Abstract
Objectives
Glucocorticoids such as dexamethasone have pleiotropic effects, including desired antileukemic, anti-inflammatory or immunosuppressive effects, and undesired metabolic or toxic effects. The most serious adverse effects of dexamethasone among patients with acute lymphoblastic leukemia (ALL) are osteonecrosis and thrombosis. To identify inherited genomic variation involved in these serious adverse effects, we performed genome-wide association studies (GWAS) by analyzing 14 pleiotropic glucocorticoid phenotypes in 391 patients with ALL.
Methods
We used the Projection Onto the Most Interesting Statistical Evidence (PROMISE) integrative analysis technique to identify genetic variants associated with pleiotropic dexamethasone phenotypes, stratifying for age, sex, race, and treatment, and compared results to conventional single-phenotype GWAS. The phenotypes were: osteonecrosis, central nervous system toxicity, hyperglycemia, hypokalemia, thrombosis, dexamethasone exposure, body mass index, decreased growth trajectory, and levels of cortisol, albumin, asparaginase antibodies, and change in cholesterol, triglycerides, and low density lipoproteins after dexamethasone.
Results
The integrative analysis identified more pleiotropic SNP variants (p = 1.46 × 10−215), and these variants were more likely to be in gene regulatory regions (p = 1.22×10−6), than traditional single-phenotype GWAS. The integrative analysis yielded genomic variants (rs2243057 & rs6453253) in F2RL1, a receptor that functions in hemostasis, thrombosis, and inflammation, which were associated with pleiotropic effects, including osteonecrosis and thrombosis, and were in regulatory gene regions.
Conclusions
The integrative pleiotropic analysis identified risk variants for osteonecrosis and thrombosis not identified by single-phenotype analysis that may have importance for patients with underlying sensitivity to multiple dexamethasone adverse effects.
Keywords: Dexamethasone, Glucocorticoids, Acute Lymphoblastic Leukemia, Adverse Effects, Pleiotropy, Osteonecrosis, Thrombosis, Asparaginase
Introduction
Glucocorticoids have been used for decades in the treatment of many diseases, including acute lymphoblastic leukemia (ALL). Dexamethasone has become the preferred glucocorticoid over prednisone due to its greater antileukemic efficacy [1–3]. However, at current doses, more toxicity is observed in ALL patients treated with dexamethasone than prednisone, including infections [3], osteonecrosis [4], myopathy [5, 6], and central nervous system (CNS) toxicity [7].
Osteonecrosis is one of the most common and serious toxicities associated with ALL therapy, with frequencies ranging from 1 – 25% depending on the clinical trial [4, 8–14]. Osteonecrosis is defined as bone death resulting from poor blood supply [15]. Risk of osteonecrosis has been associated with the intensity of the glucocorticoid exposure, type of glucocorticoid (dexamethasone > prednisone) [4], age, sex [16], race [14], asparaginase antibodies [17], and genetic polymorphisms [8, 18–21]. Additionally, our group demonstrated that the top single nucleotide polymorphisms (SNPs) associated with osteonecrosis were also associated with other dexamethasone-related phenotypes, including low serum albumin and high cholesterol levels,[8] and lack of allergy to asparaginase [21]. These findings suggest that a comprehensive analysis of genetic predisposition to pleiotropic glucocorticoid-induced adverse effects might improve the power to identify genetic variants associated with glucocorticoid-induced osteonecrosis and other adverse effects. Moreover, association of the same variants with multiple phenotypes, e.g. osteonecrosis in addition to other phenotypes, might suggest common pathways that underlie more than one glucocorticoid-induced phenotype.
Herein we have implemented a method to use prior pharmacologic knowledge to perform an integrated analysis of SNPs associated with pleiotropic glucocorticoid-induced biological and clinical endpoints [22–24], with particular emphasis on the clinically important phenotypes of osteonecrosis and thrombosis, both of which can necessitate substantial clinical interventions [25–29]. We compared the results of the pleiotropic analysis with those obtained with a single-phenotype genome-wide association study (GWAS) approach.
Materials & Methods
Patients
Informed consent was obtained from the parents or the patient, and assent from the patient, where appropriate, and the study was approved by the Institutional Review Board. Children with newly diagnosed ALL (n = 498) were enrolled on a frontline protocol, St. Jude Total XV (ClinicalTrials.gov ID NCT00137111) [29] (Supplemental Digital Content Figure 1). This cohort has been previously reported in single phenotype association studies [8, 30].
Phenotypes
Patients were prospectively screened for osteonecrosis by MRI, as described previously [8]. Toxicities were prospectively graded using the National Cancer Institute Common Terminology Criteria for Adverse Events, Version 3.0 (Supplemental Digital Content Text, Supplemental Digital Content Table 1). We evaluated all reported adverse events and identified those that were likely at least partly due to glucocorticoids; these included osteonecrosis, CNS toxicity (not related to high-dose or intrathecal methotrexate), hyperglycemia, hypokalemia, and thrombosis. Because glucocorticoids are known to cause weight gain and decreased growth trajectory, [31–34] we used measures of height and weight (at least monthly per patient) to estimate body mass index (BMI) and the decrease in growth trajectory. In addition, each patient had serial blood samples collected on day 1 of week 7 and week 8 (after 7 continuous days of dexamethasone) of the continuation phase, for assessment of plasma dexamethasone pharmacokinetics, serum lipids, albumin and cortisol levels (See Supplemental Digital Content Text for details) [8, 17, 30]. We have previously shown an association of dexamethasone and asparaginase exposure on osteonecrosis [8, 17, 30], and thus we also analyzed the AUC of antibodies against Elspar (asparaginase) during the first 35 weeks of therapy. There were no other pharmacologic measures routinely available for these patients that were related to glucocorticoids or these likely glucocorticoid-induced adverse effects.
