Summary
The protein 53BP1 plays a central regulatory role in DNA double-strand break repair. 53BP1 relocates to chromatin by recognizing RNF168-mediated mono-ubiquitylation of histone H2A Lys15 in the nucleosome core particle dimethylated at histone H4 Lys20 (NCP-ubme). 53BP1 relocation is terminated by ubiquitin ligases RNF169 and RAD18 via unknown mechanisms. Using NMR spectroscopy and biochemistry, we show that RNF169 bridges ubiquitin and histone surfaces, stabilizing a pre-existing ubiquitin orientation in NCP-ubme to form a high-affinity complex. This conformational selection mechanism contrasts with the low-affinity binding mode of 53BP1 and ensures 53BP1 displacement by RNF169 from NCP-ubme. We also show that RAD18 binds tightly to NCP-ubme through a ubiquitin-binding domain that contacts ubiquitin and nucleosome surfaces accessed by 53BP1. Our work uncovers diverse ubiquitin recognition mechanisms in the nucleosome, explaining how RNF168, RNF169 and RAD18 regulate 53BP1 chromatin recruitment and how specificity can be achieved in the recognition of a ubiquitin-modified substrate.
Introduction
In mammals, the DNA double-strand break (DSB) repair protein 53BP1 promotes non-homologous end-joining (NHEJ) DNA repair and immunoglobulin class switch recombination by facilitating long-range joining of broken DNA ends (Dimitrova et al., 2008; Manis et al., 2004; Ward et al., 2004). 53BP1 also antagonizes homologous recombination (HR) DSB repair by inhibiting DNA end resection, the generation of single-stranded DNA ends required for HR (Bunting et al., 2010). As such, 53BP1 plays a central role in maintaining the balance between the two main DSB repair pathways NHEJ and HR (Bouwman et al., 2010; Bunting et al., 2010). Understanding how 53BP1 association with chromatin flanking DNA breaks is regulated is of particular importance because imbalance in the NHEJ and HR pathways promotes genetic instability and cancer (Bouwman et al., 2010; Bunting et al., 2010).
53BP1 recruitment to DSBs is triggered by RNF168-mediated ubiquitylation of histone H2A lysine 15 in the nucleosome core particle (NCP) constitutively dimethylated at histone H4 lysine 20 (NCPH2AK15ubH4K20me2), modifications recognized by 53BP1 (Botuyan et al., 2006; Fradet-Turcotte et al., 2013; Gatti et al., 2012; Mattiroli et al., 2012; Wilson et al., 2016). 53BP1 recruitment is terminated by DNA repair factors RNF169 and RAD18 but the underlying mechanisms are not known (Chen et al., 2012; Helchowski et al., 2013; Panier et al., 2012; Poulsen et al., 2012). Here, we show that RNF169 specifically binds NCPH2AK15ubH4K20me2 with an affinity two orders of magnitude higher than that of 53BP1 for the same substrate. We found that RNF169 stabilizes ubiquitin–histone interactions in NCPH2AK15ub and also in H2AK15-ubiquitylated H2A-H2B heterodimer (H2AK15ub-H2B). We determined the structures of RNF169 bound to H2AK15ub-H2B and NCPH2AK15ub, giving an unprecedented view of how a ubiquitin reader protein recognizes its ubiquitylated substrate. Our study reveals that, unlike 53BP1, RNF169 selects a pre-existing orientation of ubiquitin in NCPH2AK15ub and H2AK15ub-H2B. This unique mechanism explains the large affinity difference between 53BP1 and RNF169 and the efficient displacement of 53BP1 from NCPH2AK15ubH4K20me2 by RNF169 leading to inhibition of 53BP1 relocation to DSBs in cells. We also show that RAD18 binds NCPH2AK15ub with high affinity and specificity. This finding unveils a recognition mechanism where a compact domain in RAD18 contacts both ubiquitin and conjugated substrate, and explains how RAD18 accumulates at DSBs (Huang et al., 2009) and interferes with 53BP1 recruitment (Helchowski et al., 2013). Finally, we show that RNF168 reads its lysines 13 and 15 ubiquitylation products, H2AK13ub and H2AK15ub, but unlike RNF169 and RAD18 cannot displace 53BP1 from the modified NCP, shedding light on the ubiquitin signal amplification mechanism that promotes 53BP1 chromatin recruitment.
Results and Discussion
RNF168 and RNF169 Specifically Bind the Nucleosome and Histone H2A-H2B Ubiquitylated at H2A Lysines 13 and 15
Because RNF169 inhibits 53BP1 recruitment to chromatin ubiquitylated by RNF168 (Chen et al., 2012; Panier et al., 2012; Poulsen et al., 2012) and because the ubiquitin ligase activity of RNF168 promotes its own accumulation to damaged chromatin (Doil et al., 2009; Panier et al., 2012; Pinato et al., 2009; Stewart et al., 2009), we examined whether RNF168 and RNF169 would recognize the NCP ubiquitylated at H2AK13 and H2AK15 (NCPH2AK13K15ub) and, as a control, at H2AK119 (NCPH2AK119ub) in reactions catalyzed by E2/E3 ligases UbcH5c/RNF168 and UbcH5c/BMI1–RING1B, respectively. The reactions give mainly mono-ubiquitylated NCPs, with UbcH5c/RNF168 also producing weak poly-ubiquitylation as observed previously (Mattiroli et al., 2012). We knew from Durocher's group that the MIU2s (motifs interacting with ubiquitin) of RNF168 and RNF169 extended at the C-termini by a segment termed LR motif (LRM) controlled the chromatin accumulation of these proteins in response to ionizing radiation (Panier et al., 2012). By testing various RNF168 and RNF169 constructs, we determined that N- and C-terminally extended MIU2 of RNF168 (aa 430-481) and RNF169 (aa 653-708), hereafter referred to as RNF168 and RNF169, were optimal for specific interaction with NCPH2AK13K15ub (Figures 1A and 1B). RNF168 and RNF169 did not bind NCPH2AK119ub (Figure 1B).
Figure 1. Specific interaction of RNF169, RNF168 and RAD18 with the nucleosome core particle ubiquitylated at histone H2A lysines 13 and 15.
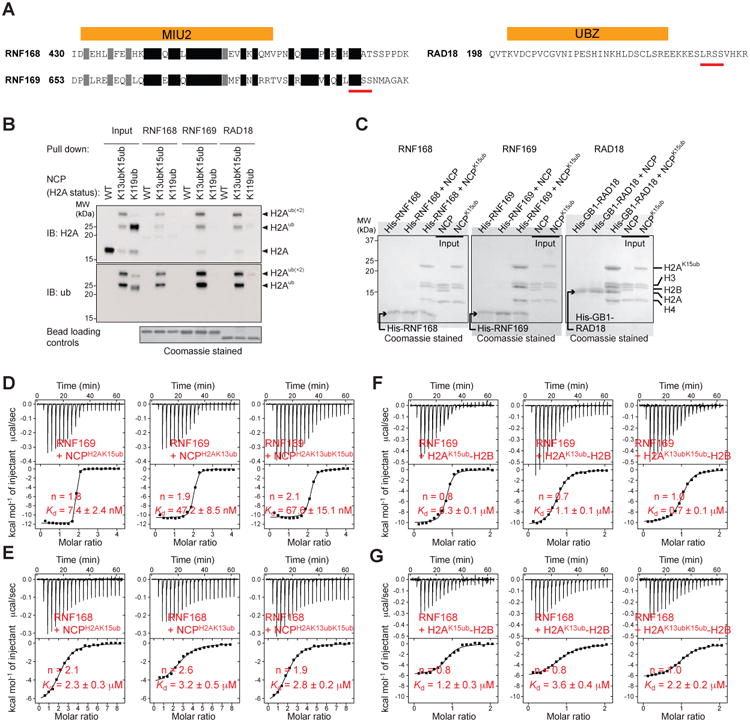
(A) Amino acid sequences of RNF168, RNF169 and RAD18 ubiquitylated nucleosome binding motifs. A stretch of 4 residues conserved in RNF169 and RAD18 in underlined.
(B) Ni2+-NTA pull-down of unmodified (WT), RNF168-ubiquitylated (H2AK13/K15) and RING1B-BMI1-ubiquitylated (H2AK119) NCPs by His6-tagged RNF168, RNF169 and RAD18, immunoblotted (IB) for histone H2A and ubiquitin (ub).
(C) Ni2+-NTA pull-down of NCP and NCPH2AK15ub (reconstituted with ubiquitin-fused H2A) by His6-tagged RNF168, RNF169 and GB1-RAD18, analyzed by SDS-PAGE.
(D) ITC results (top, raw titration data; bottom, integrated heat measurements) for RNF169 interactions with the NCP enzymatically ubiquitylated at Lys13, Lys15 or both. n is the stoichiometry of binding. Kds are reported with s.d. determined by nonlinear least-squares analysis.
(E) ITC results for RNF168 interactions with the NCP enzymatically ubiquitylated at Lys13, Lys15 or both.
(F) ITC results for RNF169 interactions with H2A-H2B enzymatically ubiquitylated at Lys13, Lys15 or both.
(G) ITC results for RNF168 interactions with H2A-H2B enzymatically ubiquitylated at Lys13, Lys15 or both.
After we developed an efficient enzymatic procedure for reconstituting ubiquitylated recombinant human H2A-H2B dimer and NCP on a scale that permits structural and quantitative binding studies (Figures S1A, S1B, S1C and S1D, and Methods), we used isothermal titration calorimetry (ITC) to further probe the interactions. With the aforementioned procedure wherein H2A is fused to the C-terminus of H2B, ubiquitin is enzymatically conjugated to H2A Lys13 or Lys15 or both via a native isopeptide bond. RNF169 bound significantly tighter to NCPH2AK15ub or NCPH2AK13ub than RNF168. The corresponding RNF169/RNF168 Kds were 7.4 nM and 2.3 mM with NCPH2AK15ub and 47.2 nM and 3.2 mM with NCPH2AK13ub (Figures 1D and 1E). The stoichiometry values are close to 2, even for NCPH2AK13K15ub, consistent with two RNF169 or RNF168 binding sites per nucleosome. RNF169 has low affinity for ubiquitin alone with a Kd of only 50.2 μM (Figure S1E).
Considering the spatial organization of histones in the NCP and the small size of RNF168 and RNF169 MIU2-LRM fragments, we surmised that these proteins would only contact histone H2A or H2B of the NCP, and not H3 or H4. Indeed, RNF168 and RNF169 also interacted with histone H2A-H2B enzymatically ubiquitylated at H2A Lys13, Lys15 or both (H2AK13/K15ub-H2B). Akin to its interaction with ubiquitylated NCP, RNF169 favored K15- over K13-ubiquitylated H2A-H2B with corresponding Kds of 0.3 μM and 1.1 μM (Figure 1F). Affinity for doubly ubiquitylated H2A-H2B was intermediate with a Kd of 0.7 μM. (Figure 1F). For RNF168, the Kds were 1.2, 2.3 and 3.6 μM for K15-, K13K15- and K13-ubiquitylated H2A-H2B, respectively (Figure 1G). The stoichiometry values are close to unity in agreement with one RNF169 or RNF168 binding site per ubiquitylated H2A-H2B dimer. Note that RNF168 has about the same affinity for ubiquitylated NCP and H2A-H2B (Figures 1E and 1G) whereas RNF169 binds tighter to ubiquitylated NCP than to ubiquitylated H2A-H2B. This is discussed below.
