86–89. doi: 10.1016/s0140-6736(96)02501-9. [DOI] [PubMed] [Google Scholar]
- Watt G, Parola P. Scrub typhus and tropical rickettsioses. Curr Opin Infect Dis. 2003;16:429–436. doi: 10.1097/00001432-200310000-00009. [DOI] [PubMed] [Google Scholar]
- Wisseman CLJ, Silverman DJ, Waddell A, Brown DT. Penicillin-induced unstable intracellular formation of spheroplasts by rickettsiae. J Infect Dis. 1982;146:147–158. doi: 10.1093/infdis/146.2.147. [DOI] [PubMed] [Google Scholar]
- Yang Y, Bhachech N, Bush K. Biochemical comparison of imipenem, meropenem and biapenem: permeability, binding to penicillin-binding proteins, and stability to hydrolysis by beta-lactamases. J Antimicrob Chemother. 1995;35:75–84. doi: 10.1093/jac/35.1.75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Live imaging of purified Orientia in water with and without 80 μg/ml mutanolysin. Bacteria were purified from host cells and labelled with 1 mM SYTO9 as described in (Atwal et al., 2016). Images are acquired every 10 minutes, and the addition of mutanolysin is indicated in the movie. The large clumps accumulating prior to mutanolysin addition reflect natural aggregation of purified bacteria over time.


