Abstract
Background & Aims
As important virological markers, serum HBsAg and HBV DNA levels show large fluctuations among chronic hepatitis B (CHB) patients. The aim of this study was to reveal the potential impact and mechanisms of amino acid (AA) substitutions in small hepatitis B surface proteins (SHBs) on serum HBsAg and HBV DNA levels.
Methods
Serum samples from 230 untreated chronic hepatitis B patients with genotype C HBV were analyzed in terms of HBV DNA levels, serological markers of HBV infection and SHBs sequences. In vitro functional analysis of the identified SHBs mutants was performed.
Results
Among 230 SHBs sequences, there were 39 (16.96%) sequences with no mutation detected (wild-type, WT) and 191 (83.04%) with single or multiple mutations. SHBs consist of 226 AAs, of which 104 (46.02%) had mutations in our study. Some mutations (e.g. sE2G, sL21R, sG24K, sT47A/K, sC69stop (sC69*), sL95W, sL98V, and sG145R) negatively correlated with serum HBsAg levels. HBsAg and HBV DNA levels from this group of patients had a positive correlation (r = 0.61, p < 0.001). In vitro analysis showed that these mutations reduced extracellular HBsAg and HBV DNA levels by restricting virion secretion and antibody binding capacity. Virion secretion could be rescued for sE2G, sC69*, and sG145R by co-expression of WT HBsAg.
Conclusion
The serum HBsAg levels were lower in untreated CHB patients with novel SHBs mutations outside the major antigenic region than those without mutations. Underlying mechanisms include impairment of virion secretion and lower binding affinity to antibodies used for HBsAg measurements.
Keywords: HBV, HBsAg mutation, virion secretion, antigenicity, ‘a’ determinant
Graphical abstract
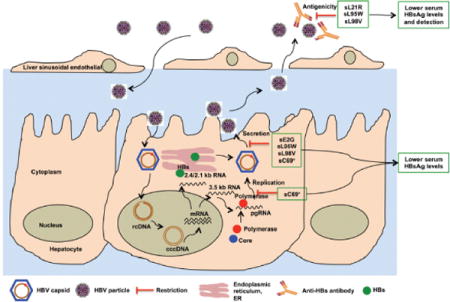
Introduction
Chronic hepatitis B virus (HBV) infection remains a severe public health problem. About 240 million people worldwide are chronically infected and have an increased risk for developing liver cirrhosis and hepatocellular carcinoma [1, 2]. The hepatitis B surface antigen (HBsAg), a major viral protein secreted into patient serum, consists of three distinct, but structurally related proteins: the large, middle and small hepatitis B surface proteins (LHBs, MHBs, and SHBs, respectively). Within the SHBs, the amino acids (AA) 99 to 169 region is termed as the major hydrophilic region (MHR), where AAs 124–147 is defined as the ‘a’ determinant, a dominant neutralizing epitope [3]. Antibodies used in commercial detection kits usually recognize and bind to this region.
Since its discovery as the Australia antigen by Blumberg et al. in 1963 [4], HBsAg has been considered an important biomarker of HBV infection. The loss of HBsAg and the development of anti-HBs antibodies (HBsAg seroconversion) are believed to indicate a clinical cure [1]. The HBsAg quantitative assay is often a better option than the qualitative tests for other biomarkers. [5]. The Architect QT assay (Abbott Laboratories) and the Elecsys HBsAg II Quant assay (Roche Diagnostic) are widely used in the clinic. Both yield quantitative HBsAg (qHBsAg) results as international units per ml (IU/ml) [6, 5].
The wide clinical application of these assays resulted in suggestions that the serum qHBsAg might serve as a surrogate marker for transcriptionally active HBV covalently closed circular DNA (cccDNA) in hepatocytces and as a useful marker in guiding antiviral therapy. A study on 26 hepatitis B e antigen (HBeAg)-positive CHB patients receiving 32-week Peg-IFN treatment followed by two-year lamivudine treatment showed that the HBsAg levels correlated with cccDNA (r = 0.54, p = 0.004) at baseline, and its reduction after treatment showed good correlation with the reduction of hepatocyte cccDNA levels (r = 0.68, p < 0.0001) [7]. A study on HBeAg-negative CHB patients showed that a decrease of HBsAg levels by 0.5 log10 IU/ml at week 12 and 1.0 log10 IU/ml at week 24 of Peg-IFN therapy was associated with the positive predictive values of 89% and 92% for HBV DNA negativity 24 weeks after drug withdrawal, supporting its role as a predictive marker for response-guided therapy in IFN-treated patients [8]. In addition, HBsAg levels could be used as a stopping rule for Peg-IFN-treated patients [8].
However, significantly different serum HBsAg levels could be found in CHB patients with the same disease progression, HBV genotype and HBeAg status [9]. Locarnini et al. showed that serum HBsAg levels only correlated with HBV DNA loads during the immune clearance phase for patients with HBV genotype B and C infection [10, 9]. A better understanding of the mechanisms behind serum HBsAg level fluctuation is of clinical importance. Theoretically, the underlying mechanisms could include the factors affecting the HBsAg expression, secretion, antigenicity and/or the immune response. The current study focused on how AA substitutions in SHBs influence qHBsAg values. It is known that new qHBsAg assays have incorporated the impact of some classical SHBs AA substitutions on qHBsAg, such as sK122I, sI126S and sG145R [11]. However, some newly discovered SHBs mutations were under quantitated by the Architect assay, whereas sP142L, sP142S and sG145K mutations yielded lower results in the Elecsys system when compared with those obtained with the Architect system [12]. In addition, Locarnini et al. reported that the rtA181T/sW172stop (sW172*) variant had a secretory defect and exerted a dominant negative effect on wild-type (WT) HBV virion secretion [13].
Beside the aforementioned mutations, SHBs could also have other naturally occurring substitutions affecting serum HBsAg levels, which are yet to be identified or poorly characterized. Thus, the aim of this study was to identify novel mutations that correlated with lower HBsAg levels by using a study cohort of 230 untreated CHB patients, who were all HBeAg-positive and infected with genotype C HBV. The impact and mechanisms of these novel AA substitutions on HBsAg levels were further studied using in vitro cell culture systems.
Materials and methods
Patients
Serum samples from 230 untreated HBeAg-positive CHB patients with genotype C HBV were collected from 23 hospitals in China during 2010 to 2012 in a registered clinical study (NCT01088009) [14]. Informed consent was obtained. The clinical diagnosis of CHB was performed according to the Chinese Society of Hepatology [15]. All patients were negative for hepatitis C virus, hepatitis D virus and human immunodeficiency virus serum markers. We selected only genotype C HBeAg-positive patients to exclude the potential impacts of HBV genotype, HBeAg status and disease progression on sequence mutation analysis. In addition, genotype C HBV is the most prevalent genotype in China and some areas of Asia [16, 10, 9, 17].