Solution NMR Structures of RNF169 in Complex with H2AK15ub-H2B and NCPH2AK15ub
To probe further the binding modes of RNF169 with NCPH2AK15ub and H2AK15ub-H2B, we resorted to 1H-13C methyl-transverse optimized nuclear magnetic resonance (NMR) spectroscopy (methyl-TROSY) (Kato et al., 2011; Tugarinov et al., 2006). We reconstituted NCP, NCPH2AK15ub and H2AK15ub-H2B with H2A-H2B or ubiquitin 1H,13C methyl-labeled at isoleucine, leucine and valine residues in an otherwise perdeuterated background (Figures S1B, S1C and S2). A protonated 147 base-pair DNA with the Widom 601 nucleosome-positioning sequence was used for the NCPs (Makde et al., 2010).
The NCP with 1H,13C methyl-labeled H2A and H2B produced methyl-TROSY signals that were uniformly sharp (Figure 2A). In contrast, the signals were markedly broadened in NCPH2AK15ub, likely reflecting transient interaction of ubiquitin with the surface of H2A-H2B (Figure 2A). In NCPH2AK15ub with 1H,13C methyl-labeled ubiquitin, signals were likewise broad, consistent with decreased ubiquitin flexibility (Figure 2B). Upon addition of RNF169 to NCPH2AK15ub, several H2A-H2B and ubiquitin signals became extremely broad, presumably due to an increase in ubiquitin population contacting H2A-H2B and probably also DNA (see below). Several other ubiquitin signals were shifted as similarly detected in the titration of H2AK15ub-H2B with RNF169 (Figure S2A). While binding of RNF169 to unmodified NCP is weak, this interaction caused several changes in H2A and H2B signals, perturbations also seen in the titration of H2AK15ub-H2B with RNF169 (Figure S2B).
Figure 2. Methyl-TROSY spectra of H2AK15ub-H2B and NCPH2AK15ub.
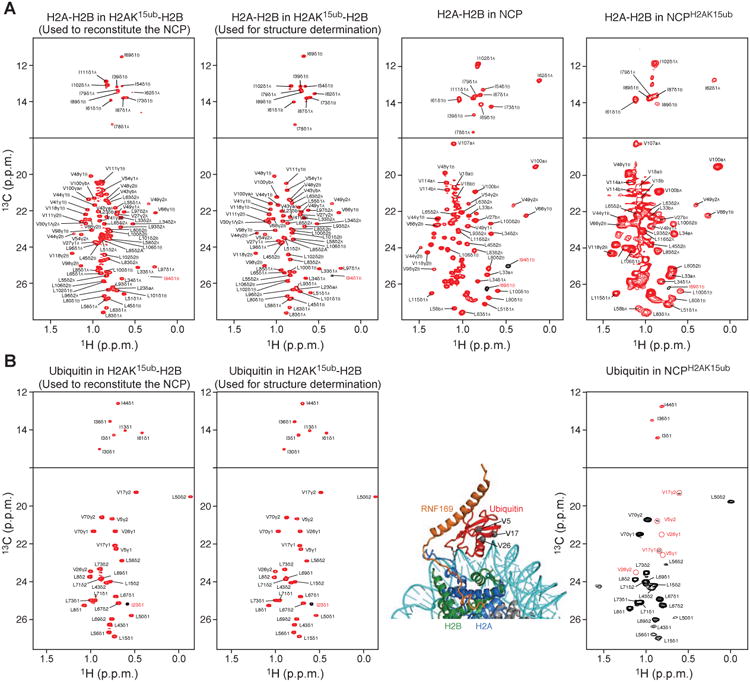
(A) Methyl-TROSY spectra of Ile, Val, Leu methyl-labeled H2A-H2B in the context of H2AK15ub-H2B, NCP and NCPH2AK15ub. From left to right: First panel corresponds to the enzymatically ubiquitylated long version of H2A-H2B used to reconstitute the NCP. Second panel corresponds to the enzymatically ubiquitylated short version of H2A-H2B used for NMR structure determination of H2AK15ub-H2B–RNF169. Third panel corresponds to the NCP reconstituted with the non-ubiquitylated long version of H2A-H2B. Fourth panel corresponds to ubiquitylated NCP reconstituted with the long version of H2AK15ub-H2B in the first panel.
(B) Methyl-TROSY spectra of Ile, Val, Leu methyl-labeled ubiquitin in the context of H2AK15ub-H2B and NCPH2AK15ub described in A. In the right spectrum, the ubiquitin methyl signals that extensively broadened in the context of NCPH2AK15ub but not in H2AK15ub-H2B (left spectra) are highlighted with red circles. The corresponding residues (Val5, Val17 and Val26) are shown as gray spheres in the NCPH2AK15ub–RNF169 complex structure.
Because of comparable chemical shift perturbations in NCPH2AK15ub and H2AK15ub-H2B and high quality NMR spectra of the latter (Figures 3A and S2C), we proceeded to determine the structure of RNF169 bound to H2AK15ub-H2B to uncover the molecular basis for complex formation (Figure 3). We prepared multiple samples of the complex where RNF169, H2A-H2B and ubiquitin were isotopically labeled, one at a time (Figures 3A, 2 and S2C, and Table S1). The structure ensemble was calculated from distance restraints derived from inter-proton nuclear Overhauser enhancement (NOE) signals, dihedral angles and paramagnetic relaxation enhancement (PRE) data (Figures 3B and S3). The structural statistics are given in Table 1. The NMR ensemble is supported by observed chemical shift changes associated with complex formation (Figures 4A and 4C).
Figure 3. NMR characterization of RNF169 in complex with H2A-H2B and the nucleosome ubiquitylated at H2AK15.
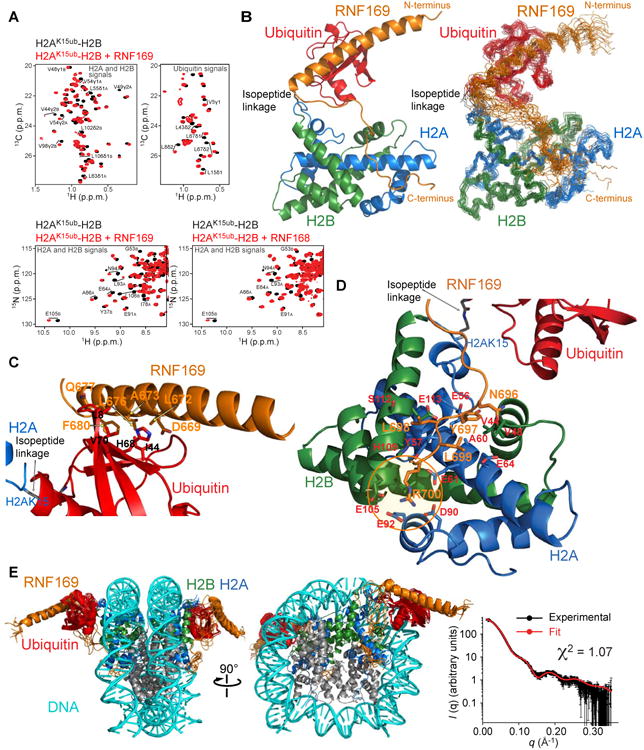
(A) Top: Regions of methyl-TROSY spectra of H2AK15ub-H2B selectively 1H-13C-labeled at methyl groups of Ile, Leu and Val residues of H2A-H2B or ubiquitin in an otherwise perdeuterated background, free (black) and bound to unlabeled RNF169 (red). Bottom: Regions of 1H-15N TROSY HSQC spectra of H2AK15ub-H2B prepared with 15N-labeled H2A-H2B and unlabeled ubiquitin, free (black) and bound to unlabeled RNF169 or RNF168 (red). Suffix A is for H2A and B for H2B.
(B) Cartoon representation and NMR structure ensemble of H2AK15ub-H2B in complex with RNF169.
(C) Cartoon representation of a region of the H2AK15ub-H2B–RNF169 complex highlighting the interaction of RNF169 α-helix and ubiquitin. Key residues are in stick representation.
(D) Cartoon representation of a region of the H2AK15ub-H2B–RNF169 complex highlighting the interaction of RNF169 LRM with H2A-H2B. Key residues are in stick representation. H2A-H2B acidic patch area binding RNF169 Arg700 is circled in orange.
(E) NMR/SAXS-based model of NCPH2AK15ub in complex with RNF169. Goodness of fit of the model to SAXS data recorded for the NCPH2AK15ub–RNF169 complex.
Table 1. NMR and refinement statistics for H2AK15ub-H2B–RNF169.
| H2AK15ub-H2B–RNF169 (653-708) | |
|---|---|
| NMR distance, dihedral and PRE restraints | |
| Distance restraints | |
| Total nuclear Overhauser effect | 4378 |
| Intra-residue | 861 |
| Inter-residue | 3074 |
| Sequential (|i – j| = 1) | 1144 |
| Medium-range (|i – j| ≤ 4) | 1164 |
| Long-range (|i – j| ≥5) | 766 |
| Intermolecular | 443 |
| H2A-H2B | 320 |
| Ub-RNF169 | 83 |
| H2A-RNF169 | 6 |
| H2B-RNF169 | 34 |
| Hydrogen bonds | 292 |
| PRE | 573 |
| Total dihedral angle restraints | 412 |
| ϕ | 206 |
| ψ | 206 |
| Structure statistics | |
| Violations | |
| Distance restraints (Å) | 0.043 ± 0.070 |
| Dihedral angle restraints (°) | 0.536 ± 0.804 |
| Max. dihedral angle violation (°) | 3.625 ± 0.358 |
| Max. distance violation (Å) | 0.380 ± 0.053 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.005 ± 0.000 |
| Bond angles (°) | 0.537 ± 0.004 |
| Impropers (°) | 0.340 ± 0.005 |
| PRE Q | 0.356 ± 0.015 |
| a Average r.m.s. deviation to mean structure (Å) | |
| Backbone | 1.02 ± 0.24 |
| Heavy | 1.39 ± 0.21 |
| a Average r.m.s. pairwise deviation (Å) | |
| Backbone | 1.42± 0.40 |
| Heavy | 1.95± 0.37 |
| a Ramachandran plot (%) | |
| Most favored regions | 97.4 |
| Additionally allowed regions | 2.6 |
| Generously allowed regions | 0.0 |
| Disallowed regions | 0.0 |
The r.m.s. deviations and Ramachandran plot parameters were calculated for residues H2A (15-97), H2B (39-123), ubiquitin (1-71) and RNF169 (654-685 and 697-700) using an ensemble of 20 structures.
Figure 4. NMR of RNF169 and RNF168 interactions with H2AK15ub-H2B.