Body mass index (BMI) was calculated from height and weight at day 15 of the consolidation phase of therapy. Height was measured each week, and growth trajectory was calculated by using a linear fit of the change in height during the first twelve months of therapy for each subject. For each subject, the slope of the linear fit was used as the phenotype in the analysis.
Genotyping
Germline DNA was extracted from blood after remission was achieved. Genotypes were generated and calls made as described [35], using GeneChip Human Mapping 500K Array sets or the Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA), Infinium HumanExome BeadChip (Illumina, Inc, San Diego, CA), and using Illumina Golden Gate platform (SNP Center, Johns Hopkins University). SNPs were filtered for call rate >95% and minor allele frequency ≥1%.
Statistical analysis
Each SNP was tested for association with each phenotype in single-phenotype analyses by Spearman-type correlation, treating genotype as AA>AB>BB. The PROMISE approach uses data defining the biological inter-relationships of the phenotypes with one another. The PROMISE statistic for each SNP was calculated as a weighted sum of each correlation statistic for that SNP (Supplemental Digital Content Figure 2) [22, 23]. The signs and magnitudes of the linear combination coefficients were predetermined according to the relationships between pairs of phenotypes. The null distributions were approximated by 4,000 permutations stratified by age (greater or less than median), sex (male vs female), risk group treatment arm (low vs standard/high risk) and ancestry group (white, black, other defined by SNP data as we have previously described) [21], for a total of 24 strata. For the pleiotropic PROMISE analyses, we used 4 different analyses, described in detail in Supplemental Digital Content Text. Our primary analysis is termed PR14; PR14 placed greatest emphasis on osteonecrosis and thrombosis, which were weighted twice as heavily as the other phenotypes (1/7 vs 1/14) in order to prioritize these clinical phenotypes. In PR14, the three measurements of changes in lipid levels were each weighted one third of other phenotypes (1/42 vs 1/14), in order to reduce the chances of finding only lipid-related associations, and PR14 included dexamethasone AUC as one of the phenotypes of interest. The second PROMISE analysis, PR13, was identical but did not include dexamethasone AUC as one of the phenotypes of interest, given that dexamethasone can be considered as either a “biomarker” phenotype or a determinant of other phenotypes. The third PROMISE analysis, EW13, was identical to PR13 but used equal weights for all 13 phenotypes; the fourth PROMISE analysis, EW14, included all 14 phenotypes and all were equally weighted. A comparison of these four analyses is shown in Supplemental Digital Content Figure 3 and the SNPs identified by any of the four PROMISE analyses are included in Supplemental Digital Content Table 2. The similarity of SNPs identified by these four analyses led us to focus on the PR14 analysis throughout the remainder of the manuscript. The p-values for each single-phenotype association statistic and the linear combination of those statistics were determined by permutation of the assignment of phenotype data to genotype data. In this study, we used the hybrid-permutation algorithm [36] to compute p-values.
The p value threshold for significance for each single-phenotype GWAS and for each pleiotropic PROMISE GWAS was determined by the profile information (Ip) criterion, which sets the p-value threshold that best balances false-positive and false-negative errors [36]. Supplemental Digital Content Table 3 contains all PR14 Ip-selected pleiotropic SNPs, as well as their associations with each individual phenotype. The Ip selection criteria was also used to generate lists of SNPs significant for each single-phenotype GWAS (shown for osteonecrosis, thrombosis, and CNS toxicity) in Supplemental Digital Content Tables 4 – 6. An R package that implements the PROMISE procedure is available from our website (http://stjuderesearch.org/site/depts/biostats/promise). A stratified Spearman-type rank-based association statistic was used to evaluate the association of each individual phenotype with the genotype of each SNP, as previously described [22, 23] (see Supplemental Digital Content Text for details). The individual phenotype association statistics were stratified by risk (standard and high vs. low), ancestry (black, white, other), gender (male/female), and age (<10, >10 years).
Results
We identified 14 clinical phenotypes likely related to glucocorticoid effects (Supplemental Digital Content Table 1), then tested the pairwise correlation between all combinations of the 14 phenotypes and found the expected associations (Figure 1). The phenotypes consisted of osteonecrosis, CNS toxicity, hyperglycemia, hypokalemia, thrombosis, plasma dexamethasone exposure, body mass index (BMI), serum cortisol level, albumin level, growth trajectory, asparaginase antibody exposure, and change in serum cholesterol, triglycerides, and low density lipoproteins during dexamethasone treatment. The strongest associations were found between changes in serum LDL and total cholesterol (p=4.9 × 10−61), albumin level and change in cholesterol (p=5.9 × 10−28), albumin level and dexamethasone AUC (p=6.3 × 10−25), and albumin level and change in LDL (p=2.3 × 10−24, Supplemental Digital Content Figure 4). Osteonecrosis was associated with the increase in serum cholesterol during dexamethasone treatment (p=0.0027), hyperglycemia (p=0.01) and thrombosis (p=0.03, Supplemental Digital Content Figure 4).
Figure 1.