PCR amplification, DNA sequencing, and sequence analysis
HBV RT sequences covering the entire S gene amplification and sequence analysis for identifying the AA substitution in SHBs have been described [18].
Plasmids
Mutants sE2G, sL21R, sG24K, sT47A, sT47K, sC69* sL95W, sL98V, and sG145R were cloned into plasmid pBB4.5 1.2/PC, which contained a 1.2-fold length HBV genome of genotype C with a G1896A mutation in the preC region to facilitate the DNA replication in the in vitro system [19]. Primers for site-directed mutagenesis and overlapping PCR are shown in Supplementary Table 1. Introduction of rtD205H mutation resulted in a polymerase-deficient HBV control plasmid, termed tyrosine-methionine-histidine-aspartate (YMHD), which corresponds to the mutated reverse transcriptase (RT) active site tyrosine-methionine-aspartate-aspartate (YMDD). Plasmids with the human influenza hemagglutinin (HA) tag at the C-terminal of SHB derivatives (WT-HA, sE2G-HA, sC69*-HA, sL95W-HA, sL98V-HA, and sG145R-HA) were cloned into pBB4.5 1.2/PC. Since the mutant sC69* has a premature stop codon at the SHBs AA position 69, we inserted a HA tag at three different SHBs locations (N terminus, before the stop codon between positions 68 and 69, and after the C terminus of WT SHB sequence). A pBluescript II KS (+) plasmid (Addgene, USA) was used as the vector for HBsAg expression. In brief, the preS1, preS2 and S HBV coding regions were inserted into the KpnI and Sac I sites of this vector.
Cell cultures and transfection
HepG2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 1% non-essential amino acids (NEAA). HepG2 cells were seeded 24 hours before transfection, then cells were transfected with plasmids in the presence of X-tremeGENE 9 at a ratio of plasmids and transfection reagent 1 μg :3 μl (Roche diagnostics, Mannheim, Germany). After overnight incubation, the cells were washed with Dulbecco’s phosphate-buffered saline (DPBS, Life Technologies, USA) five times and fresh media (DMEM+10%FBS+1%NEAA) was added. Transfected cells were harvested and supernatant was collected 72 hours post-transfection. Transfection efficiency was assessed by co-transfecting a reporter plasmid expressing enhanced green fluoresence protein (EGFP). All transfections were performed in triplicate in at least three independent experiments.
Western blot
The protocol for Western blotting has been described [20]. Briefly, HBV-transfected cells from 6-well plates were washed three times with PBS and lysed with RIPA buffer in the presence of a cocktail of proteinase inhibitors (Roche, Mannheim, Germany). Supernatants were loaded on a 4–12% SDS-PAGE gradient gel (Life technologies) and transferred to a polyvinylidene fluoride (PVDF) membrane. Antibodies against HBV S protein: polyclonal horse anti-HBs (Abcam, USA) and monoclonal mouse anti-HBV preS2 (Abcam, USA) and anti-HA-tag (Sigma, USA), were used at 1:500 dilutions. SuperSignal® West Femto Maximum Sensitivity Chemiluminescent Substrate (Thermo Scientific, USA) was used for imaging.
Quantification of HBV DNA titers
The qPCR for HBV DNA quantification was performed as previously described [18]. Briefly, serum HBV DNA was quantitated by using the COBAS® AmpliPrep/COBAS® TaqMan® HBV test version 2.0 (Roche Diagnostics, Switzerland). In addition, HBV DNA was extracted by using QIAamp DNA Blood Mini Kit (Qiagen, Germany). HBV DNA in in vitro experiments was quantitated by qPCR using the TaqMan Universal PCR Master Mix (Applied Bio systems, Foster) [20]. The primers and probe for qPCR are 5′-CCGTCTGTGCCTTCTCATCTG-3′ (sense), 5′-AGT-CCAAGAGTCCTCTTATGTAAGACCTT-3′ (antisense) and 5-/56- FAM/CCG TGT GCA/ZEN/CTT CGCTTC ACCTCT GC/3IABkFQ/-3 (probe). The qPCR conditions were: (i) denaturation at 50 °C for 5 min followed by 95 °C for 10 min (one cycle); (ii) qPCR at 95 °C for 15 s, 56 °C for 40 s, and 72 °C for 20 s (40 cycles); (iii) melting at 65 °C for 10 s, followed by 95 °C (continuous).
Detection of intracellular HBV total mRNAs and pgRNA in cell culture system
Total HBV RNA was isolated with the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), and quantified by using a NanoDrop (Thermo Scientific). First strand cDNA was synthesized by using the SuperScript III RT kit (Invitrogen). The qPCR for HBV total mRNA and pgRNA was performed as previously described [20]. DNase I (NEB) was used to degrade HBV DNA during RNA extraction and ribosomal protein S11 mRNA was used to normalize qRT-PCR results for HBV transcripts.
Detection of HBsAg and HBeAg
Serum HBsAg levels were detected by the Architect HBsAg QT (Abbott) according to the manufacturer’s protocols. HBsAg and HBeAg levels from cell culture supernatants were detected with commercial enzyme-linked immunosorbent assay (ELISA) kits (Autobio diagnostic Co., China), according to the manufacturer’s protocols. For intracellular HBsAg, cells were lysed with lysis buffer containing 1% of Nonidet P40, 150 mM NaCl and 50 mM trishydroxym-ethylaminomethane-HCl (pH 7.5). The samples were also detected with an ELISA kit (Autobio diagnostic Co., Zhengzhou, China) according to the manufacturer’s protocol.
Statistical analyses
Statistical analysis was performed using the SAS version 9.1 software package. The interdependence between numerical variables was performed using the ANOVA test and Spearman rank correlation test. Continuous and categorical variables were compared between groups using the student’s T test and Fisher’s exact test, respectively. Values of p < 0.05 were considered as statistically significant.
Results
Patient characteristics
230 untreated CHB patients were recruited. All of the patients were Chinese with HBV genotype C infection. Demographic, biochemical and virological characteristics are shown in Table 1. All samples were HBeAg-positive. The median HBV DNA level was 7.97 log10 IU/ml (range: 4.12–9.80 log10 IU/ml). However, the median HBsAg level was 4.15 log10 IU/ml (range: 2.37–5.50 log10 IU/ml). Anti-HBs and anti-HBe were negative.