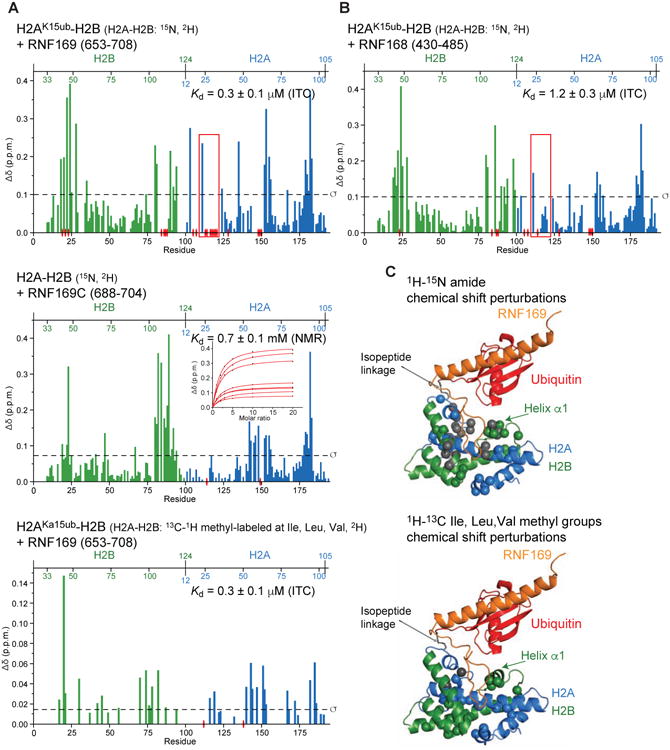
(A) Top: Magnitude of 1H-15N chemical shift changes in H2A (blue) and H2B (green) in the context of H2AK15ub-H2B after adding 3-fold molar excess RNF169 (aa 653-708). The Kd determined using ITC is indicated. Isotope labeling schemes are specified in this and subsequent diagrams. Residues for which signals disappear due to exchange broadening are indicated with red elliptical disks on the x axis. The red rectangle highlights the main difference with the H2AK15ub-H2B–RNF168 interaction (see Figure 4B). Middle: Magnitude of 1H-15N chemical shift changes in H2A and H2B in the context of H2A-H2B after adding 10-fold molar excess RNF169 (aa 688-704). The Kd calculated from NMR chemical shift changes of selected H2A-H2B residues caused by interaction with RNF169C is indicated. Bottom: Magnitude of 1H-13C chemical shift changes in Ile, Val and Leu methyl groups of H2A and H2B in the context of H2AK15ub-H2B after adding 3-fold molar excess RNF169.
(B) Magnitude of 1H-15N chemical shift changes in H2A (blue) and H2B (green) in the context of H2AK15ub-H2B after addition of 3-fold molar excess RNF168. The ITC-derived Kd is indicated. The red rectangle highlights the main difference with the H2AK15ub-H2B–RNF169 interaction (see Figure 4A).
(C) Top: Cartoon representation of the H2AK15ub-H2B–RNF169 structure highlighting H2A and H2B residues (spheres) for which there are marked changes in 1H-15N chemical shifts (larger than the s.d. of the shift for all residues). Spheres are colored gray when interaction with RNF169 causes signal disappearance. Bottom: Like in Top but marked changes are in 1H-13C chemical shifts of Ile, Leu and Val methyl groups.
The N-terminal segment of RNF169 forms a long 31-residue α-helix extending from Pro654 to Arg684 that contacts ubiquitin via Gln665, Glu666, Asp669, Arg670, Leu672, Ala673, Leu676, Gln677 and Phe680 (Figure 3C). This α-helix may be stabilized by a salt bridge between Asp681 and Arg684 close to its C-terminus. These two residues as well as the nine residues interacting with ubiquitin are conserved in RNF169 across species (data not shown). As in the prototypical MIU domain of guanine nucleotide exchange factor Rabex-5 (Lee et al., 2006; Penengo et al., 2006), an alanine (Ala673 in RNF169) interacts with the ubiquitin surface comprised of Leu8, Ile44, His68 and Val70 (Figure 3C).
Explaining the specific recognition of H2AK15ub or H2AK13ub in ubiquitylated H2A-H2B and NCP, the C-terminal segment of RNF169 encompassing residues Tyr697 to Ser701 in the LRM contacts the surface of H2A-H2B through both coulombic and hydrophobic interactions (Figure 3D). Noticeably, the guanidinium group of RNF169 Arg700 is in the vicinity of the acidic side chains of Glu61, Asp90 and Glu92 in H2A and Glu105 and Leu106 in H2B. These and additional acidic residues are often referred to as the acidic patch of H2A-H2B, an interaction node for nucleosome binding proteins (Armache et al., 2011; Barbera et al., 2006; Kato et al., 2013; Makde et al., 2010; McGinty et al., 2014; Morgan et al., 2016). Another key interaction supported by 18 intermolecular NOEs involves RNF169 Tyr697, which contacts a hydrophobic surface formed by Val44 and Val48 near Lys43 and Gln47 in the N-terminal α-helix of H2B and H2A Glu56. Intermolecular NOEs, albeit weaker, also implicate RNF169 Leu698 and Leu699 in intermolecular contacts. Leu698 interacts with His109, Ser112 and Glu113 of H2B, and H2A Tyr57. Leu699 interacts with H2B Val48 and H2A Glu64. Sample intermolecular NOEs defining the interaction of RNF169 with H2AK15ub-H2B are shown in Figure S3A.
The intervening region of RNF169 from Ser688 to Asp695 is poorly defined in the NMR ensemble due to lack of NOEs (Figure 3B). Nevertheless, extreme signal broadening for several H2A-H2B residues closest to RNF169 intervening region makes a case for transient intermolecular contacts (Figures 4A and 4C). Consistent with this possibility, mutation of selected residues in H2B (Lys120) and H2A (Arg20, Gly22 and Leu23) close to RNF169 intervening region significantly reduced the affinity of RNF169 for H2AK15ub-H2B (see below).
Similarity in chemical shift perturbations argues for comparable binding modes for RNF168 and RNF169 (Figure 3A), in line with sequence conservation (Figure 1A). Eight of the nine ubiquitin-interacting residues and three key H2A-H2B anchor residues in RNF169 (Tyr697, Leu699 and Arg700) are conserved in RNF168. In the intervening region, however, the radical substitution of two valines in RNF169 by prolines in RNF168 suggests that the ∼5-fold tighter binding of RNF169 to H2AK15ub-H2B originates from transient contacts between this region and H2A-H2B, contacts that are likely less populated in H2AK15ub-H2B–RNF168. Supporting this interpretation, RNF168 binding to H2AK15ub-H2B does not cause significant resonance broadening for H2A-H2B residues closest to the intervening region of RNF169 (Figures 4A and 4B).
To further evaluate the H2AK15ub-H2B–RNF169 structure, we also performed NMR structure calculations in the restrained environment of the human NCP crystal structure (Tachiwana et al., 2010) and with inclusion of small-angle X-ray scattering (SAXS) data obtained for the NCPH2AK15ub–RNF169 complex (Figures 3E and S4). The model derived from these calculations is compatible with the H2AK15ub-H2B–RNF169 structure (Figure 3B). Importantly, we note that RNF169 Arg684 and Arg685 may contact the DNA in NCPH2AK15ub, which likely explains the higher affinity of RNF169 for NCPH2AK15ub than for H2AK15ub-H2B, and the absence of affinity enhancement for RNF168, which has a glutamine and a methionine in place of RNF169 Arg684 and Arg685 (Figure 1A).
Mutations in H2A, H2B, Ubiquitin and RNF169 Validate the H2AK15ub-H2B–RNF169 and NCPH2AK15ub–RNF169 Structures
We introduced mutations in H2A, H2B and ubiquitin to assess the H2AK15ub-H2B–RNF169 structure. Testing mutations in the acidic area of H2A-H2B presented a challenge because this region is also critical for RNF168-mediated ubiquitylation of histone H2A (Leung et al., 2014; Mattiroli et al., 2014), the approach we used to generate the NMR samples. We therefore designed an alternative system wherein ubiquitin was genetically tethered to the N-terminus of H2A using appropriate linker lengths to mimic ubiquitylation of H2A Lys13 or Lys15. We validated the system by quantifying the RNF169 interaction in the context of H2A-H2B and NCP reconstituted with ubiquitin-fused H2A (Figures 1C, S1F and S1G). Affinities were only slightly reduced for the alternative system. The Kds for the H2AK15 and H2AK13 H2A-H2B ubiquitylation mimics are 0.4 μM and 1.7 μM compared to 0.3 μM and 1.1 μM for native ubiquitylation, respectively (Figures 1F and S1F). Agreeing well with the H2AK15ub-H2B–RNF169 contact interfaces described above, 10 of 11 mutations tested (6 in H2A, 4 in H2B and 1 in ubiquitin) led to significantly decreased affinity of H2AK15ub-H2B for RNF169 (Figure S5A). These mutations are in the vicinity of RNF169 binding sites and do not affect folding of H2AK15ub-H2B. Kd increase ranged from 3.2-fold for the H2B Q47A mutation to over 40-fold for the H2A E61A mutation in the acidic region.
Mutations were also introduced in RNF169. RNF169 A673G and R700A mutations prevented interaction with H2AK15ub-H2B while Y697A and L699A mutations caused a marked decrease in affinity with 8.2- and 11.2-fold increases in Kd as shown using ITC (Figure S5B). The R689A mutation in the intervening region increased the Kd by 3.5 fold (Figure S5B). These results rationalize the effect of point mutations previously designed based on conserved sequence elements in RNF168 and RNF169 LRMs (Panier et al., 2012). The R689A, Y697A and L699A/R700A mutants impaired the recruitment of RNF169 to DNA damage sites as shown by the abrogation of irradiation-induced foci (IRIF) formation in U2OS cells. Corresponding to RNF169 L699A/R700A is the RNF168 L476A/R477A mutation, which impaired RNF168 recruitment to DSBs (Panier et al., 2012).
RNF169 Stabilizes a Pre-existing Conformation of Ubiquitin in H2AK15ub-H2B and NCPH2AK15ub
As one could have anticipated, the conformational flexibility of ubiquitin in H2AK15ub-H2B decreases upon interaction with RNF169. This is readily apparent from the qualitative analysis of NMR relaxation data showing increased values of the spectral density function at zero angular frequency (Figure S6). Nonetheless, ubiquitin assumes a preferred orientation relative to H2A-H2B, even in the absence of RNF169. We came to this conclusion based on the marked 1H and 15N chemical shift differences in the spectra of H2AK15ub-H2B versus isolated ubiquitin and H2A-H2B (Figure 5A). For comparison, this observation contrasts with the minimal changes in the NMR spectra of ubiquitin upon covalent attachment to Lys164 of proliferating cell nuclear antigen (PCNA), the archetype system for ubiquitin or small-ubiquitin like modifier flexibly tethered to a target protein (Armstrong et al., 2012; Hibbert and Sixma, 2012) (Figure 5A). The only marked changes in the spectrum of ubiquitin caused by ubiquitylation of PCNA are restricted to the C-terminus of ubiquitin near the isopeptide bond.
Figure 5. Ubiquitin adopts a preferred orientation relative to H2A-H2B.
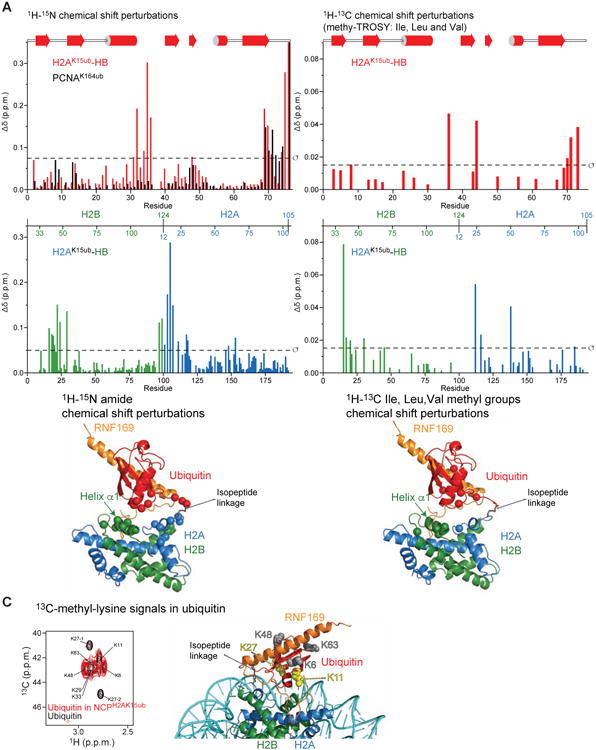
(A) Top: Magnitude of 1H-15N chemical shift changes in ubiquitin caused by ubiquitylation of H2A Lys15 in H2A-H2B (red) versus Lys164 in PCNA (black). Middle: Magnitude of 1H-15N chemical shift changes in H2A (blue) and H2B (green) caused by ubiquitylation of H2A Lys 15 in H2A-H2B. Bottom: Cartoon representation of the H2AK15ub-H2B–RNF169 structure highlighting residues (spheres) for which there are marked changes in 1H-15N chemical shifts (larger than the s.d. of the shift for all residues) in H2AK15ub-H2B caused by ubiquitylation of H2A Lys15. Note the shifts in helix α1 of H2B indicating a preferred orientation of ubiquitin in the absence of RNF169.