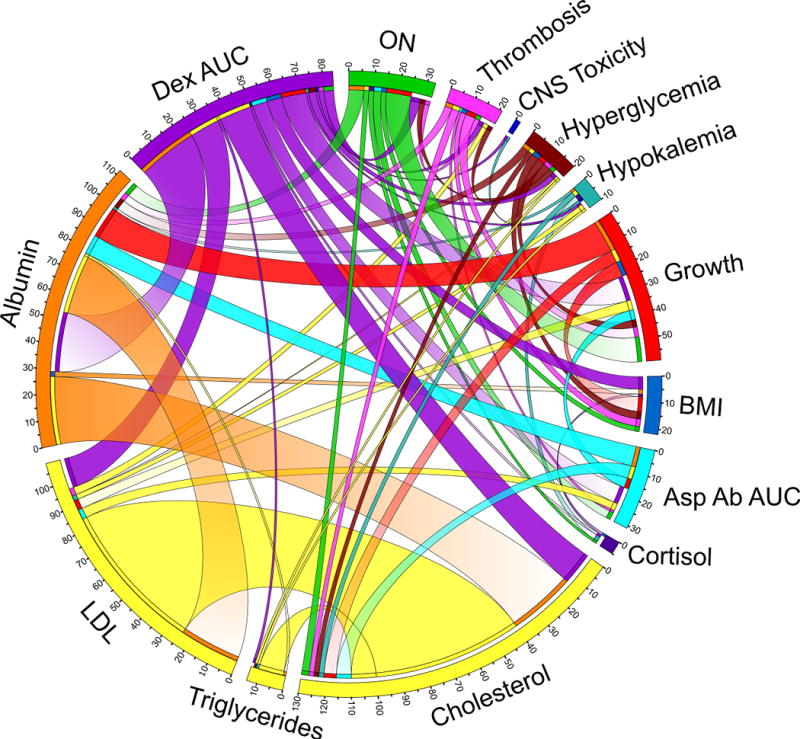
Circos plot of pleiotropic phenotypes. Connections shown between two phenotypes have a Spearman rank correlation p<0.2. The width of the connection corresponds to the −log10(p-value) for the correlation, which is represented by numbers around the circle. The segment is faded if the correlation is inverse (e.g. higher dexamethasone AUC with lower albumin). ON, osteonecrosis. CNS, central nervous system. Growth, decreased growth trajectory. BMI, body mass index. Asp Ab AUC, asparaginase antibody area under the curve. LDL, low density lipoproteins. Dex AUC, dexamethasone area under the curve. Detailed descriptions of the phenotypes are included in the Materials and Methods and in Supplemental Digital Content Figure 5.
There was substantial variability in the distributions and frequencies of these phenotypes (Supplemental Digital Content Figure 5). Each SNP was tested for association with each phenotype in conventional single-phenotype GWAS, and the Projection onto the Most Interesting Statistical Evidence (PROMISE) technique [22, 23] was used to select SNPs associated with pleiotropic phenotypes, weighted toward finding SNPs associated with osteonecrosis and CNS toxicity (the two most clinically important phenotypes) in our main analysis, termed PR14 (see Materials and Methods). A total of 610 SNPs met the PROMISE information profile (Ip) p-value threshold of less than 7.3×10−4 in the PR14 analysis (Figure 2, Supplemental Digital Content Table 3). Five of the top ten SNPs (Table 1) are on chromosome 12 near keratin genes, of these five, four were in linkage disequilibrium (LD) with SNPs in a glucocorticoid receptor binding site (rs830376, rs389523, rs112746594, r2≥0.8, Haploreg v4.1).
Figure 2.
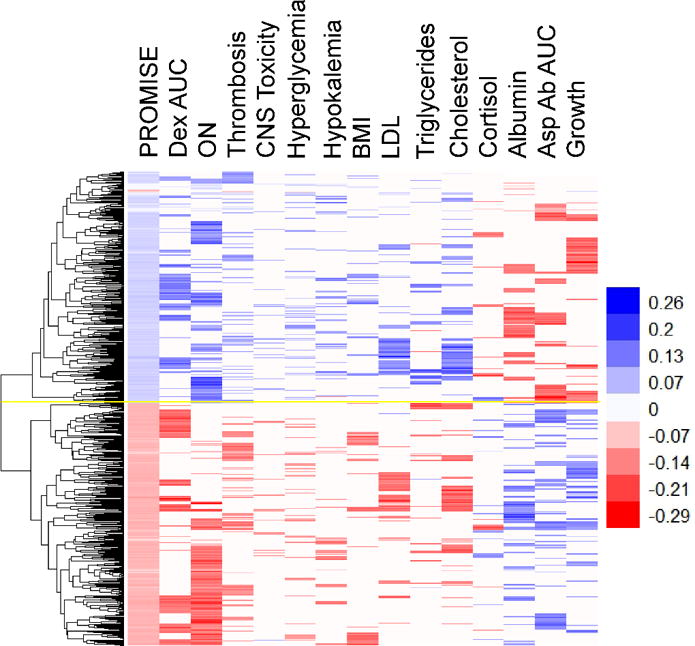
Heat map of 610 PROMISE SNPs (with p<7.3×10−4), selected as significant in the PR14 analysis (see Materials and Methods for details). Each row is a SNP (n=610) and each column is a phenotype. Associations of SNP variants with the indicated phenotype with p<0.05 are shown in color, with red indicating an effect size of less than one and blue indicating an effect size greater than one. SNPs with similar associations across phenotypes are clustered by the dendogram at left. Yellow line indicates separation of negative associations (bottom) and positive associations (top). PROMISE (Projection onto the Most Interesting Statistical Evidence) statistic. ON, osteonecrosis. CNS, central nervous system toxicity. Growth, decreased growth trajectory. BMI, body mass index. Asp Ab AUC, asparaginase antibody area under the curve. LDL, concentration of low density lipoproteins in serum. Dex AUC, plasma dexamethasone area under the curve (week 7). See Supplemental Digital Content Table 1, Supplemental Digital Content Figure 5, and the Materials and Methods section for more details on each phenotype.
Table 1.