Table 1.
Demographic, biochemical and virological characteristics of CHB patients without treatment.
| Characteristics | Patients (N = 230) |
|---|---|
| Age (years), median (range) | 31.50 (18.00–64.00) |
| Male sex (%) | 70.94 |
| ALT (U/L), median (range) | 122.00 (0.00–1150.00) |
| AST (U/L), median (range) | 75.00 (0.00–985.00) |
| HBV DNA (log10 IU/ml), median (range) | 7.97 (4.12–9.80) |
| HBsAg (log10 IU/ml), median (range) | 4.15 (2.37–5.50) |
| HBeAg (% positive) | 100.00 |
CHB, chronic hepatitis B; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HBsAg, hepatitis B surface antigen; HBeAg, Hepatitis B e antigen.
SHBs mutations and their influence on circulating HBsAg and HBV DNA levels
To estimate the possible influence of S gene mutations on the fluctuations of the quantitative detection of both serum HBsAg and HBV DNA levels, we performed sequence analysis on full-length SHBs fragments amplified from 230 serum samples. Mutation frequencies at each individual AA sites were determined for all 226 positions of the SHBs fragments based on the excel software. Among the 230 patient sequences, 39 (16.96%) were WT without any mutation and 191 (83.04%) harbored mutations at one or more AA positions. In total, 104 AA positions within SHBs (104/226, 46.02 %) had detectable mutations with a total mutation frequency of 1.06 % [549/(230 × 226)]. The mutation frequency at individual AA positions is displayed in Fig. 1A.
Fig. 1. Mutational profile of SHBs among 230 CHB patients with genotype C HBV infection.
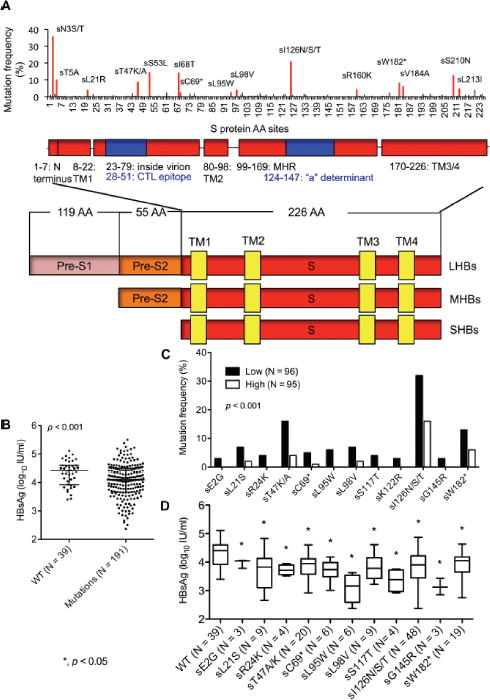
(A) Map of amino acid substitution frequencies at a total of 226 SHBs sites [9]. The lower schematic highlights functional regions of SHBs and three HBV envelope proteins (LHBs, MHBs and SHBs) share a common C terminus. (B) Comparison of HBsAg levels between WT and mutant groups stratified by the median in 230 samples. Dots represent individual values and horizontal lines as median value with interquartile range. (C) The identified individual mutations occurred mostly in the Low HBsAg group (p < 0.05). The group Low and High was stratified by the median HBsAg level of 4.10 log10 IU/ml in 191 patients harboring SHBs substitutions. (D) Comparison of HBsAg levels between the identified mutants and WT. Boxes represent values and bars as median with range. AA: amino acid; CHB: chronic hepatitis B; SHBs: small hepatitits B surface protein; MHBs: middle hepatitis B surface protein; LHBs: large hepatitis B surface protein; TM: transmembrane; MHR: major hydrophilic region; CTL: cytotoxic T lymphocytes; WT, wild-type. ANOVA test (B and D) and Fisher’s exact test (C) were employed.
We then compared the HBsAg levels in patients carrying WT versus mutated SHBs. Interestingly, HBsAg levels in the patients with WT HBV were significantly higher than those in patients with HBV bearing mutations in SHBs (p < 0.001) (Fig. 1B). Thus, we divided the 191 patients having SHBs mutations into two subgroups based on their median HBsAg level 4.10 log10 IU/ml (range: 2.37–5.50 log10 IU/ml), and analyzed the SHBs mutations in the low (N = 96) and high (N = 95) HBsAg value groups, with median HBsAg levels 3.67 log10 IU/ml (interquartile range (IQR): 3.35–3.90 log10 IU/ml) and 4.51 log10 IU/ml (IQR: 4.25–4.76 log10 IU/m), respectively. In-depth data mining (Fig. 1C) revealed previously identified immune escape mutations within the ‘a’ determinant region (sS117T, sK122R, sI126N/S/T, and sG145R), carboxyl terminal truncation mutations (sC69* and sW182*) and additional rarely described mutations (sE2G, sL21S, sR24K, sT47K/A, sL95W and sL98V). The mutation frequency was significantly higher in the group with low HBsAg levels compared to the group with high HBsAg levels (p < 0.05) (Fig. 1C). HBsAg levels in mutants were significantly lower than that in WT (Fig. 1D). Twelve samples were found with HBsAg level < 1000 IU/ml, but their median HBV DNA level was 6.77 log10 IU/ml (range: 4.65–8.22 log10 IU/ml). To note, 75.00% (9/12) of their sequences harbor one or two of our newly identified SHBs mutations (sL21R, sT47K, sL95W, sK122R, sI126T, sG145R and sW182*) (Supplementary Table 2). The overall prevalence of the identified SHBs substitutions is shown in Supplementary Table 3.
A significant positive correlation was observed between HBsAg and HBV DNA levels in the overall study population (r = 0.70; p < 0.001) (Fig. 2A). However, the correlation coefficients were lower in the mutant group (SHB sequences with the above selected mutations, N = 99) (r = 0.61; p < 0.001) (Fig. 2B). Interestingly, patients within multiple or single mutations had significantly lower HBsAg levels compared to WT group (p < 0.001) (Fig. 2C). Similarly, HBV DNA levels in the multiple and single mutation groups were also significantly lower than the WT group (p < 0.001) (Fig. 2D).
Fig. 2. SHBs mutations affect serum HBsAg and HBV DNA levels in HBeAg-positive CHB patients.