(B) Top: Magnitude of 1H-13C chemical shift changes in Ile, Val and Leu methyl groups of ubiquitin caused by ubiquitylation of H2A Lys 15 in H2A-H2B. Middle: Magnitude of 1H-13C chemical shift changes in Ile, Val and Leu methyl groups of H2A (blue) and H2B (green) caused by ubiquitylation of H2A Lys 15 in H2A-H2B. Bottom: Cartoon representation of the H2AK15ub-H2B–RNF169 structure highlighting residues (spheres) for which there are marked changes in 1H-13C chemical shifts (larger than the s.d. of the shift for all residues) in H2AK15ub-H2B caused by ubiquitylation of H2A Lys15. Note the shifts in helix a1 of H2B indicating a preferred orientation of ubiquitin in the absence of RNF169.
(C) Left: Comparison of 13C-methyl-lysine signals of ubiquitin in the free state (black) and in NCPH2AK15ub (red). Right: Cartoon representation of the NMR/SAXS-based model of NCPH2AK15ub–RNF169 in which ubiquitin Lys11 and Lys27 are highlighted in yellow and Lys6, Ly48 and Lys63 highlighted in gray. Note that there are two signals for Lys27.
The preferred orientation of ubiquitin relative to H2A-H2B is further corroborated by the observed differences in the 1H and 13C chemical shifts in the methyl-TROSY spectra of ubiquitin, H2A-H2B and H2AK15ub-H2B (Figure 5B). Remarkably, these chemical shift perturbations support an orientation of ubiquitin approaching that in the H2AK15ub-H2B–RNF169 complex where a negatively charged surface of ubiquitin (including Asp32 and Glu34) faces a positively charged area centered on the N-terminal helix (α1) of histone H2B (including Lys43 and Lys46) (Figures 5A, 5B and 4C). The chemical shift perturbations at the H2A-H2B– ubiquitin interface in unliganded H2AK15ub-H2B are enhanced in the presence of RNF169 suggesting a population shift toward the conformation where ubiquitin contacts the H2A-H2B surface near H2B helix α1 (Figures 4A and 4C). Thus, although the H2AK15ub-H2B–RNF169 complex retains flexibility (Figure S6), RNF169 binding is likely facilitated by selection of a pre-existing conformational substate of unliganted H2AK15ub-H2B. The low entropic cost of complex formation enabled by conformational selection (Boehr et al., 2009) is consistent with the high affinity of RNF169 for NCPH2AK15ub or H2AK15ub-H2B with respective Kds of 7.4 nM and 0.3 mM (Figures 1D and 1F).
In the methyl-TROSY spectra of ubiquitin in NCPH2AK15ub, there is preferential broadening of the methyl groups (Val17 and Val26) that are closest to the DNA in NCPH2AK15ub–RNF169, supporting a preferred orientation for ubiquitin in the nucleosomal context (Figure 2B). To further probe the orientation of ubiquitin, we prepared an NCPH2AK15ub sample in which ubiquitin was 13C-dimethylated at lysine residues (Figure 5C). In comparison to free ubiquitin, the 1H-13C methyl-lysine TROSY signals assigned to ubiquitin Lys11 and Lys27 became selectively weaker in NCPH2AK15ub (Figure 5C). Lys11 and Lys27 are close to the NCP surface in the NCPH2AK15ub–RNF169 model (Figure 5C), another indication that the preferred orientation of ubiquitin in the NCPH2AK15ub–RNF169 complex is significantly populated in the absence of RNF169, even if ubiquitin is flexible in H2AK15ub-H2B as shown from the NMR relaxation data (Figure S6). The signals of ubiquitin Lys6, Lys48 and Lys63 remain strong in NCPH2AK15ub suggesting that these residues do not contact the NCP surface, consistent with the NCPH2AK15ub– RNF169 structure (Figure 5C). No conclusion could be drawn for ubiquitin Lys29 and Lys33 because of signal overlap. We obtained similar results with NCPH2AK13ub prepared with 13C-lysine-dimethylated ubiquitin (data not shown).
RNF169 But Not RNF168 Displaces 53BP1 From NCPH2AK15ubH4K20me2
While both RNF169 and 53BP1 recognize the NCP ubiquitylated at H2A Lys15, the orientation of ubiquitin relative to H2A-H2B in the recent cryo-EM structure of chemically dimethylated NCPH2AK15ubH4Kc20me2 in complex with a dimeric fragment of 53BP1 (Wilson et al., 2016) differs radically from that in the H2AK15ub-H2B–RNF169 and NCPH2AK15ub–RNF169 complexes (Figure 6A). This difference implies a high entropic barrier for ubiquitin reorientation and reduced flexibility in the 53BP1 complex, which correlates with the low affinity of 53BP1. Binding to NCPH2AK15ubH4Kc20me2 is only detectable for a dimeric form of 53BP1 obtained by fusion to GST of a 53BP1 construct composed of the tandem Tudor domain and the ubiquitin-dependent recruitment (UDR) motif (GST-53BP1 (aa 1484-1635)) (Fradet-Turcotte et al., 2013; Wilson et al., 2016). Using ITC, we derived a Kd of 1.9 μM for the interaction of GST-53BP1 with NCPH2AK15ubH4Kc20me2 (Figure 6B). Under identical conditions, binding of GST-53BP1 to NCPH2AK15ub is too weak for the affinity to be determined, demonstrating the key role of H4K20me2 in the interaction of 53BP1 with the NCP (Figure 6A). As expected, RNF169 binds tightly to NCPH2AK15ubH4Kc20me2 with a Kd of 15.1 nM while RNF168 binds more weakly to NCPH2AK15ubH4Kc20me2 with a Kd of 2.3 mM (Figure 6B).
Figure 6. Effects of RNF168, RNF169 and RAD18 on the association of 53BP1 with the nucleosome ubiquitylated at H2AK15 and dimethylated at H4K20.
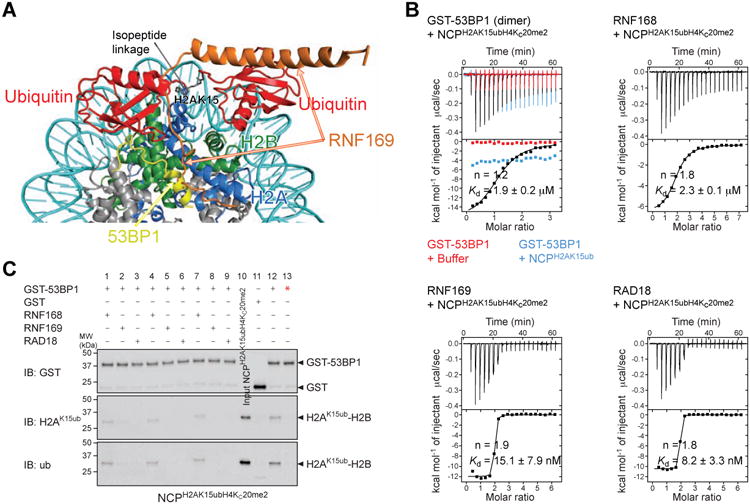
(A) Overlay of the cryo-EM structure of NCPH2AK15ubH4Kc20me2–53BP1 (aa 1611-1631) and NMR/SAXS-based model of NCPH2AK15ub–RNF169 (aa 653-708).
(B) ITC results for the interactions of GST-53BP1 (aa 1484-1635) dimer, RNF168, RNF169 and RAD18 with NCPH2AK15ubH4Kc20me2. The GST-53BP1–NCPH2AK15ub interaction was also probed. n is the stoichiometry of binding. Kds are reported with s.d. determined by nonlinear least-squares analysis.
(C) GST pull-down assays of NCPH2AK15ubH4Kc20me2 in the absence (lane 12) and presence of equimolar (lanes 1-3), 2-fold (lanes 4-6) and 4-fold (lanes 7-9) molar excess of RNF168, RNF169 and RAD18 with GST-53BP1 (aa 1484-1635), immunoblotted (IB) for GST, K15-ubiquitylated H2A (H2AK15ub) and ubiquitin (ub). Pull-downs of NCPH2AK15ubH4Kc20me2 with GST (lane 11) and GST-53BP1 (aa 1484-1635) T1609E/S1618E mutant (lane 13, red star) were done as negative controls. 53BP1 T1609E/S1618E is unable to bind NCPH2AK15ubH4Kc20me2 as reported (Lee et al., 2014; Orthwein et al., 2014). Input NCPH2AK15ubH4Kc20me2 is in lane 10.
That RNF169 contacts almost the same NCP surface as 53BP1 but with approximately two orders magnitude smaller Kd offers a plausible explanation for the efficient inhibition of 53BP1 recruitment to DSBs by RNF169. To test this possibility, we probed the effectiveness of RNF169 in displacing 53BP1 from NCPH2AK15ubH4Kc20me2 (Figure 6C). Even with just an equimolar amount of RNF169 with respect to the two ubiquitin sites in NCPH2AK15ubH4Kc20me2, RNF169 completely prevented the interaction of GST-53BP1 with NCP H2AK15ubH4Kc20me2, suggesting that RNF169 inhibits 53BP1 recruitment by blocking access to H2AK15-ubiquitylated NCP (Figure 6C). These findings clarify the antagonistic cellular functions of 53BP1 and RNF169, with RNF169 stimulating HR DNA repair and inhibiting NHEJ (Poulsen et al., 2012). While our work reveals the mechanism underlying the functional competition between 53BP1 and RNF169 for recruitment to RNF168-ubiquitylated chromatin, we note that the cellular function of RNF169 remains unclear. Our studies imply that the recruitment of RNF169 to DNA damage sites is mediated by its interaction with the NCP ubiquitylated at H2A Lys15 or Lys13. Since RNF169 bears a RING domain and has auto-ubiquitylation activity, this protein likely contributes to ubiquitylation of an unknown target near DNA damage sites. Displacement of 53BP1 from chromatin might therefore not be the primary function of RNF169.
In contrast to RNF169, RNF168 did not displace 53BP1 from NCPH2AK15ubH4Kc20me2 (Figure 6C), a result readily explained by the lower affinity of RNF168 compared to RNF169 for NCPH2AK15ub (Figures 1D and 1E). This observation makes sense from a physiological perspective since 53BP1 requires the E3 ubiquitin ligase activity of RNF168 to accumulate at damaged chromatin (Doil et al., 2009; Panier et al., 2012; Pinato et al., 2009; Stewart et al., 2009). That RNF168 directly reads its ubiquitylation products H2AK13ub and H2AK15ub, as we showed, explains a signal amplification loop that promotes chromatin spreading of H2AK13ub and H2AK15ub and recruitment of 53BP1. One would therefore expect 53BP1 to be able to displace RNF168 from NCPH2AK15ubH4K20me2 and not the opposite, which agrees well with our results. 53BP1 and RNF168 have almost identical affinities for NCPH2AK15ubH4Kc20me2 (Figure 6B).