Top 10 SNPs associated with pleotropic glucocorticoid effects ranked by PROMISE PR14 p-value.
| PR rank | SNP | Ch | Gene | Location | Alleles | MAF | RDB score | PR14 p-value | PR14 stat | ON p-value | ON stat |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs4703882 | 5 | ATP6AP1L(dist=100900) | intergenic | A/T | 17.05% | 3a | 2.54×10−7 | 0.084 | 0.019 | 0.115 |
| 2 | rs381518 | 12 | KRT75 | upstream | C/A | 37.63% | 5 | 2.01×10−6 | 0.078 | 0.109 | 0.078 |
| 3 | rs3743712 | 16 | TK2 | UTR3 | A/G | 9.82% | 1f | 2.30×10−6 | 0.072 | 0.167 | 0.064 |
| 4 | rs401926 | 12 | KRT75 | SYN | G/A | 37.94% | 4 | 2.74×10−6 | 0.077 | 0.122 | 0.075 |
| 5 | rs7743593 | 6 | SLC16A10 | DS | T/C | 22.95% | 6 | 8.79×10−6 | 0.071 | 0.021 | 0.115 |
| 6 | rs10869729 | 9 | PCSK5 | UTR3 | T/C | 10.27% | No Data | 9.76×10−6 | 0.069 | 0.003 | 0.141 |
| 7 | rs298109 | 12 | KRT75 | NS | C/T | 38.26% | 5 | 1.17×10−5 | 0.073 | 0.125 | 0.077 |
| 8 | rs13218329 | 6 | KIF25-AS1 | DS | A/G | 14.57% | 5 | 1.26×10−5 | 0.071 | 0.038 | 0.100 |
| 9 | rs4107159 | 12 | KIF25-AS1(dist=5019) | intergenic | G/A | 49.10% | 6 | 1.46×10−5 | 0.072 | 0.010 | 0.124 |
| 10 | rs298120 | 12 | KRT6A | intronic | C/T | 39.44% | 5 | 1.67×10−5 | 0.070 | 0.106 | 0.078 |
PR14, main PROMISE analysis (see Methods for details). Ch, chromosome. UTR3, 3′ untranslated region. SYN, synonymous. NS, nonsynonymous. The major and minor alleles are listed in the Alleles column, (major is first, minor is second, and risk allele is in bold). MAF, minor allele frequency. Stat, statistic (effect size) for the minor allele. A negative statistic means the minor allele is associated with less ON, while a positive statistic means the minor allele is associated with more ON. ON, osteonecrosis. RDB, RegulomeDB.
As expected, the top-ranked SNPs by PROMISE (PR14) included a greater number of pleiotropic SNPs than did the individual phenotype GWASs (Supplemental Digital Content Table 7, Supplemental Digital Content Figure 6). PROMISE (PR14) exhibited a greater power to identify pleiotropic SNPs according to the pleiotropic enrichment index (PLEI, see Supplemental Digital Content Text, Supplemental Digital Content Figure 7), which was 3309.9, significantly exceeding the PLEI for all of the single-phenotype analyses, which ranged from 895.3 to 2506.3 (p = 1.46 × 10−215). For example, among the 5000 top ranked SNPs, PR14 identified 2,354 SNPs of pleiotropy degree 2 (SNPs associated with two or more phenotypes in single-phenotype GWASs), 621 SNPs of pleiotropy degree 3 (SNPs associated with three or more phenotypes in single-phenotype GWASs), 50 SNPs of pleiotropy degree 4 (SNPs associated with 4 or more phenotypes), and 7 SNPs of degree 5 (SNPs associated with 5 or more phenotypes). At the same level of top 5000 SNPs, none of the single-phenotype GWASs identified more than 1,744 SNPs, 211 SNPs, 16 SNPs, or 1 SNP of pleiotropy degrees 2, 3, 4, and 5, respectively (Supplemental Digital Content Table 7). PROMISE (PR14) showed greater enrichment of SNPs predicted to be eQTLs (RegulomeDB [37] score of 1) than did single-phenotype GWAS for ON, thrombosis or CNS toxicity (p = 1.22 × 10−6 Figure 3).
Figure 3.
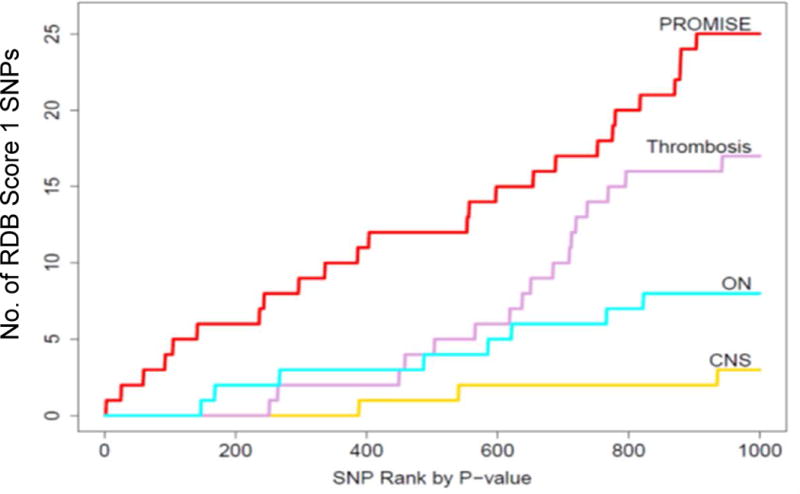
PROMISE identified more SNPs with a RegulomeDB score of 1 than single phenotype analyses (p = 1.22 × 10−6). Results shown for PR14 (see Methods for details). Cumulative number of SNPs with a RegulomeDB score of 1 for each analysis (PROMISE, red; ON, blue; thrombosis, pink; CNS toxicity, yellow. See Supplemental Digital Content Text for statistical details.
We used the RegulomeDB score to prioritize the 610 PR14 SNPs selected by the Ip criterion. The top RegulomeDB score (1b) was annotated to a SNP downstream of F2RL1 (rs6453253), which was nearby another PR14-selected SNP in the intron of F2RL1 with a score of 1f (rs2243057, Table 2). Both of these SNPs were in the regulatory regions indicated by H3K27 acetylation in osteoblast and HUVEC cell lines (Supplemental Digital Content Figure 8), which were the most relevant cell lines available for the osteonecrosis phenotype. One of the F2RL1 SNPs, rs6453253, was also in a glucocorticoid receptor (NR3C1) binding site (Supplemental Digital Content Figure 8). The A allele of rs2243057 was associated with an increased risk of ON (p=0.0069, Figure 4A) and thrombosis (p=0.01, Figure 4B), with lower albumin levels (p=0.04), and a greater increase in cholesterol (p=0.003) and triglycerides (p=0.0002) from week 7 to week 8 of continuation therapy (Figure 4C–F). The G allele of rs6453253 was also associated with increased risk of osteonecrosis (p=0.042), greater increase in cholesterol (p=0.01) and higher dexamethasone exposure (p=0.006). Both SNPs have high minor allele frequencies (MAF) of 48% in our patients and were in positive LD (R2= 0.84). Both F2RL1 SNPs were eQTLs for F2RL1 expression in liver and whole blood, and for F2R, which encodes the receptor for thrombin in skeletal muscle (Supplemental Digital Content Figure 9) [38].