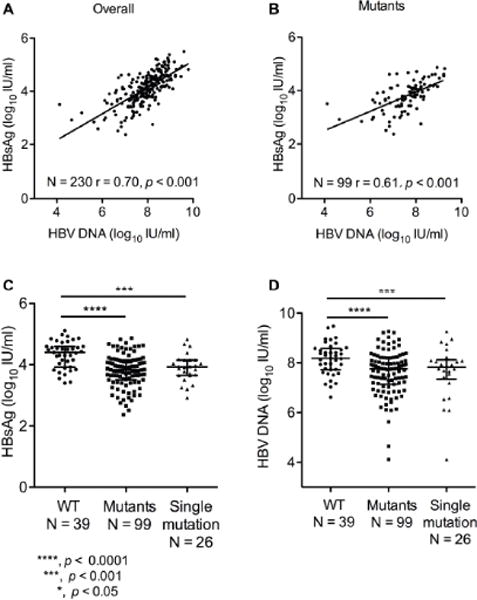
Correlation of HBsAg and HBV DNA levels in (A) whole, (B) mutants groups of the samples. Comparison of (C) HBsAg levels and (D) HBV DNA levels among wild type, mutants and single mutation groups. Mutants group indicates that the sequences with the selected mutations are grouped. Single mutation group indicates that the sequences with only one mutation occurred from the selected mutation sequences are grouped. Dots represent individual values and horizontal lines as median value with interquartile range. ***, p < 0.0001; **, p < 0.001; *, p < 0.05. Spearman rank correlation test and ANOVA test were employed.
There was no correlation of HBsAg levels with ALT and AST levels in the study population. No significant differences were observed in terms of age, ALT and AST between the groups with or without SHBs mutations (Supplementary Fig. 1).
In vitro validation of the influence of S mutations on HBsAg levels
To study whether the S mutants influence HBsAg levels, we introduced individual mutations (sE2G, sL21R, sG24K, sT47A, sT47K, sC69*, sL95W, sL98V, and sG145R) in the background of HBV plasmid pBB4.5 1.2/PC. HBsAg levels were determined by ELISA 72 hours after transfection of HepG2 cells. The in vitro experiments showed a similar trend with the clinical study. Specifically, mutations sE2G, sC69*, sL95W, sL98V and sG145R resulted in lower extracellular HBsAg levels compared to WT (Fig. 3A). The intracellular HBsAg levels from these mutants were also lower than that of WT (Fig. 3B). However, the sG24K and sT47A/K mutations did not significantly affect the extracellular and intracellular HBsAg levels compared to WT (Fig. 3A, 3B).
Fig. 3. Comparison of WT and mutant HBsAg expression in transfected HepG2 cells.
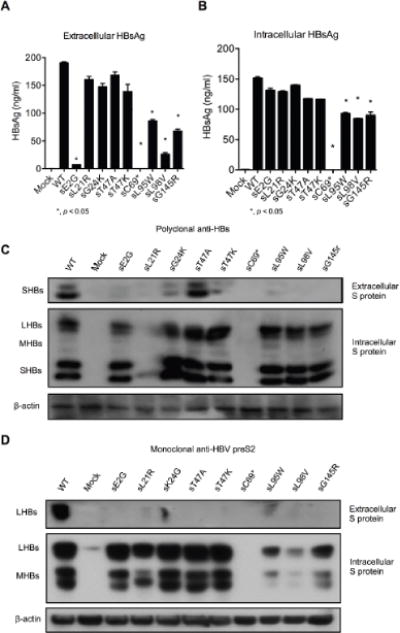
WT and the indicated mutant derivatives were constructed on the pBB4.5 1.2/PC HBV background and used to transfect HepG2 cells. Supernatant and cells were harvest 72 hours post transfection. (A) Extracellular and (B) Intracellular HBsAg levels were measured by ELISA and Western blot with (C) polyclonal anti-HBs and (D) monoclonal anti-pre-S2 antibody. *, p < 0.05. Student’s T test was employed.
To further determine whether these mutations influence HBsAg levels due to impaired HBsAg detection and/or secretion, Western blot analysis with polyclonal anti-HBs antibody and monoclonal anti-HBs antibody was used. Most of the mutants showed lower extracellular HBsAg than WT except for sT47K/A. In addition, most mutants showed similar intracellular HBsAg band intensities to WT except for sL21R and sC69*, which were undetectable (Fig. 3C). Similarly, when a monoclonal anti-preS2 antibody was used, most mutants exhibited low or undetectable extracellular HBsAg compared to WT. However, intracellular HBsAg could be detected for all mutants except for sC69*, although the band intensities varied among different mutants. Specifically, sL95W, sL98V and sG145R showed much lower intracellular HBsAg than WT (Fig. 3D).
Based on the ELISA and Western blot results, sE2G, sC69*, sL95W, sl98V and sG145R appeared to express significantly decreased HBsAg levels. To rule out possible confounding effects due to the antibodies used for detection, we constructed plasmids encoding HA-tagged S protein sequences for each of these substitutions and transfected HepG2 cells (Fig. 4A). Western blot analysis using a monoclonal anti-HA antibody showed similar results to the anti-HBs antibodies. Taken together, these results show that the mutations decrease HBsAg levels by influencing either detection (Fig. 3C, sL21R) or secretion (sE2G, sC69*, sL95W, sl98V and sG145R) (Fig. 4B).
Fig. 4. Comparison of HBsAg expression of HBV WT and mutants fused with HA-tag in transfected HepG2 cells.
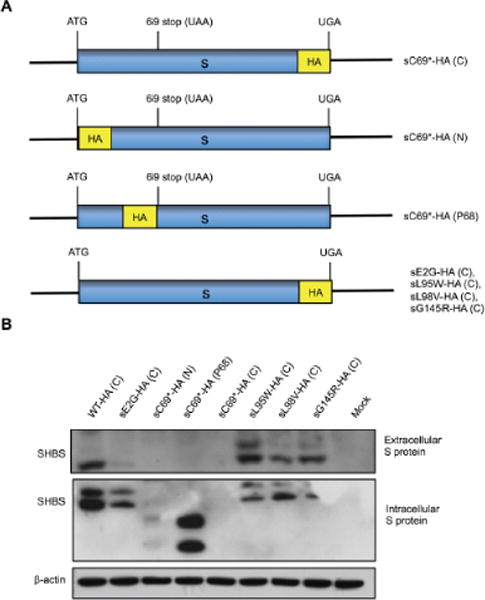
To abolish the influence of HBsAg antigenicity, a HA-tag was fused to S protein for detection with Western blot. (A) Scheme of construction of HBV S gene with HA-tag. (B) Western blot was performed to detect extracellular and intracellular WT and mutant HBsAg levels using anti-HA antibody.