RAD18 Forms a Tight Complex With NCPH2AK15ub and Displaces 53BP1 From the Nucleosome
It was recently shown that ectopic expression of the DNA damage response E3 ubiquitin ligase RAD18 or its ubiquitin-binding zinc finger (UBZ) domain in mammalian cells inhibited the recruitment of 53BP1 and BRCA1 to DSBs (Helchowski et al., 2013). These observations and in vivo evidence that RAD18 requires chromatin ubiquitylation for localization to DNA damage sites (Huang et al., 2009; Panier et al., 2012) prompted us to probe a possible interaction between RAD18 UBZ and the NCP ubiquitylated by RNF168. Indeed, a version of UBZ extended at its C-terminus (aa 198-240), hereafter referred to as RAD18, pulled-down NCPH2AK13K15ub or NCPH2AK15ub but not NCPH2AK119ub, demonstrating a specific interaction (Figures 1B and 1C). We showed that akin to RNF169, RAD18 efficiently displaced 53BP1 from NCP H2AK15ubH4Kc20me2, explaining its inhibition of 53BP1 recruitment in cells (Helchowski et al., 2013) (Figure 6C). Although RAD18 binds free ubiquitin relatively tightly (Kd = 2.3 μM) compared to RNF169, affinity is dramatically enhanced in the context of NCPH2AK15ub or NCPH2AK15ubKc20me2 with Kd values of 25.2 nM and 8.2 nM (Figures 7A and 6B). The C-terminal extension of RAD18 UBZ is important for tight interaction with NCPH2AK15ub as its deletion increases the Kd to 0.9 μM (Figure 7A). This extension shares limited similarity with RNF169 LRM sequence (Figure 1A). Four residues are conserved (LRSS), with RAD18 Arg234 in this motif corresponding to RNF169 Arg700 that interacts with the acidic patch. Based on NMR chemical shift perturbations, we showed that RAD18 and RNF169 interact with approximately the same surface of H2AK15ub-H2B (Figures 7B, S7A and S7B). We also detected intermolecular NOEs between Arg234 and residues in the acidic region of H2A-H2B (Figure S7C). These similarities between RAD18 and RNF169 notwithstanding, distinct binding modes must specify their interactions with H2AK15ub-H2B and NCPH2AK15ub since RAD18 and RNF169 recognize ubiquitin in nearly opposite orientations (Figure S7D). In addition, RAD18 UBZ C-terminal extension is markedly shorter than that of RNF169 MIU2 implying a reorientation of ubiquitin in H2AK15ub-H2B upon interaction with RAD18. In agreement, using NMR spectroscopy we showed that not only the C-terminal extension but also RAD18 UBZ domain contacts the histone surface in H2AK15ub-H2B (Figures 7B and S7A). In an NMR-based model that we calculated for the NCPH2AK15ub–RAD18 complex, the UBZ domain is sandwiched between ubiquitin and the H2A-H2B surface (Figure 7C). Although RAD18 and RNF169 have almost opposite orientations relative to ubiquitin, Lys218 near the N-terminus of RAD18 helix points toward DNA in the NCP like the arginines at the C-terminus of RNF169 helix. These charged residues may contribute to the high affinity of these two proteins for NCPH2AK15ub.
Figure 7. Interaction of RAD18 with the nucleosome ubiquitylated at H2AK15.
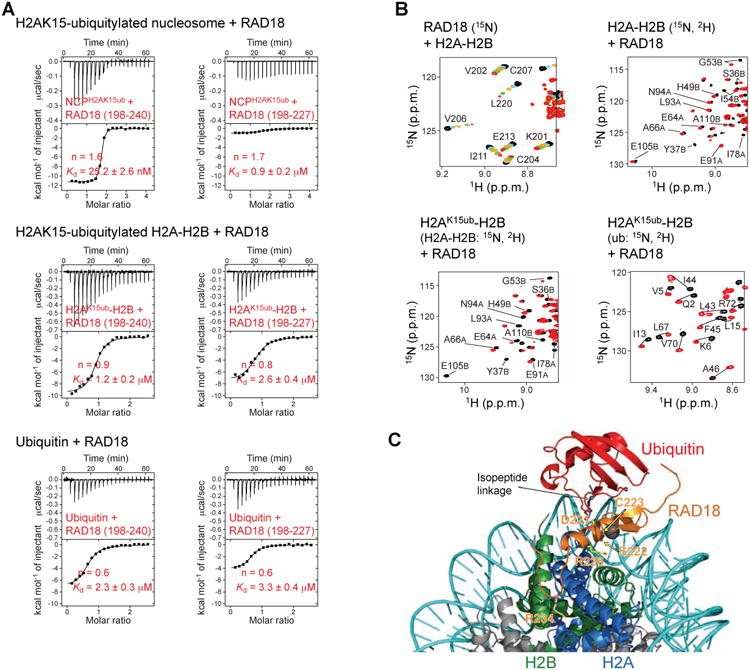
(A) ITC results for the interactions of RAD18 (aa 198-240; aa 198-227) with ubiquitin and with the NCP and H2A-H2B enzymatically ubiquitylated at H2AK15. n is the stoichiometry of binding. Kds are reported with s.d. determined by nonlinear least-squares analysis.
(B) Regions of 1H-15N TROSY HSQC titration spectra of RAD18, H2A-H2B and H2AK15ub-H2B illustrating the interaction of RAD18 with H2A-H2B and ubiquitin in H2AK15ub-H2B. Different isotope labeling schemes (15N or 15N, 2H) were used as indicated.
(C) Cartoon representation of the NMR-based model of NCPK15ub in complex with RAD18 (aa 198-240). The zinc atom is shown as a gray sphere. Side chains of RAD18 for which NMR signals were most affected (i.e. exchange broadened) by interaction with H2A-H2B are shown in yellow sticks. RAD18 R234 side chain interacting with H2A-H2B acidic patch is also shown.
Our finding that RAD18 recognizes H2AK15ub and the H2A-H2B surface near the acidic patch in the NCP, akin to 53BP1 but with higher affinity than 53BP1, explains by a simple competition mechanism the strong inhibition of 53BP1 IRIF formation in response to overexpression of RAD18 in mammalian cells (Helchowski et al., 2013). There is no evidence that RAD18 displaces 53BP1 from chromatin under physiological conditions, and it was even reported that RAD18 ubiquitylates 53BP1 and enables 53BP1 retention on chromatin (Watanabe et al., 2009). It is nonetheless noteworthy that RAD18 promotes HR (Huang et al., 2009; Saberi et al., 2007), the DNA repair pathway inhibited by 53BP1, and that the E3 ubiquitin ligase activity of RAD18 is not needed for its HR function while its UBZ domain is required (Huang et al., 2009). How RAD18 mediates HR remains unknown but it could be related to the direct association of RAD18 with the RAD51C recombinase in mammalian cells (Huang et al., 2009). However, considering the large affinity difference between RAD18 and 53BP1 for the ubiquitylated NCP, one cannot totally exclude that displacement of 53BP1 from chromatin by RAD18 could occur under physiological conditions and contribute to the HR promoting activity of RAD18.
Concluding Remarks
In summary, we showed that RNF168, RNF169 and RAD18 specifically read chromatin ubiquitylation at H2A Lys13 and Lys15, with a preference of RNF168 and RNF169 for the latter site. While RNF168 and RNF169 recognize the same posttranslational modifications in the NCP and share similar amino acid sequences in their modified histone recognition motifs, they evolved different binding affinities, explained by dissimilarities in structures and dynamics, that determine their distinct functions in cells. We found that RNF169 recognizes NCPH2AK15ubH4Kc20me2 with much higher affinity than 53BP1, which explains previous observations that RNF169 terminates 53BP1 chromatin recruitment and promotes HR, a DNA repair pathway suppressed by 53BP1. In contrast to RNF169, RNF168 binds NCPH2AK15ubH4Kc20me2 with the same or even slightly lower affinity than 53BP1, supporting a signal amplification loop promoting chromatin spreading of H2AK15 ubiquitylation catalyzed by RNF168 in response to DSBs and associated chromatin recruitment of 53BP1.
Like RNF169, RAD18 binds tightly to NCPH2AK15ub or NCPH2AK15ubH4Kc20me2 and displaces 53BP1 from NCPH2AK15ubH4Kc20me2, explaining why overexpression of RAD18 in mammalian cells impairs the formation of irradiation-induced 53BP1 foci and increases HR. While RAD18 is known to promote HR under physiological conditions, at this time there is no experimental evidence that it does so by blocking the recruitment of 53BP1 to DNA damage sites.
Different from RNF168 and RNF169, RAD18 harbors a canonical ubiquitin-binding domain that not only contacts ubiquitin but also the surface of its targeted substrate. This hitherto unobserved feature contributing to specificity for a particular ubiquitylated ligand may apply to other ubiquitin-binding domains.
Contact For Reagent and Resource Sharing
Further information and requests for reagents should be directed to Lead Contact Georges Mer (mer.georges@mayo.edu).
Experimental Model And Subject Details
Plasmid DNA for protein expression was amplified in E. coli DH5a strain in Luria Bertani (LB) medium at 37 °C overnight. Recombinant proteins were overexpressed in E. coli BL21 (DE3) strain. Cells were grown at 37 °C in LB or isotope-enriched M9 media to an OD600 of ∼0.6 and then induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) for 3 h at 37 °C or 16 h at 15 °C.
Method Details
Expression and Purification of Proteins
All protein cDNAs were inserted in pET-based bacterial expression vectors. Human RNF168 (aa 430-481), RNF169 (aa 653-708), RNF169N (aa 653-687) and RNF169C (aa 688-708) were cloned with an N-terminal His6-tag cleavable by tobacco etch virus (TEV) protease. Human RAD18 (aa 198-240) was cloned with a TEV protease-cleavable N-terminal His6-tag, a TEV protease-cleavable N-terminal His6-GB1-tag and with a non-cleavable C-terminal His6-tag and a tryptophan to facilitate protein quantification. A shorter RAD18 (aa 198-227) with a TEV protease-cleavable N-terminal His6-tag was also cloned. Human ubiquitin (aa 1-76) constructs were prepared without a tag or with an N-terminal His6-tag, non-cleavable or cleavable by PreScission protease. Human 53BP1 (aa 1484-1635) corresponding to the tandem Tudor domains and ubiquitylation-dependent recruitment motif (UDR) (Botuyan et al., 2006; Fradet-Turcotte et al., 2013) was cloned with an N-terminal GST tag. Mutations were introduced by QuikChange (Stratagene). All proteins were produced in BL21(DE3) E. coli cells grown at 37 °C in LB or isotope-enriched M9 media to an OD600 of ∼0.6 prior to induction with 0.5 mM IPTG. Except for 53BP1, which was expressed at 15 °C for ∼16 h, all other proteins above were expressed at 37 °C for 3 h. For RAD18, 100 μM ZnCl2 was added to the culture media prior to induction. Harvested cells were lyzed using an Avestin Emulsiflex C5 homogenizer. Untagged ubiquitin was purified as previously reported (Sundd et al., 2002). All His6-tagged proteins were purified by Ni2+-nitrilotriacetic acid (NTA) agarose chelation chromatography (Qiagen) using solutions of 50 mM sodium phosphate (NaPi), pH 7.5, 300 mM NaCl with 5, 20 and 250 mM imidazole to bind, wash and elute the proteins, respectively. GST-tagged 53BP1 was bound to a GSTPrep FF 16/10 column (GE Healthcare), washed with PBS, pH 7.3, and eluted with PBS, pH 8.0, 20 mM glutathione. After elution, before and after removal of tags by overnight incubation with TEV or PreScission protease at 4 °C, the proteins were purified by size exclusion chromatography using HiLoad 16/60 Superdex 75 or 200 columns (GE Healthcare) and running buffer of 50 mM NaPi, pH 7.5, 300 mM NaCl. RNF168, RNF169 and ubiquitin were further purified by reversed-phase chromatography using a Jupiter 5 μm C18 300 Å preparative column (Phenomenex). UbcH5c, Uba1, RNF168-RING and RING1B-BMI1 were purified according to published protocols (Benirschke et al., 2010; Bentley et al., 2011; Berndsen and Wolberger, 2011; Zhang et al., 2013).