Table 2.
Top PR14-selected SNPs associated with pleiotropic glucocorticoid effects ranked by RegulomeDB score.
| rsID | Gene | Location | Alleles | MAF | RDB score | PR14 p-value | PR14 stat | PR14 rank | ON p-value | ON stat |
|---|---|---|---|---|---|---|---|---|---|---|
| rs6453253 | F2RL1 | DS | C/G | 47.45% | 1b | 2.77 ×10−4 | 0.060 | 257 | 0.043 | 0.098 |
| rs2425047 | EIF6 | intronic | A/C | 20.81% | 1b | 8.67 ×10−5 | 0.063 | 91 | 0.063 | 0.091 |
| rs12412962 | SFXN4 | intronic | T/A | 30.15% | 1d | 7.63 ×10−5 | 0.064 | 84 | 0.21 | 0.061 |
| rs3743712 | TK2 | UTR3 | A/G | 9.82% | 1f | 2.30 ×10−6 | 0.072 | 3 | 0.17 | 0.064 |
| rs2243057 | F2RL1 | intronic | A/G | 47.92% | 1f | 2.68 ×10−5 | −0.069 | 20 | 0.007 | −0.132 |
| rs13334391 | TK2 | intronic | A/G | 11.47% | 1f | 4.81 ×10−5 | 0.062 | 54 | 0.31 | 0.047 |
| rs17668734 | SFXN4 | intronic | G/A | 29.46% | 1f | 1.01 ×10−4 | 0.062 | 108 | 0.21 | 0.060 |
| rs6060341 | MMP24 | UTR3 | A/G | 21.07% | 1f | 1.17 ×10−4 | 0.061 | 123 | 0.063 | 0.091 |
| rs4680340 | CCNL1 | DS | T/C | 28.81% | 1f | 1.47 ×10−4 | 0.062 | 146 | 0.10 | 0.083 |
| rs11545619 | ZNF83 | NS | T/C | 9.10% | 1f | 3.13 ×10−4 | 0.058 | 290 | 0.012 | 0.122 |
| rs1736959 | intergenic | C/T | 25.07% | 1f | 3.69 ×10−4 | 0.057 | 332 | 0.10 | 0.082 | |
| rs9912773 | STAT3 | intronic | C/G | 25.76% | 1f | 3.90 ×10−4 | 0.057 | 346 | 0.044 | 0.098 |
| rs10190226 | PLCL1 | intronic | C/T | 33.12% | 1f | 4.31 ×10−4 | −0.058 | 381 | 0.002 | −0.150 |
| rs3935870 | DIP2B | intronic | C/T | 34.29% | 1f | 4.39 ×10−4 | 0.056 | 387 | 0.022 | 0.112 |
| rs2891577 | UBE2S | intronic | T/C | 9.31% | 1f | 5.41 ×10−4 | 0.055 | 480 | 0.23 | 0.059 |
| rs1632988 | intergenic | G/A | 25.27% | 1f | 5.47 ×10−4 | 0.056 | 482 | 0.15 | 0.072 | |
| rs2297789 | EIF6 | intronic | A/T | 20.41% | 1f | 5.89 ×10−4 | 0.055 | 521 | 0.10 | 0.080 |
| rs3007421 | PLEKHG5 | intronic | G/A | 16.67% | 1f | 6.57 ×10−4 | 0.054 | 573 | 0.47 | 0.036 |
| rs4665328 | OTOF | intronic | T/C | 44.80% | 1f | 7.05 ×10−4 | 0.055 | 599 | 0.31 | 0.050 |
The major and minor alleles are listed in the Alleles column, (major is first, minor is second, and risk allele is in bold). DS, downstream. NS, nonsynonymous. UTR3, 3′ untranslated region. MAF, minor allele frequency. PR14, PROMISE, based on PR14 analysis (see Methods for details). ON, osteonecrosis. Stat, statistic (effect size) for the minor allele. A negative statistic means the minor allele is associated with less ON, while a positive statistic means the minor allele is associated with more ON. RDB, RegulomeDB
Figure 4.
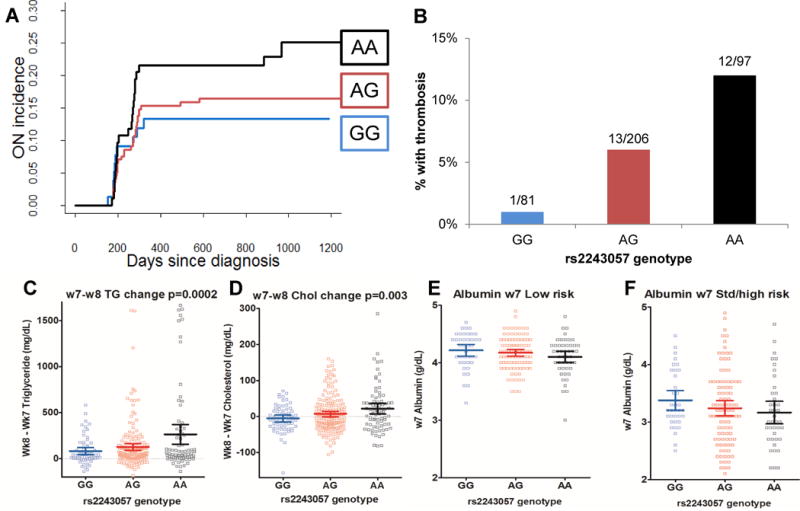
The A allele of F2RL1 rs2243057 is associated with pleotropic phenotypic effects phenotypes. A, Cumulative incidence of osteonecrosis (ON) plotted by rs2243057 genotype. B, Percentage of patients with thrombosis by rs2243057 genotype. Numbers above the column indicate cases over total number of patients with the indicated genotype. C, Change in serum triglycerides (TG) from week 7 to week 8 by rs2243057 genotype. D, Change in serum cholesterol (Chol) from week 7 to week 8 by rs2243057 genotype. E, Week 7 serum albumin by rs2243057 genotype in low risk patients. F, Week 7 serum albumin by rs2243057 genotype in standard/high risk (Std/high) patients. Mean is indicated by dark horizontal line, whiskers indicate 95% confidence interval, squares indicate raw data.