Impact of SHBs mutations on HBV replication and virion secretion
Due to the overlapping nature of the S and RT gene regions, mutations in the S gene may result in concomitant mutations in RT, such as sC69*/rtS78T, sL98V/rtS106C and sG145R/rtR153Q, potentially affecting RT activity and HBV replication. However, some S gene mutations, e.g., sE2G/rtG10G and sL95W/rtV103V, do not result in AA changes in the RT due to the degeneracy of the genetic code (Fig. 5A). In this study, sG145R was used as a control for a mutation known to inhibit virion secretion and antibody detection and the YMHD RT mutation, to abolish RT activity and control for input plasmid DNA background [21, 22]. Seventy two hours after transfection, the supernatants were collected and the cells were lysed. Both the supernatants and the lysates were treated with DNase I for two hours to remove input DNA. Subsequently, the capsid-protected HBV DNA from intracellular replicative intermediates and extracellular viral particles was analyzed by Taqman qPCR.
Fig. 5. Comparison of HBV replication between HBV WT and mutants in transfected HepG2 cells.
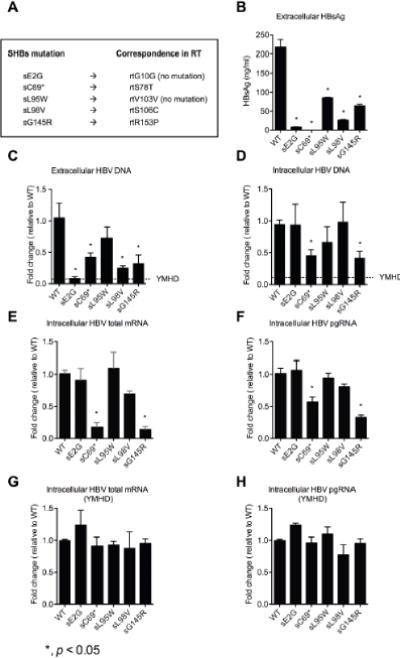
(A) The concomitant AA mutations in SHBs and RT. Analysis of (B) extracellular HBsAg levels, (C) extracellular, (D) intracellular HBV DNA levels, (E) total mRNA, (F) pgRNA, (G) total mRNA in YMHD and (H) pgRNA in YMHD of HBV WT and mutants 72 hours after transfecting HepG2 cells. YMHD RT mutation was used as a control for input plasmid DNA background. Data shown as fold change relative to WT. *, p < 0.05. Student’s T test was employed.
Our results showed that there was significantly less extracellular HBV DNA for sE2G, sC69*, sL98V and sG145R compared to WT (p < 0.05) (Fig. 5C) and also less extracellular HBsAg levels detected by ELISA (Fig. 5B). The intracellular HBV DNA from the mutants sC69* and sG145R but not sE2G, sL95W and sL98V was significantly lower than WT, respectively (Fig. 5D). HBV total RNA and pgRNA levels from sC69* and sG145R were significantly lower than WT (Fig. 5E and Fig. 5F). But, when restricting HBV reverse transcription by RT mutation (YMHD), HBV total RNAs and pgRNA from all of the mutations are similar to those from WT (Fig. 5G and Fig. 5H)”. These results indicated that AA substitutions did not affect HBV transcription. However, in the presence of a functionally active RT (Fig. 5E and Fig. 5F), the reduced amounts of HBV mRNAs from sC69* and sG145R might reflect the net effects of many factors. The potential factors might include the altered RT activities of rtS78T/sC69* and rtR153Q/sG145R, the altered stabilities of mutant mRNAs, the replenishment efficacies of cccDNA pool and so on [23, 21, 22]. Combined with the results in Fig. 3 and Fig. 4, we conclude that mutants sE2G, sL95W and sL98V might negatively influence HBV virion secretion and mutants sC69* and sG145R may influence HBV replication.
Rescue of virion secretion by co-expression of WT envelope proteins
To spread in a chronic infection and persist, defective mutants must presumably coexist with a small amount of WT to supply functional HBs [24, 21]. To determine whether WT surface protein could trans-complement and rescue mutant virus secretion, open reading frames encoding the LHBs, MHBs and SHBs proteins were cloned into the pBluescript (+) KS vector (named pLMS). Co-transfection of mutant and pLMS plasmid DNAs was conducted at ratios of 5:1 (500:100 ng), 1:1 (250:250 ng) and 1:5 (100:500 ng) into HepG2 cells, respectively. The extracellular HBsAg levels for mutants with co-transfection were higher than those without co-transfection (Fig. 6A and Fig. 6C). Interestingly, co-transfection of sE2G, sC69* and sG145R with pLMS showed higher extracellular HBV DNA levels as well (Fig. 6B and Fig. 6D). When the ratio of mutant to pLMS was adjusted to 1:5 (100:500 ng), supplementing with WT HBsAg by pLMS helped both WT and mutant replication-compatible pBB4.5 1.2/PC plasmids to achieve a significantly higher extracellular HBsAg levels, indicating that the HBsAg detected originates largely from pLMS (Fig. 6E). Interestingly, compared to pBB4.5 1.2/PC transfection alone, the co-transfections of the mutants and pLMS increased extracellular HBV DNA to WT levels, but did not enhance extracellular HBV DNA for WT (Fig. 6F). These results suggest that both replication and secretion of the mutants can be rescued by WT HBsAg.
Fig. 6. Co-expression of WT HBsAg and mutant HBV plasmids restores virion secretion.
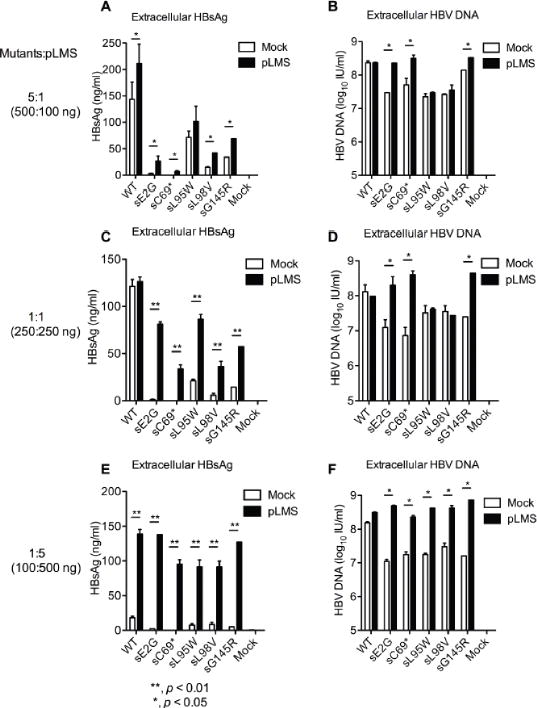
ELISA analysis of extracellular HBsAg and qPCR analysis of extracellular HBV DNA upon co-transfection of WT or mutant replication constructs together with HBsAg expressing plasmid pLMS at different ratios. (A and B) at a 5:1 ratio of replication construct to pLMS (500 ng to 100 ng), (C and D) at a 1:1 ratio (500 ng to 500 ng) and (E and F) at a 1:5 ratio (100 ng to 500 ng). The pBluescript II KS (+) plasmid was used as a control (Mock). **, p < 0.01; *, p < 0.05. Student’s T test was employed.