Preparation of Histones and Nucleosomes
Full-length human histone proteins H2A (aa 1-129), H2B (aa 1-125), H3.1 (aa 1-135) and H4 (aa 1-102) codon-optimized for expression in E. coli were cloned in a modified pET vector encoding an N-terminal His6-tag and a PreScission protease cleavage site. Fusion constructs of H2A linked to the C-terminus of H2B were also cloned in the aforementioned vector. The first construct includes H2B residues 1-123 and H2A residues 12-129 with a serine between the two proteins. A linker serine was chosen because it is small and hydrophilic and was expected to provide maximum flexibility between H2B and H2A. The second and shorter construct includes H2B residues 33-123 and H2A residues 12-105 also tethered via a serine residue. Fusion constructs of ubiquitin and H2A with a TEV protease-cleavable N-terminal His6-tag were also engineered wherein the C-terminus of ubiquitin is genetically linked to the N-terminus of H2A. In these ubiquitin-H2A fusions there are two intervening glycine residues between ubiquitin Gly76 and H2A Ala14 or Thr16, mimicking H2AK13ub and H2AK15ub, respectively. Histone proteins were expressed in BL21(DE3) E. coli cells grown at 37 °C in LB or isotope-enriched M9 media to an OD600 of ∼0.6 and induced for 3 h with 0.5 mM IPTG, and purified as previously reported (Tachiwana et al., 2010) by Ni2+-NTA agarose chelation chromatography (Qiagen) using solutions described above but with 6-8 M de-ionized urea added. Histones were then extensively dialyzed in water with 5 mM β-mercaptoethanol, lyophilized and kept at -80 °C until needed.
To prepare the histone octamer, lyophilized H2A, H2B, H3.1 and H4, or the longest version of H2A-H2B fusion construct in lieu of individual H2A and H2B were each dissolved in 20 mM Tris-HCl, pH 7.5, 8 M de-ionized urea and mixed at equimolar amounts to a final concentration of 1 mg total histones per mL. The mixture was serially dialyzed in 20 mM Tris-HCl, pH 7.5, 2 M NaCl, 5 mM DTT containing 4, 3, 2, 1, and 0 M urea. The His6-tags of the refolded octamer were next cleaved with PreScission protease at 4 °C overnight and the octamer purified by size exclusion chromatography using HiLoad 16/60 Superdex 200 column (GE Healthcare) and 20 mM Tris-HCl, pH 7.5, 2 M NaCl, 10 mM DTT running buffer. The NCP was reconstituted from the histone octamer employing the reported salt gradient protocol (Luger et al., 1999) in the presence of a Widom 601 147 base-pair DNA (Makde et al., 2010). DNA was prepared from HB101 E. coli cells as published (Makde et al., 2010), with an additional final ion exchange chromatography step using a Resource Q column (GE Healthcare) and running buffers 10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, with 0 and 1 M NaCl. The NCP was purified by Resource Q anion-exchange chromatography (GE Healthcare), concentrated and dialyzed using appropriate storage buffers (Luger et al., 1999). To prepare the ubiquitylated NCP, the longest version of fused H2A-H2B was first enzymatically ubiquitylated (see below) before octamer incorporation and subsequent NCP reconstitution. Prior to enzymatic ubiquitylation, Ni2+-NTA-eluted fused H2A-H2B was refolded in 50 mM NaPi, pH 6.0, 300 mM NaCl and purified by size exclusion chromatography using the same buffer and HiLoad 16/60 Superdex 75 column (GE Healthcare). To prepare the ubiquitylated and methylated NCP, H4 with K20C mutation was chemically methylated using a published procedure (Cui et al., 2009; Simon et al., 2007) before octamer refolding with ubiquitylated H2A-H2B and H3, and subsequent NCP reconstitution. The same approach was used to generate NCPH2AK15ub and NCPH2AK13ub with ubiquitin 13C-methylated at lysine residues. 13C-methylated ubiquitin was prepared by reductive alkylation using 13C-enriched formaldehyde as published (Hattori et al., 2013). For the Ni+2-NTA pull-down assays, two other versions of ubiquitylated NCPs were prepared. One is prepared by K13/K15 or K119 enzymatic ubiquitylation of NCP. Another is prepared by using an octamer formed from ubiquitin-H2A fusion (H2AK13ub or H2AK15ub mimics), H2B, H3.1 and H4.
Ubiquitylation Reactions
While both K13 and K15 of H2A in wild type linked H2A-H2B can be ubiquitylated, one lysine can be selectively ubiquitylated by changing the other lysine into an unreactive serine or arginine. Ubiquitylation was carried out in 50 mM Tris-HCl, pH 7.5 containing 100 mM NaCl, 10 mM MgCl2, 1 μM ZnCl2, 1 mM TCEP, 1.6-3.2 μM Uba1 (E1), 8 μM UbcH5c (E2), 8 μM Rnf168-RING (E3), 80 μM linked H2A-H2B, 120 μM ubiquitin and 3 mM ATP. The reaction was allowed to proceed at 32 °C for 90 min and then quenched by addition of 5 mM EDTA, pH 8.0. Ubiquitylated linked H2A-H2B was next dialyzed in 50 mM NaPi, pH 6.0, 300 mM NaCl before it was concentrated and purified using the same buffer and HiLoad 16/60 Superdex 75 column (GE Healthcare). While the NCP or H2A-H2B dimer can be mono-ubiquitylated in vitro, efficiency is quite low (Mattiroli et al., 2014; Wilson et al., 2016). In contrast and to our surprise, we were able to achieve mono-ubiquitylation (at H2A K13, K15 or both) of the two linked versions of H2A-H2B with enhanced efficiency (100% completion) after optimizing reaction conditions (Figure S1A).
NMR Spectroscopy
All NMR experiments were carried out on a Bruker Avance III 700 MHz spectrometer equipped with a triple-resonance cryoprobe. The NMR spectra were processed with NMRPipe/Draw (Delaglio et al., 1995) and analyzed using Sparky 3.115 (T.D. Goddard and D.G. Kneller, University of California) and NMRView (Johnson and Blevins, 1994). For structure determination of the H2AK15ub-H2B–RNF169 complex, multiple samples (0.2-1 mM) were prepared including free H2A-H2B and H2AK15ub-H2B as well as RNF169-titrated H2AK15ub-H2B and ubiquitin in 25 mM MES–Bis-Tris, pH 6.0, 50 mM KCl, 4 mM EDTA and 10% D2O/90% H2O or 100% D2O. What we refer to as H2A-H2B and H2AK15ub-H2B have H2B (aa 33-123) fused to the N-terminus of H2A (aa 12-105) with an intervening serine residue bridging H2B and H2A (see above). Various labeling schemes were used for component proteins in the complexes including [15N], [13C,15N], [U-2H,15N], [U-2H,13C, 15N], partial deuteration (30% and 70%) and selective [1H, 13C]-labeling of methyl groups of isoleucine, leucine and valine residues in an otherwise perdeuterated background (i.e., Ile8l-[13CHD2]; Leu,Val-[13CHD2; 12CD3]) (Botuyan et al., 2004; Kato et al., 2011; Tugarinov et al., 2006). The NMR signals for H2AK15ub-H2B–RNF169 were assigned using established three-dimensional experiments (Ferentz and Wagner, 2000) recorded on several samples prepared with multiple isotope labeling strategies (Table S1). Some of the published methyl resonance assignments for H2A and H2B in Drosophila melanogaster NCP (Kato et al., 2011) and some from human H2AK15ub-H2B (this study) could be transferred to human NCP and NCPH2AK15ub Amide 15N relaxation data were acquired and analyzed using established methods (Botuyan et al., 2001; Lefevre et al., 1996; Mer et al., 1996). For chemical shift mapping experiments, variously labeled H2A-H2B, H2AK15u-H2B and NCPH2AKi5ub with concentrations from 0.04 to 0.3 mM were incrementally titrated with nonlabeled RNF168, RNF169, RAD18 or RNF169C up to 3- to 20-fold molar excess. NCPH2AK15ub in these experiments contains H2A K15-ubiquitylated H2A-H2B in which H2B (aa 1-123) is fused to the N-terminus of H2A (aa 12-129) with an intervening serine residue bridging H2B and H2A (see above). The normalized chemical shift changes Δδ for 1H-15N correlation signals were calculated using equation . For He, Leu and Val 1H-13C methyl signals, Δδ was calculated using equation , where N= 1 for Ile and N = 2 for Leu and Val (Kato et al., 2011). All NMR experiments were recorded at 25 °C or 30 °C except those involving selectively [1H,13C]-methyl labeled H2AK15ub-H2B (30 °C and 40 °C) and NCPH2AK15ub (40 °C and 45 °C).
Distance and Torsion Angle NMR Restraints
Intermolecular NOEs were initially identified using 3D 15N-filtered, 15N-edited and 13C-filtered, 13C-edited NOESY experiments (Zwahlen et al., 1997) recorded on the ubiquitin-RNF169 (aa 653-708) complex with a 120 ms mixing time and H2A-H2B-RNF169C complex with 200 and 150 ms mixing times. In each complex, one component was uniformly [15N, 13C]-labeled, with partial (30% and 70%) or without deuteration, and the other component was unlabeled, and vice versa. These NOEs were complemented and validated by 15N-edited NOESY-HSQC and 13Cedited NOESY-HSQC or NOESY-HMQC experiments (150 ms mixing time) recorded on the aforementioned complexes and additionally on H2AK15ub -H2B–RNF169 (aa 653-708), including complexes in which H2A-H2B and ubiquitin were each at a time methyl-labeled at isoleucine, leucine and valine residues while RNF169 was not labeled. Among all the complexes used, a total of 123 unambiguous intermolecular NOEs involving RNF169 were detected, of which 83 were between RNF169 and ubiquitin, 6 were between RNF169 and H2A, and 34 were between RNF169 and H2B. Additionally, 320 NOEs were identified between H2A and H2B in the linked H2A-H2B construct. NOE-based restraints were categorized into seven distance ranges with upper limits of 3.0, 3.5, 4.0, 4.5, 5.0, 5.5 and 6.0 Å. For all distance restraints, a lower limit of 1.8 Å was used. In totality, 4,378 NOE-based distance restraints were used for structure calculations (Table 1).
Backbone dihedral angle φ and ψ restraints were obtained from the analysis of 1H, HN, 13Cα, 13Cβ, 13CO and 15N chemical shifts using TALOS+ (Shen et al., 2009). Hydrogen bonds were identified based on 1H/2H exchange rates and converted into distance restraints of 1.8-2.3 Å and 2.8-3.3 Å for the HN–O and N–O bonds, respectively. Together with NOE-derived restraints, angle restraints and hydrogen bond distances were included in the structure calculations (Table 1).