Another SNP highly ranked in the PR14 PROMISE analysis was rs10869729 in the 3′ UTR of PCSK5. This non-synonymous variant was associated with ON (p=0.003), lower albumin (p=0.003), decreased growth trajectory (p=0.0002), and hyperglycemia (p=0.02). Variants in this gene have previously been associated with many traits, including levels of LDL, cholesterol, and triglycerides [39], glucose tolerance in diabetics [40] and height [41–43].
An intriguing PR14 PROMISE variant (Supplemental Digital Content Table 3) associated with dexamethasone pharmacokinetics but not osteonecrosis was rs709063, near ZNF467 and SSPO. The T allele was associated with six phenotypes with p<0.05: increased dexamethasone AUC (p=3.77×10−4), lower albumin (p=0.008), lower asparaginase antibody AUC (p=0.02), thrombosis (p=0.02), CNS toxicity (p=0.03), and hyperglycemia (p=0.03). The ZNF467 gene encodes a cofactor that promotes adipocyte differentiation and suppresses osteoblast differentiation [44], while the SSPO gene encodes a glycoprotein of the thrombospondin family secreted in the cerebrospinal fluid that interacts with LDL during brain development [45].
Using a single-phenotype GWAS with osteonecrosis as the primary phenotype, 645 SNPs were selected at the Ip threshold of p<6.4×10−4, 49 (7.6%) of which were also selected in the PR14 PROMISE analysis (Supplemental Digital Content Table 4). A single-phenotype GWAS for thrombosis revealed 794 SNPs that were selected at the Ip threshold of p<5.6×10−4, 11 (1.4%) of which were also selected in the PR14 PROMISE analysis (Supplemental Digital Content Table 5). There were 22 SNPs that reached the conventional GWAS p-value threshold of p<5×10−8. In a single-phenotype GWAS for CNS toxicity, there were 1146 SNPs selected with the information profile threshold p<5.5×10−4, none of which were selected in the PR14 PROMISE analysis (Supplemental Digital Content Table 6). This absence of overlap may be attributable to the fact that the PR14 PROMISE analysis is driven by multiple phenotypes that show relatively weak association of CNS toxicity (Figure 1). There were 38 SNPs that reached the conventional GWAS p-value threshold of p<5×10−8 for CNS toxicity. The top two SNPs were located in an intron of the RERE gene, which encodes a nuclear receptor expressed in the brain; deficiency of this gene in a mouse model leads to abnormal brain development [46].
Discussion
Pleiotropy, the effect of one variant on multiple traits, is commonly observed in complex diseases [47]. Cancer clinical trials, in which multiple related toxicity phenotypes are measured prospectively and genome-wide genetic analysis is performed, provide an excellent opportunity to discover pleiotropic SNPs. Among several available methods for assessing pleiotropy [22, 23, 43, 48–52], we selected the PROMISE technique, which incorporates prior biological knowledge, to identify SNPs associated with pleiotropic adverse effects of dexamethasone.
Using PROMISE, we were able to discover a larger number of pleiotropic SNPs for glucocorticoid effects that were in regulatory regions than we could using traditional single-phenotype GWAS. PROMISE leveraged information from multiple phenotypes that may be related to many proposed mechanisms of osteonecrosis, including intravascular thrombotic occlusion, marrow fat hypertrophy [53–55], osteocyte and/or endothelial cell apoptosis, hypercoagulability, arteriolar occlusion via arteriopathy [56], and vasoconstriction of specific arteries and arterioles supplying bone [57]. Dexamethasone may contribute to osteonecrosis via its effects on lipids, coagulation, fibrinolysis, and direct toxic effects on vasculature and bone [58, 59]. It should be noted that correlations between some of these putatively related phenotypes have been observed previously [60, 61]. By accounting for pleiotropic effects, we have enhanced the probability of selecting for variants that exert their effect via common mechanisms, such as via glucocorticoid-responsive transcriptional machinery or via systemic pharmacokinetics of dexamethasone, and might be expected to impact on multiple downstream clinical phenotypes. In contrast, we acknowledge that not all adverse effects of glucocorticoids are expected to occur via common mechanisms, and the PROMISE approach may be less able to detect such non-pleiotropic adverse drug effects than do single-phenotype GWAS approaches. The fact that single-phenotype GWAS for CNS toxicity resulted in many SNPs that had genome-wide significance, with no overlap with PR14 PROMISE SNPs, suggest that common mechanisms are less likely to underlie CNS toxicity and other dexamethasone-related adverse effects. We weighted the phenotypes in the PR14 PROMISE analysis in order to prioritize our biological question of interest (i.e. to find genetic variants that underlie the risk of glucocorticoid adverse effects). Our choice of weights influenced the SNPs identified, as expected, but there was high concordance between the weighted (PR14) and unweighted (EW14) analysis (Supplemental Digital Content Figure 3 and Supplemental Digital Content Table 2). Thus, our primary reported results (PR14) are relatively robust against modifications (PR13, EW13, and EW14) of the PROMISE analysis
Use of the PROMISE technique enabled us to discover variants more likely to have biological function than SNPs identified by single phenotype GWASs (Figure 3). It has been demonstrated that GWAS hits for biomedical phenotypes are enriched within regulatory regions [62, 63]. Lower RegulomeDB scores indicate that a SNP is in a regulatory region, such as a DNAse hypersensitivity site, transcription factor binding site, promoter region, or is an expression quantitative trait locus (eQTL) [37]. SNPs associated with complex traits and pharmacologic phenotypes are more likely to be eQTLs than allele frequency-matched SNPs [64].