Discussion
Serum HBsAg quantification is an important clinical marker for antiviral response during IFN treatment. It is therefore important to understand factors that affect serum HBsAg levels and detection. It has been previously reported that several factors can influence serum HBsAg levels. These include the HBV genotype, AA substitutions within the ‘a’ determinant of SHBs, and mutations within the HBV core promoter [17]. In this study, several SHBs AA substitutions that affect serum HBsAg levels were identified in a cohort of 230 HBeAg-positive CHB patients infected with genotype C HBV.
We identified several new HBV mutations (sE2G, sL21S, sR24K, sC69*, sL95W and sL98V) with higher prevalence in the HBsAg low group than those in high group stratified by the median HBsAg levels (Fig. 1C). Moreover, we discovered that HBV mutants sS117T, sK122R, sI126N/S/T, sG145R, and sW182*, reported in previous studies, were also associated with lower HBsAg levels. [21, 25]. It has been suggested that HBsAg level < 1000 IU/ml and HBV DNA level < 2000 IU/ml were sufficient to identify inactive carriers with low cccDNA activity [26]. In this study, we found that 12 samples with HBsAg level < 1000 IU/ml with median HBV DNA level 6.77 log10 IU/ml and 75.00% (9/12) of their sequences were found to harbor one or two our newly identified SHBs mutations (sL21R, sT47K, sL95W, sK122R, sI126T, sG145R and sW182*) (Supplementary Table 2). This suggested that serum HBsAg < 1000 IU/ml in this study did not predict a low metabolic cccDNA activity. Instead, SHBs mutation was highly likely to be an important mechanism leading to a low qHBsAg value. Therefore, our results indicate that circulating HBsAg levels can be significantly influenced by HBsAg mutations.
Interestingly, the newly identified mutations (sE2G, sL21S, sR24K, sC69*, sL95W and sL98V) localize outside the ‘a’ determinant. Before this report, it was not understood how such mutations might influence HBsAg expression, secretion, detection and HBV replication. We therefore studied the influence of these mutations on HBsAg levels in vitro. To measure intracellular and extracellular HBsAg we used commercially available ELISA assays or Western blot detection with polyclonal anti-HBs antibody for LHBs, MHBs and SHBs or a monoclonal anti-preS2 antibody for virion detection. Anti-HA was used to detect engineered HA-tagged HBsAg mutant forms. Taken together, our results suggest that these AA substitutions outside the ‘a’ determinant decrease circulating HBsAg levels through three possible mechanisms.
One potential mechanism is interruption of HBV particle secretion leading to retention and accumulation of HBsAg within cells. These included sE2G (near the N terminus), and sL95W and sL98V (in TM2) (Fig. 1A, Fig. 3C and Fig. 4B). Huang et al. reported that major hydrophilic region (MHR) mutations in the “a” determinant region (G119R, C124Y, I126S, Q129R, S136P, C139R, T140I, K141E, D144A, and G145R) can significantly decrease virion, HBsAg detection and apparent occult HBV infection [27]. Moreover, Ye et al. reported that the sE164G substitution, driven by vaccine escape and outside of the ‘a’ determinant, was also associated with occult HBV infection [28]. Our data suggest that AA substitutions outside the ‘a’ determinant may also affect HBsAg conformation or secretion. sE2G resides in the SHBs signal peptide sequence and may affect S protein translocation efficiency and secretion. The sL95W and sL98V mutations are in a transmembrane segment and AA substitutions in this region could alter the conformation of S protein resulting in lower HBsAg secretion.
A second potential mechanism is that mutations could influence the antigenicity of HBsAg to decrease the binding capacity of anti-HBs antibodies. A number of previous reports demonstrated that mutations in the ‘a’ determinant (e.g., sG145R) influence antigenicity [29, 30]. Similarly, our results showed that substitutions sL21R, sL95W and sL98V, which unlike previous reports are located outside the ‘a’ determinant, affected extracellular and intracellular HBsAg levels by ELISA and Western blot with two different antibodies. This suggests that these mutations result in the lower binding affinity to some of the anti-HBs antibodies. Our data showed for the first time that AA substitutions located outside the ‘a’ determinant could also impair the antibody binding capacity to SHBs, which was evidenced not only in in vitro experiments but also in patients carrying these mutants with lower serum HBsAg levels.
A third potential mechanism is that some mutants, especially the truncated mutations (e.g., sW182*), may lead to ER stress-induced oxidative DNA damage triggered by the unfolded protein response, which in turn could result in HBV genomic instability and reduction/loss of HBV DNA levels. Pollicino et al. reported that the sW182* mutation induced retention of truncated S protein in the perinuclear ER and was associated with lower HBV transcript levels due to decreased stability, but without impact on HBV replication [25]. Accordingly, our study on patients showed that the sW182* mutant was more prevalent in the HBsAg low group than that in the high group (Fig. 1C)
We also found that most newly identified mutations were associated with lower extracellular HBV DNA levels. However, such mutants usually coexisted with low levels of WT sequence as evidenced by heterogeneity at mutational sites. This raises the possibility that WT S protein in co-infected cells could trans-complement and rescue HBV mutant virion secretion. This possibility is supported by experimental evidence from several previous reports showing that the mutant virion secretion could be rescued efficiently (i.e., sP127S) or moderately (i.e., sW172*) by co-expression of WT surface protein [13]. We also showed that impaired virion secretion for several mutants (sE2G, sC69* and sG145R) can be efficiently rescued by trans-complementation of the WT S protein (Fig. 6). Hence coexistence of WT and defective mutants may be a general survival requirement for some mutant variants. Lee et al. reported that sW182* was associated with enhanced cell growth, indicating that HBV might also gain survival benefit by up-regulating proliferation of infected cells [31]. However, additional studies are needed to unravel the driving forces that allow the emergence and survival of these mutants in vivo.