Paramagnetic Relaxation Enhancement Restraints
Initial structures calculated using distance and dihedral angle restraints were used to select strategic sites in H2AK15ub-H2B for spin labeling with the paramagnetic probe MTSL (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate) (Toronto Research Chemicals Inc. and Santa Cruz Biotechnology). Thr9, Thr14, Glu34, Asp39 and Ser57 in ubiquitin, and H2B Ser56, Ser87, Ser112, Ser123 and H2A Gly37, Lys74 in H2A-H2B were selected for spin labeling and thus individually mutated to a cysteine. Wild type proteins were 15N-labeled while mutants were not. In all cases, H2A-H2B was deuterated. Prior to reaction with 10- to 15-fold molar excess of MTSL (0.5 M stock in acetonitrile) at 4 °C in the dark overnight, each purified H2AK15ub-H2B was treated with 10 mM DTT and then passed twice through a Superdex 75 10/300 GL column (GE Healthcare) using running buffers of 50 mM NaPi, pH 7.8, 300 mM NaCl, with 10 mM DTT initially and then without DTT. After reaction, excess MTSL was removed by a third passage using a running buffer of 50 mM NaPi, pH 6.0, 300 mM NaCl. Samples were buffer exchanged in the final NMR buffer: 25 mM MES–Bis-Tris, pH 6, 50 mM KCl and 10% D2O/90% H2O; and concentrated between 0.2 and 0.5 mM. 1H-15N HSQC TROSY spectra were recorded on the different H2AK15ub-H2B samples, first with the oxidized (paramagnetic) form of MTSL, and next with the reduced (diamagnetic) form of MTSL, both in the absence and presence of 3-fold molar excess of nonlabeled RNF169 (aa 653-708). Oxidized MTSL was reduced by addition of ∼ 6 mM ascorbic acid to the NMR samples (Battiste and Wagner, 2000). To ensure complete reduction of MTSL, 1H-15N HSQC TROSY spectra were collected at least 1 h after ascorbic acid was added.
The ratios of oxidized and reduced peak intensities (Iox/Ire) in the NMR spectra are related to distances as formalized in equations 1 and 2 (Battiste and Wagner, 2000; Chen et al., 2010; Iwahara et al., 2007).
| [Equation 1] |
| [Equation 2] |
In equations 1 and 2, PRE is the paramagnetic rate enhancement, t is the total evolution time of the INEPT in the 1H-15N HSQC, R2 is the intrinsic transverse relaxation rate, ωh is the Larmor frequency of the proton, τc is the correlation time of the electron-proton interaction [τc=(τr-1+ τs-1)-1], and r is the distance between the electron in MTSL and nuclear spin of interest. The value of τc can be approximated by that of the protein rotational correlation time (τr) because the electron spin relaxation time of a nitroxide spin label (τs) is much longer than τr. K is a constant equal to 1.23 × 10-32 cm6 s-2.
For structure calculations, the Iox/Ire peak ratios were converted into three sets of distance restraints as previously reported (Battiste and Wagner, 2000; Chen et al., 2010). Protons with Iox/Ire < 0.15, including those whose resonances were not detected in the paramagnetic spectra, were assigned distance restraints in the range of 1.8 to 15 A. Protons with Iox/Ire between 0.15 and 0.8 were assigned distance restraints of 15 to 23.5 A, and those with Iox/Ire > 0.8 were assigned a lower limit of 23 A and no upper limit. The PRE-based distances were restrained from the nitrogen atom of the MTSL ring and the amide protons of affected residues. A total of 573 PRE restraints were used for structure calculations (Table 1).
Structure Calculations
Structure calculations were performed using XPLOR-NIH (version 2.44) (Schwieters et al., 2006). Initial structures were generated using distance geometry and simulated annealing with NOE-derived distance restraints and torsion angle restraints from TALOS+. In the first simulated annealing, a starting temperature of 3500 °K was decreased to 100 °K in 25 increments. Two hundred structures with lowest energies were then used as initial templates for further refinement using simulated annealing with NOE-, PRE- and hydrogen bond-based distance restraints and torsion angle restraints. During the simulated annealing refinement, a starting temperature of 3000 °K was decreased to 25 °K in 12,500 increments. The 20 calculated structures with the lowest energies were retained. The MTSL-labeled cysteines were replaced by the corresponding original amino acids and the resulting structures were used for analysis and statistics (Table 1). The quality factor for PRE restraints was calculated from where Rcalc and Rexp are the back-calculated and experimental PREs derived using equations 1 and 2 above (Battiste and Wagner, 2000; Chen et al., 2010; Iwahara et al., 2007). Rexp values from the very flexible regions of the complex are not included in the calculation of Q.
SAXS Data Collection and Analysis
Small-angle X-ray scattering (SAXS) data (Rambo and Tainer, 2013) were collected on samples of NCP (0.7, 1.4 and 2 mg/mL), NCPH2AK15ub (0.8, 1.6 and 2.4 mg/mL), and NcpH2AK15ub– RNF169 (0.7, 1.5 and 2.2 mg/mL) in 20 mM NaPi, pH 7.0, 50 mM NaCl, 5% glycerol. The SAXS data were collected at the SIBYLS Beamline 12.3.1, Advanced Light Source, Lawrence Berkeley National Laboratory. For each sample, the scattering intensities were measured at three different protein concentrations. Exposures of 0.5, 1.0 and 5.0 s were used for each sample and data were monitored for radiation-dependent aggregation. Scattering data were plotted as a function of q = 4π[sin(θ/2)]/λ, where θ is the scattering angle and λ is the X-ray wavelength. The buffer scattering curve was subtracted from the sample scattering curves and the difference curves scaled for solute concentration and extrapolated to infinite dilution. Data were analyzed using programs from the ATSAS 2.4.2 software package including PRIMUS, GNOM, DAMMIF, SUPCOMB and CRYSOL (Petoukhov et al., 2012). PRIMUS version 3.0 was used for initial data processing. GNOM was used to generate the pair distribution function (P(r)) from which maximum particle dimension (Dmax) was estimated. The radius of gyration (Rg) was estimated using the Guinier plot. Divergent low-q data points exhibiting artifacts from beam-stop scattering and data points for which q > 0.25 Å-1 were not included in Guinier and P(r) analysis. The ab initio reconstruction of a molecular envelope for the NCPH2AK15ub–RNF169 complex was done using DAMMIF. Ten independent runs were performed and the normalized spatial discrepancy (NSD) was used to evaluate the results. The molecular envelope was superimposed to the NMR/SAXS-based model of NCPH2AK15ub–RNF169 using SUPCOMB. CRYSOL was used to back-calculate the scattering curve for the NCPH2AK15ub–RNF169 structural model and evaluate the goodness of fit to experimental data.
NMR- and SAXS-Based Modeling of NCPH2AK15ub–RNF169
An initial model of NCPH2AK15ub–RNF169 complex was constructed based on the crystal structure of human NCP (PDB 3AFA) (Tachiwana et al., 2010). Using PRE data of H2AK15ub-H2B and SAXS data of NCPH2AK15ub–RNF169, the model was refined following a reported protocol (Schwieters et al., 2010). In the modeling, the folded domains of the 8 histone molecules, the C-terminal domain of RNF169 (aa 697-700) and the DNA were held fixed in space while two ubiquitin molecules (aa 1-71) and the N-terminal domain of RNF169 (aa 650-684) were treated as rigid bodies. The linker region of RNF169 (aa 685-696) and the N- and C-tails of all histones were given torsional and Cartesian degrees of freedom. Simulated annealing calculations were done using X-PLOR-NIH at an initial temperature of 3000 °K with PRE and SAXS force constants set to 0.05 kcal mol-1 s2 and 400 kcal mol-1 s2, respectively. The system was then cooled to 25 °K in 25,000 steps, fixing the SAXS force constant and ramping the PRE constant to 1 kcal mol-1 s2. Final Cartesian and torsion angle space minimizations were conducted in 2,000 iterations each. Ten of the 100 simulated annealing structures with the lowest energies and best agreement with PRE and SAXS data were then used for analysis.
NMR-Based Modeling of NCPH2AK15ub–RAD18
We first determined the solution NMR structure of RAD18 (aa 198-240)–ubiquitin complex using NOE-based and hydrogen bond-based distance and TALOS+-based dihedral angle restraints plus 1H-15N residual dipolar couplings for RAD18 and ubiquitin measured in a 5% alkyl-polyethylene glycol (C12E5)/n-hexanol mixture (Rückert and Otting, 2000). Structures were calculated using a simulated annealing protocol in XPLOR-NIH (version 2.44). Out of 200 structures, the 20 structures with lowest energies define the ensemble used for structural analysis and statistics (Table S2). The RAD18 (aa 198-240)–ubiquitin structure differs slightly from the previously reported NMR structure of RAD18 (aa 198-227)–ubiquitin (Rizzo et al., 2014) in that it harbors a longer RAD18 α-helix (aa 213-230 instead of 213-224).
The RAD18 (aa 198-240)–ubiquitin NMR structure was used in a rigid body simulated annealing protocol in XPLOR-NIH to generate a model of the NCPH2AK15ub–RAD18 complex. Similar to the method used for modeling NCPH2AK15ub–RNF169, the folded domains of the 8 histone molecules were fixed in space while two ubiquitin molecules (aa 1-71) and the N-terminal domain of RAD18 (aa 200-230) were treated as rigid bodies with full rotational and translational freedom. Ambiguous restraints derived from chemical shift perturbation experiments were used to restrain RAD18–ubiquitin relative to H2A-H2B (Figure S7). NOE-derived restraints between RAD18 C-terminal LRM (R234) and the acidic patch of H2A-H2B (E105 and L106 of H2B) were also included. Based on the analysis of ITC data, we inferred that the interaction between DNA from NCP and RAD18–ubiquitin could contribute to the high affinity of the NCPH2AK15ub–RAD18 complex. Thus, ambiguous restraints between DNA and RAD18–ubiquitin (residues 72, 74 of ubiquitin and 201, 215 and 218 of RAD18) were also included in the docking calculations. Subsequent simulated annealing steps were performed as described above (see Model of NCPH2AK15ub–RNF169). Ten of the 100 simulated annealing structures with the lowest energies were then used for analysis.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) experiments were carried out at 10 °C using VP-ITC or iTC200 microcalorimeters (MicroCal, Malvern). Samples were buffer-exchanged extensively in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl. RNF169, RNF168, RAD18 or 53BP1 in the calorimeter injection syringe at concentrations ranging from 0.1 to 0.4 mM were delivered as a series of 2 μl injections every 3 min (iTC200) and 5 min (VP-ITC) to the reaction cell containing 5-10 μM NCP, NCPH2AK13ubK15ub, NCPH2AK13ub, NCPH2AK15ub or NCPH2AK15ubH4Kc20me2. For titrations of H2A-H2B, H2AK13ubK15ub-H2B, H2AK13ub-H2B, H2AK15ub-H2B and their mutants with RNF168, RNF169 (wild type and mutants) or RAD18, concentrations of 10-40 μM and 0.2-0.5 mM were used for samples in the reaction cell and injection syringe, respectively. The measurements were paired with control experiments for heat of mixing and dilution. Data were analyzed using a one-site model with Levenberg-Marquardt nonlinear regression programmed in Origin 7.0 software (OriginLab Corporation).