The variants identified by PROMISE (PR14) in or near F2RL1 may be tissue-specific eQTLs for both F2RL1 (PAR2) expression and F2R (PAR1) expression. PAR1 is activated by thrombin, and when thrombin binds the receptor on platelets, it stimulates clot formation. PAR2 is activated by trypsin and trypsinogen, and when activated, has proliferative and angiogenic effects on endothelial cells, which are key players in the arteriopathy that can initiate glucocorticoid-induced osteonecrosis in mice [56]. F2RL1 (PAR2) is expressed in bone, and F2rl1−/− mice have reduced repair in response to bone biopsy [65]. In a meta-analysis for platelet count that included more than 44,000 individuals, a SNP in the region between the F2RL1 and F2R genes was among the top-ranked SNPs (rs17568628, p=9.61×10−10) [66]; this low-frequency SNP (MAF 2.8%) was not associated with osteonecrosis or thrombosis in our cohort, but was associated with lower cortisol (p=0.007) and albumin levels (p=0.04).
We acknowledge several limitations of our study. First, there is a lack of a similarly characterized clinical trial cohort elsewhere, thus we lack an external replication group. Secondly, there may be contributions of other medications that the ALL patients receive during treatment to the phenotypes we attribute to dexamethasone. We have attempted to minimize this limitation by including the asparaginase exposure and restricting our analysis to toxicities that are most likely associated with dexamethasone exposure (i.e., we excluded CNS toxicities likely due to methotrexate). Finally, there is currently no method to compute the power of the PROMISE procedure, thus we have not included a power calculation for the technique.
In summary, we identified SNPs associated with the pleiotropic effects of dexamethasone that were enriched for regulatory regions of the genome. These variants may be of importance for identifying patients with underlying sensitivity to dexamethasone-induced effects prior to treatment.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health [P50 GM115279 to M.V.R., R01 CA142665 to M.V.R.], and P30 CA21765 to M.V.R.; and ALSAC.
Source of Funding: This work was supported by National Institutes of Health P50 GM115279 to M.V.R., R01 CA142665 to M.V.R., and P30 CA21765 to M.V.R.; and ALSAC.
Footnotes
Conflicts of Interests: The authors declare no conflicts of interest.
References
- 1.Veerman AJP, Hahlen K, Kamps WA, Van Leeuwen EF, De Vaan GAM, Solbu G, et al. High cure rate with a moderately intensive treatment regimen in non-high-risk childhood acute lymphoblastic leukemia: results of protocol ALL VI from the Dutch Childhood Leukemia Study Group. J Clin Oncol. 1996;14(3):911–8. doi: 10.1200/JCO.1996.14.3.911. [DOI] [PubMed] [Google Scholar]
- 2.Silverman LB, Gelber RD, Dalton VK, Asselin BL, Barr RD, Clavell LA, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Dana-Farber Consortium Protocol 91-01. Blood. 2001;97(5):1211–8. doi: 10.1182/blood.v97.5.1211. [DOI] [PubMed] [Google Scholar]
- 3.Moricke A, Reiter A, Zimmermann M, Gadner H, Stanulla M, Dordelmann M, et al. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008;111(9):4477–89. doi: 10.1182/blood-2007-09-112920. [DOI] [PubMed] [Google Scholar]
- 4.Vrooman LM, Stevenson KE, Supko JG, O’Brien J, Dahlberg SE, Asselin BL, et al. Postinduction dexamethasone and individualized dosing of Escherichia Coli L-asparaginase each improve outcome of children and adolescents with newly diagnosed acute lymphoblastic leukemia: results from a randomized study–Dana-Farber Cancer Institute ALL Consortium Protocol 00-01. J Clin Oncol. 2013;31(9):1202–10. doi: 10.1200/JCO.2012.43.2070. Epub 2013/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell CD, Richards SM, Kinsey SE, Lilleyman J, Vora A, Eden TO. Benefit of dexamethasone compared with prednisolone for childhood acute lymphoblastic leukaemia: results of the UK Medical Research Council ALL97 randomized trial. Br J Haematol. 2005;129(6):734–45. doi: 10.1111/j.1365-2141.2005.05509.x. [DOI] [PubMed] [Google Scholar]
- 6.Bostrom BC, Sensel MR, Sather HN, Gaynon PS, La MK, Johnston K, et al. Dexamethasone versus prednisone and daily oral versus weekly intravenous mercaptopurine for patients with standard-risk acute lymphoblastic leukemia: a report from the Children’s Cancer Group. Blood. 2003;101(10):3809–17. doi: 10.1182/blood-2002-08-2454. Epub 2003/01/18. [DOI] [PubMed] [Google Scholar]
- 7.Waber DP, Carpentieri SC, Klar N, Silverman LB, Schwenn M, Hurwitz CA, et al. Cognitive sequelae in children treated for acute lymphoblastic leukemia with dexamethasone or prednisone [see comments] J Pediatr Hematol Oncol. 2000 May-Jun;22(3):206–13. doi: 10.1097/00043426-200005000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Kawedia JD, Kaste SC, Pei D, Panetta JC, Cai X, Cheng C, et al. Pharmacokinetic, pharmacodynamic, and pharmacogenetic determinants of osteonecrosis in children with acute lymphoblastic leukemia. Blood. 2011;117(8):2340–7. doi: 10.1182/blood-2010-10-311969. Epub 2010/12/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhlen M, Moldovan A, Krull K, Meisel R, Borkhardt A. Osteonecrosis in paediatric patients with acute lymphoblastic leukaemia treated on Co-ALL-07-03 trial: a single centre analysis. Klinische Padiatrie. 2014;226(3):154–60. doi: 10.1055/s-0033-1358723. Epub 2014/04/09. [DOI] [PubMed] [Google Scholar]