A limitation of our study is that we focused exclusively on HBeAg-positive genotype C HBV infected patients. However, some of the mutations we studied also occur in other genotypes. For example, in our previous study of 143 HBV genotype B sequences, two samples have the sL95W substitution (Supplementary Table 4) [9]. The sC69* also occurs in genotype D [32]. Several mutations including sC69*, sL95W and sL98V were also identified in 60 samples of HBeAg-negative CHB patients (Supplementary Table 5) [3]. Another limitation is that we focused on the S region of the HBV genome. Mutations in other regions such as preC/basal core promoter (BCP) and preS1/S2 could also influence serum HBsAg levels. Indeed, Yan et al, reported that HBV BCP A1762T/G1764A mutations might be associated with low HBsAg levels [17], and Pollicino et al. reported that the deletion and point mutations in preS1/S2 could restrict HBsAg secretion [25]. More investigations will be needed to further explore these factors.
In conclusion, we have identified several new HBV S protein substitutions (sE2G, sL21S, sR24K, sC69*, sL95W and sL98V) outside of the ‘a’ determinant of SHBs that are associated with lower serum HBsAg and HBV DNA levels. Our in vitro experiments suggest that mutations within the S protein may impair virion secretion (sE2G, sL95W and sL98V), change antigenicity (sL21R, sL95W and sL98V), or impact HBV replication (sC69*) thereby decreasing detectable levels of HBsAg. Mutants unable to produce functional HBsAg can be rescued by WT protein expression, which presumably reflects a requirement for their spread and survival in vivo. Hence, the factors affecting circulating HBsAg level and HBsAg detection are varied and caution is warranted when interpreting clinical significance of serum HBsAg levels.
Supplementary Material
Lay summary.
HBsAg is a major viral protein of hepatitis B virus (HBV) secreted into patient serum and its quantification value serves as an important virological marker for the evaluation of chronic HBV infection and antiviral response. We found a few novel amino acid substitutions in HBsAg associated with lower serum HBsAg and HBV DNA levels. These different substitutions might impair virion secretion, change HBsAg antigenicity or impact HBV replication, which all could result in decreased detectable levels of serum HBsAg. The factors affecting circulating HBsAg level and HBsAg detection are varied and caution is needed when interpreting clinical significance of serum HBsAg levels.
Acknowledgments
This study was supported by the financial grants from Major Science and Technology Special Project of China Thirteenth and Twelfth Five-year Plans (2016ZX10002003 and 2016ZX10002011) and the Robertson Foundation (to E.M., V.L.D.T. and X.W.). We thank the fellowship from China Scholar Council (to K.X.), a Merck Postdoctoral Fellowship (to E.M.), NIH fellowship (5 F32 DK107164-02) (to E.M.), Bristol-Myers Squibb Postdoctoral Fellowship (X.W) and German Research Foundation (to V. L. D.T.). We also thank Ying-pu Yu for discussion and excellent technical assistance and Dr. Xue-qing Qian from Shanghai Runda Rongjia Bio-technology Company Ltd. for the valuable discussion on qHBsAg assays.
List of abbreviations
- HBV
Hepatitis B virus
- HBsAg
hepatitis B surface antigen
- CHB
chronic hepatitis B
- LHBs
large hepatitis B surface protein
- MHBs
middle hepatitis B surface protein
- SHBs
small hepatitis B protein
- MHR
major hydrophilic region
- AA
amino acid
- qHBsAg
quantitative hepatitis B surface antigen
- WT
wild type
- RT
reverse transcriptase
- YMHD
tyrosine-methionine-histidine-aspartate
- HA
hemagglutinin
- ELISA
enzyme-linked immunosorbent assay
- IU/ml
international units per milliliter
- TM
transmembrane
- IQR
interquartile range
- BCP
basal core promoter
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: none of the authors has any conflict of interest.
Author contributions:
Study concept and design: Tong Li, Kuan-hui Xiang
Acquisition of data (sample collection, processing and database establishment etc): Kuan-hui Xiang, Hai Ding, Ya-qin Peng, Ming-ze Su, Yao Li, Xue-en Liu
Analysis and interpretation: Kuan-hui Xiang, Tong Li, Eleftherios Michailidis, Viet Loan Dao Thi, Xian-fang Wu, William M. Schneider
Statistical analysis: Kuan-hui Xiang
Drafting of the manuscript: Kuan-hui Xiang
Critical revision of the manuscript for important intellectual content: Tong Li, Eleftherios Michailidis, Hui Zhuang, William M. Schneider, Viet Loan Dao Thi, Xian-fang Wu, Charles M. Rice
References
- 1.European Association For The Study Of The L. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Liang TJ, Block TM, McMahon BJ, Ghany MG, Urban S, Guo JT, et al. Present and future therapies of hepatitis B: From discovery to cure. Hepatology. 2015;62:1893–1908. doi: 10.1002/hep.28025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding H, Liu BM, Zhao CY, Yang JX, Yan CH, Yan L, et al. Amino acid similarities and divergences in the small surface proteins of genotype C hepatitis B viruses between nucleos(t)ide analogue-naive and lamivudine-treated patients with chronic hepatitis B. Antiviral Res. 2014;102:29–34. doi: 10.1016/j.antiviral.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg BS, Alter HJ, Visnich S. A “New” Antigen in Leukemia Sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 5.Whitehead TP, Thorpe GHG, Carter TJN, Groucutt C, Kricka LJ. Enhanced Luminescence Procedure for Sensitive Determination of Peroxidase-Labeled Conjugates in Immunoassay. Nature. 1983;305:158–159. [Google Scholar]
- 6.Deguchi M, Yamashita N, Kagita M, Asari S, Iwatani Y, Tsuchida T, et al. Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J Virol Methods. 2004;115:217–222. doi: 10.1016/j.jviromet.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Chan HLY, Wong VWS, Tse AML, Tse CH, Chim AML, Chan HY, et al. Serum hepatitis B surface antigen quantitation can reflect hepatitis B virus in the liver and predict treatment response. Clinical Gastroenterology and Hepatology. 2007;5:1462–1468. doi: 10.1016/j.cgh.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Alavian SM, Carman WF, Jazayeri SM. HBsAg variants: diagnostic-escape and diagnostic dilemma. J Clin Virol. 2013;57:201–208. doi: 10.1016/j.jcv.2012.04.027. [DOI] [PubMed] [Google Scholar]