Ni2+-NTA Pull-Down Assays
For the Ni2+-NTA pull-down assay in Figure 1B, 16 μmol of the His6-tagged bait added to 75 μL of Ni2+-NTA resin, pre-equilibrated with 50 mM NaPi, pH 7.5, 300 mM NaCl, were mixed and incubated for 1 h at 4 °C on a nutator. Baits were RNF168 (aa 430-481) and RNF169 (aa 653-708) with N-terminal His6-tag, and RAD18 (aa 198-240) with C-terminal His6-tag. Bait-loaded beads were washed 3 times with 50 mM NaPi, pH 7.5, 150 mM NaCl (1 mL, 5 min), centrifuging (21,000 g, 2 min) between washes, and then resuspended in 150 μL of this buffer. To each 50 μL of this slurry, no NCP, 12 mg unmodified NCP, 50 μL of K13/K15 NCP ubiquitylation reaction, and 50 μL of K119 NCP ubiquitylation reaction were added. Mixtures were incubated for 1 h at 4 °C on a nutator before beads were washed 3 times with 50 mM NaPi, pH 7.5, 150 mM NaCl, 20 mM imidazole, 0.02% NP-40 (1 mL, 5 min). To the “dry” resin, 25 μL of 2× Laemli dye was added. Samples were boiled for 2 min and 5 μL of the supernate was loaded onto a gel (15% TGX, Biorad). Protein bands were transferred onto a nitrocellulose membrane (Trans-Blot Turbo System, Biorad) and processed for Western blot analysis. The membrane was blocked (5% non fat milk in TBS (50 mM Tris-Cl, pH 7.6; 150 mM NaCl), 1 h, r.t.), incubated with primary antibody (1:1000 dilution in 1% BSA in TBS, overnight 4 °C), washed 5 × 5 min with TBST (TBS with 0.1% Tween 20), incubated with HRP-conjugated secondary antibody (1:10,000 dilution in 1% non fat milk in TBS, 1 h, r.t.), washed 5 × 5 min with TBST, and developed with an ECL reagent for imaging using a ChemiDoc MP system (Biorad).
The K13/K15 NCP ubiquitylation reaction contained 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 1 μM ZnCl2, 1 mM TCEP, 1.6 μM Uba1 (E1), 8 μM UbcH5c (E2), 8 μM Rnf168-RING (E3), 1.2 μM NCP, 50 μM ubiquitin and 3 mM ATP. In K119 NCP ubiquitylation reaction, RING1B-BMI1 replaced RNF168 and MgCl2 concentration was 10 mM. The reactions proceeded for 90 min at 32 °C before Ni2+-NTA beads were added.
For the Ni2+-NTA pull-down in Figure 1C, similar procedure as above was performed with some changes: RAD18 (aa 198-240) bait used has N-terminal His6-GB1-tag, K15-ubquitylated NCP was obtained from DNA/histone reconstitution using H2AK15ub mimic, NP-40 was not used in the wash buffer, and 18-20 μg of NCP and NCPK15ub baits were used.
GST Pull-Down Assays
GST pull-down assay was carried out by first incubating 40 μL of 50% GSH slurry (Clontech) in buffer 1 (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% NP-40, 0.1% BSA) with a bait (3 μg GST or equimolar amounts of GST-53BP1 wild-type or T1609E/S1618E mutant) on a nutator for 1 h at 4 °C. Beads were then washed 3 times with buffer 1 (1 mL, 5 min), centrifuging (21,000 g, 2 min) between washes. Input NCPH2AK15ubH4Kc20me2 (36 mg), without or with equimolar, 2-fold and 4-fold molar excess of RNF168, RNF169 and RAD18, were added to the immobilized baits and mixed on a nutator for 2 h at 4 °C. Beads were washed 5 × 5 min with buffer 1, the last wash omitting NP-40 and BSA, and excess liquid removed prior to adding 40 μL of 2× Laemli dye. Beads were boiled for 2 min and 10 μL of the supernate was loaded onto a 4-20% TGX gel (Biorad). Next, Western blot was done as described above.
Antibodies
Antibodies used for Western blot analysis were anti-ubiquitin (Cell Signaling P4D1, 1:1000 dilution), anti-H2A (Millipore 07146, 1:1000 dilution), anti-K15 ubiquitylated H2A (a gift from Dr. Zhiguo Zhang, 1:500 dilution) (Wang et al., 2016), anti-GST (Santa Cruz sc-138, 1:1000 dilution), anti-mouse HRP-conjugated (Cell Signaling 7076, 1:10,000 dilution) and anti-rabbit HRP-conjugated (BioRad 172-1019, 1:10,000 dilution).
Data and Software Availability
The atomic coordinates and NMR resonance assignments have been deposited to the Protein Data Bank (PDB) and Biological Magnetic Resonance Data Bank (BMRB) under accession codes PDB 5VEY and BMRB 30275 for H2Aub-H2B–RNF169 (653-708) and PDB 5VF0 and BMRB 30276 for Ubiquitin–RAD18 (198-240), respectively. Original gel images have been deposited to Mendeley Data, http://dx.doi.org/10.17632/ttr7y2ht2f.1.
Supplementary Material
Acknowledgments
We are extremely grateful to Daniel Durocher for stimulating discussions; Hitoshi Kurumizaka and Song Tan for advice on nucleosome reconstitution; Charles Schwieters for advice on structure refinement with PRE -derived restraints; John Tainer and the staff of the SYBILS beamline for help with SAXS. We thank Christine Canman, Andrea Cochran, Daniel Durocher, Michael Huen, Hitoshi Kurumizaka, Titia Sixma, Song Tan, Youlin Xia, Wolfgang Wende, Jianye Zang and Zhiguo Zhang for providing reagents. This research was supported by NIH grants GM116829 and CA132878 to G.M., US Department of Defense grant W81XWH-16-1-0391 to M.V.B., Mayo Clinic Eagles funds to Q.H., G.C. and M.V.B., and an Edward C. Kendall Fellowship in Biochemistry to Q.H.
Footnotes
Supplemental Information: Supplemental information includes seven figures and two tables.
Author Contributions: Q.H., M.V.B. and G.M. conceptualized the study and wrote the manuscript. Q.H. prepared proteins, performed NMR, SAXS and ITC experiments and determined the structures. M.V.B. designed all constructs, prepared proteins and performed the biochemical assays. G.C. contributed to structure determination. D.Z. prepared NCPs together with M.V.B. and Q.H. G.M. supervised the research and contributed to data collection and analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- Benirschke RC, Thompson JR, Nomine Y, Wasielewski E, Juranic N, Macura S, Hatakeyama S, Nakayama KI, Botuyan MV, Mer G. Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure. 2010;18:955–965. doi: 10.1016/j.str.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C. A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal Biochem. 2011;418:102–110. doi: 10.1016/j.ab.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Mer G, Yi GS, Koth CM, Case DA, Edwards AM, Chazin WJ, Arrowsmith CH. Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide. J Mol Biol. 2001;312:177–186. doi: 10.1006/jmbi.2001.4938. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Nominé Y, Yu X, Juranic N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12:1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Feng W, Jiang J, Deng Y, Huen MS. Ring finger protein RNF169 antagonizes the ubiquitin-dependent signaling cascade at sites of DNA damage. J Biol Chem. 2012;287:27715–27722. doi: 10.1074/jbc.M112.373530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Botuyan MV, Mer G. Preparation of recombinant peptides with site- and degree-specific lysine 13C-methylation. Biochemistry. 2009;48:3798–3800. doi: 10.1021/bi900348z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 2012;11:2538–2544. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Furuita K, Ohki I, Ikegami T, Fukada H, Shirakawa M, Fujiwara T, Kojima C. Utilization of lysine 13C-methylation NMR for protein-protein interaction studies. J Biomol NMR. 2013;55:19–31. doi: 10.1007/s10858-012-9675-9. [DOI] [PubMed] [Google Scholar]
- Helchowski CM, Skow LF, Roberts KH, Chute CL, Canman CE. A small ubiquitin binding domain inhibits ubiquitin-dependent protein recruitment to DNA repair foci. Cell Cycle. 2013;12:3749–3758. doi: 10.4161/cc.26640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert RG, Sixma TK. Intrinsic flexibility of ubiquitin on proliferating cell nuclear antigen (PCNA) in translesion synthesis. J Biol Chem. 2012;287:39216–39223. doi: 10.1074/jbc.M112.389890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Tang C, Clore GM. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRView: a computer program for visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, van Ingen H, Zhou BR, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci USA. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Acharya SS, Kwon M, Drané P, Guan Y, Adelmant G, Kalev P, Shah J, Pellman D, Marto JA, et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol Cell. 2014;54:512–525. doi: 10.1016/j.molcel.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre JF, Dayie KT, Peng JW, Wagner G. Internal mobility in the partially folded DNA binding and dimerization domains of GAL4: NMR analysis of the N-H spectral density functions. Biochemistry. 1996;35:2674–2686. doi: 10.1021/bi9526802. [DOI] [PubMed] [Google Scholar]
- Leung JW, Agarwal P, Canny MD, Gong F, Robison AD, Finkelstein IJ, Durocher D, Miller KM. Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS Genet. 2014;10:e1004178. doi: 10.1371/journal.pgen.1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat Commun. 2014;5:3291. doi: 10.1038/ncomms4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer G, Dejaegere A, Stote R, Kieffer B, Lefèvre JF. Structural dynamics of PMP-D2: an experimental and theoretical study. J Phys Chem. 1996;100:2667–2674. [Google Scholar]
- Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, Escribano-Diaz C, Durocher D. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- Panier S, Ichijima Y, Fradet-Turcotte A, Leung CC, Kaustov L, Arrowsmith CH, Durocher D. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol Cell. 2012;47:383–395. doi: 10.1016/j.molcel.2012.05.045. [DOI] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol Biol. 2009;10:55. doi: 10.1186/1471-2199-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M, Lukas C, Lukas J, Bekker-Jensen S, Mailand N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J Cell Biol. 2012;197:189–199. doi: 10.1083/jcb.201109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Super-Resolution in Solution X-Ray Scattering and Its Applications to Structural Systems Biology. Annu Rev Biophys. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- Rizzo AA, Salerno PE, Bezsonova I, Korzhnev DM. NMR structure of the human Rad18 zinc finger in complex with ubiquitin defines a class of UBZ domains in proteins linked to the DNA damage response. Biochemistry. 2014;53:5895–5906. doi: 10.1021/bi500823h. [DOI] [PubMed] [Google Scholar]
- Rückert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, Taniguchi Y, Murakawa Y, Zeng W, Yokomori K, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol Cell Biol. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Progr NMR Spec. 2006;48:47–62. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Suh JY, Grishaev A, Ghirlando R, Takayama Y, Clore GM. Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering. J Am Chem Soc. 2010;132:13026–13045. doi: 10.1021/ja105485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Sundd M, Iverson N, Ibarra-Molero B, Sanchez-Ruiz JM, Robertson AD. Electrostatic interactions in ubiquitin: stabilization of carboxylates by lysine amino groups. Biochemistry. 2002;41:7586–7596. doi: 10.1021/bi025571d. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, Kimura H, Kurumizaka H. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc Natl Acad Sci USA. 2010;107:10454–10459. doi: 10.1073/pnas.1003064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang H, Liu J, Cheruiyot A, Lee JH, Ordog T, Lou Z, You Z, Zhang Z. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 2016;30:946–959. doi: 10.1101/gad.271841.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res. 2009;37:2176–2193. doi: 10.1093/nar/gkp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D. The structural basis of modified nucleosome recognition by 53BP1. Nature. 2016;536:100–103. doi: 10.1038/nature18951. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen J, Wu M, Wu H, Arokiaraj AW, Wang C, Zhang W, Tao Y, Huen MS, Zang J. Structural basis for role of ring finger protein RNF168 RING domain. Cell Cycle. 2013;12:312–321. doi: 10.4161/cc.23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwahlen C, Legault P, Vincent SJF, Greenblat J, Konrat R, Kay LE. Methods of measurements of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage N-peptide/box B RNA complex. J Am Chem Soc. 1997;119:6711–6721. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