- 9.Su MZ, Xiang KH, Li Y, Li YT, Deng J, Xu XZ, et al. Higher detection rates of amino acid substitutions in HBV reverse transcriptase/surface protein overlapping sequence is correlated with lower serum HBV DNA and HBsAg levels in HBeAg-positive chronic hepatitis B patients with subgenotype B2. Infection Genetics and Evolution. 2016;40:275–281. doi: 10.1016/j.meegid.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen T, Thompson AJ, Bowden S, Croagh C, Bell S, Desmond PV, et al. Hepatitis B surface antigen levels during the natural history of chronic hepatitis B: a perspective on Asia. J Hepatol. 2010;52:508–513. doi: 10.1016/j.jhep.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Thibault V, Laperche S, Akhavan S, Servant-Delmas A, Belkhiri D, Roque-Afonso AM. Impact of hepatitis B virus genotypes and surface antigen variants on the performance of HBV real time PCR quantification. J Virol Methods. 2009;159:265–270. doi: 10.1016/j.jviromet.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Verheyen J, Neumann-Fraune M, Berg T, Kaiser R, Obermeier M. The detection of HBsAg mutants expressed in vitro using two different quantitative HBsAg assays. Journal of Clinical Virology. 2012;54:279–281. doi: 10.1016/j.jcv.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Warner N, Locarnini S. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology. 2008;48:88–98. doi: 10.1002/hep.22295. [DOI] [PubMed] [Google Scholar]
- 14.Liang X, Cheng J, Sun Y, Chen X, Li T, Wang H, et al. Randomized, three-arm study to optimize lamivudine efficacy in hepatitis B e antigen-positive chronic hepatitis B patients. J Gastroenterol Hepatol. 2015;30:748–755. doi: 10.1111/jgh.12835. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang H. Guideline on prevention and treatment of chronic hepatitis B in China (2005) Chinese Medical Journal. 2007;120:2159–2173. [PubMed] [Google Scholar]
- 16.Jaroszewicz J, Calle Serrano B, Wursthorn K, Deterding K, Schlue J, Raupach R, et al. Hepatitis B surface antigen (HBsAg) levels in the natural history of hepatitis B virus (HBV)-infection: a European perspective. J Hepatol. 2010;52:514–522. doi: 10.1016/j.jhep.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Yan CH, Zhao CY, Ding H, Peng YQ, Jin PY, Yan L, et al. Hepatitis B virus basal core promoter mutations A1762T/G1764A are associated with genotype C and a low serum HBsAg level in chronically-infected HBeAg-positive Chinese patients. Antiviral Res. 2012;96:108–114. doi: 10.1016/j.antiviral.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Hao R, Xiang KH, Peng YQ, Hou JL, Sun J, Li Y, et al. Naturally occurring deletion/insertion mutations within HBV whole genome sequences in HBeAg-positive chronic hepatitis B patients are correlated with baseline serum HBsAg and HBeAg levels and might predict a shorter interval to HBeAg loss and seroconversion during antiviral treatment. Infection Genetics and Evolution. 2015;33:261–268. doi: 10.1016/j.meegid.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Li WP, Warner N, Sozzi V, Yuen L, Colledge D, Li T, et al. Hepatitis B virus genotype C encoding resistance mutations that emerge during adefovir dipivoxil therapy: in vitro replication phenotype. Hepatology International. 2013;7:443–450. doi: 10.1007/s12072-012-9411-2. [DOI] [PubMed] [Google Scholar]
- 20.Shlomai A, Schwartz RE, Ramanan V, Bhatta A, de Jong YP, Bhatia SN, et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci USA. 2014;111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwei K, Tang X, Lok AS, Sureau C, Garcia T, Li J, et al. Impaired virion secretion by hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J Virol. 2013;87:2352–2357. doi: 10.1128/JVI.02701-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai DW, Mills C, Yu WQ, Yan R, Aldrich CE, Saputelli JR, et al. Identification of Disubstituted Sulfonamide Compounds as Specific Inhibitors of Hepatitis B Virus Covalently Closed Circular DNA Formation. Antimicrob Agents Chemother. 2012;56:4277–4288. doi: 10.1128/AAC.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aragri M, Alteri C, Battisti A, Di Carlo D, Minichini C, Sagnelli C, et al. Multiple Hepatitis B Virus (HBV) Quasispecies and Immune-Escape Mutations Are Present in HBV Surface Antigen and Reverse Transcriptase of Patients With Acute Hepatitis B. J Infect Dis. 2016;213:1897–1905. doi: 10.1093/infdis/jiw049. [DOI] [PubMed] [Google Scholar]
- 25.Pollicino T, Amaddeo G, Restuccia A, Raffa G, Alibrandi A, Cutroneo G, et al. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology. 2012;56:434–443. doi: 10.1002/hep.25592. [DOI] [PubMed] [Google Scholar]
- 26.Locarnini S, Bowden S. Hepatitis B surface antigen quantification: not what it seems on the surface. Hepatology. 2012;56:411–414. doi: 10.1002/hep.25732. [DOI] [PubMed] [Google Scholar]
- 27.Huang CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, et al. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J Hepatol. 2012;57:720–729. doi: 10.1016/j.jhep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Ye Q, Shang SQ, Li W. A new vaccine escape mutant of hepatitis B virus causes occult infection. Hum Vaccin Immunother. 2015;11:407–410. doi: 10.4161/21645515.2014.994461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locarnini S, Shouval D. Commonly Found Variations/Mutations in the HBsAg of Hepatitis B Virus in the Context of Effective Immunization Programs: Questionable Clinical and Public Health Significance. J Virol. 2014;88:6532–6532. doi: 10.1128/JVI.00234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu CC, Deng WY, Deng L, Cao L, Qin B, Li SX, et al. Amino Acid Substitutions at Positions 122 and 145 of Hepatitis B Virus Surface Antigen (HBsAg) Determine the Antigenicity and Immunogenicity of HBsAg and Influence In Vivo HBsAg Clearance. J Virol. 2012;86:4658–4669. doi: 10.1128/JVI.06353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SA, Kim K, Kim H, Kim BJ. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J Hepatol. 2012;56:63–69. doi: 10.1016/j.jhep.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 32.Betz-Stablein BD, Topfer A, Littlejohn M, Yuen L, Colledge D, Sozzi V, et al. Single-molecule sequencing reveals complex genomic variation of hepatitis B virus during 15 years of chronic infection following liver transplantation. J Virol. 2016 doi: 10.1128/JVI.00243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