- 10.te Winkel ML, Pieters R, Hop WC, de Groot-Kruseman HA, Lequin MH, van der Sluis IM, et al. Prospective study on incidence, risk factors, and long-term outcome of osteonecrosis in pediatric acute lymphoblastic leukemia. J Clin Oncol. 2011;29(31):4143–50. doi: 10.1200/JCO.2011.37.3217. [DOI] [PubMed] [Google Scholar]
- 11.Hyakuna N, Shimomura Y, Watanabe A, Taga T, Kikuta A, Matsushita T, et al. Assessment of corticosteroid-induced osteonecrosis in children undergoing chemotherapy for acute lymphoblastic leukemia: a report from the Japanese Childhood Cancer and Leukemia Study Group. Journal of pediatric hematology/oncology. 2014;36(1):22–9. doi: 10.1097/MPH.0000000000000039. Epub 2013/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rytting ME, Thomas DA, O’Brien SM, Ravandi-Kashani F, Jabbour EJ, Franklin AR, et al. Augmented Berlin-Frankfurt-Munster therapy in adolescents and young adults (AYAs) with acute lymphoblastic leukemia (ALL) Cancer. 2014 doi: 10.1002/cncr.28930. Epub 2014/07/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel B, Richards SM, Rowe JM, Goldstone AH, Fielding AK. High incidence of avascular necrosis in adolescents with acute lymphoblastic leukaemia: a UKALL XII analysis. Leukemia. 2008;22(2):308–12. doi: 10.1038/sj.leu.2405032. [DOI] [PubMed] [Google Scholar]
- 14.Mattano LA, Jr, Sather HN, Trigg ME, Nachman JB. Osteonecrosis as a complication of treating acute lymphoblastic leukemia in children: a report from the Children’s Cancer Group. J Clin Oncol. 2000;18(18):3262–72. doi: 10.1200/JCO.2000.18.18.3262. [DOI] [PubMed] [Google Scholar]
- 15.Lee JA, Farooki S, Ashman CJ, Yu JS. MR patterns of involvement of humeral head osteonecrosis. Journal of computer assisted tomography. 2002;26(5):839–42. doi: 10.1097/00004728-200209000-00030. Epub 2002/11/20. [DOI] [PubMed] [Google Scholar]
- 16.Arico M, Boccalatte MF, Silvestri D, Barisone E, Messina C, Chiesa R, et al. Osteonecrosis: An emerging complication of intensive chemotherapy for childhood acute lymphoblastic leukemia. Haematologica. 2003;88(7):747–53. [PubMed] [Google Scholar]
- 17.Liu C, Kawedia JD, Cheng C, Pei D, Fernandez CA, Cai X, et al. Clinical utility and implications of asparaginase antibodies in acute lymphoblastic leukemia. Leukemia. 2012;26(11):2303–9. doi: 10.1038/leu.2012.102. Epub 2012/04/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vora A. Management of osteonecrosis in children and young adults with acute lymphoblastic leukaemia. Br J Haematol. 2011;155(5):549–60. doi: 10.1111/j.1365-2141.2011.08871.x. Epub 2011/11/15. [DOI] [PubMed] [Google Scholar]
- 19.French D, Hamilton LH, Mattano LA, Jr, Sather HN, Devidas M, Nachman JB, et al. A PAI–1 (SERPINE1) polymorphism predicts osteonecrosis in children with acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2008;111(9):4496–9. doi: 10.1182/blood-2007-11-123885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Relling MV, Yang W, Das S, Cook EH, Rosner GL, Neel M, et al. Pharmacogenetic risk factors for osteonecrosis of the hip among children with leukemia. J Clin Oncol. 2004;22(19):3930–6. doi: 10.1200/JCO.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Karol SE, Yang W, Van Driest SL, Chang TY, Kaste S, Bowton E, et al. Genetics of glucocorticoid-associated osteonecrosis in children with acute lymphoblastic leukemia. Blood. 2015;126(15):1770–6. doi: 10.1182/blood-2015-05-643601. Epub 2015/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pounds S, Cao X, Cheng C, Yang JJ, Campana D, Pui CH, et al. Integrated analysis of pharmacologic, clinical and SNP microarray data using Projection Onto the Most Interesting Statistical Evidence with Adaptive Permutation Testing. International journal of data mining and bioinformatics. 2011;5(2):143–57. doi: 10.1504/IJDMB.2011.039174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pounds S, Cheng C. PROMISE: A tool to identify genomic features with a specific biologically interesting pattern of correlations with multiple endpoint variables. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp357. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao X, Crews KR, Downing JR, Lamba J, Pounds S. CC-PROMISE Effectively Integrates Two Forms of Molecular Data with Multiple Biologically Related Endpoints. BMC Bioinformatics. 2016 doi: 10.1186/s12859-016-1217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattano LA, Jr, Devidas M, Nachman JB, Sather HN, Hunger SP, Steinherz PG, et al. Effect of alternate-week versus continuous dexamethasone scheduling on the risk of osteonecrosis in paediatric patients with acute lymphoblastic leukaemia: results from the CCG-1961 randomised cohort trial. Lancet Oncol. 2012;13(9):906–15. doi: 10.1016/S1470-2045(12)70274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karol SE, Mattano LA, Jr, Yang W, Maloney KW, Smith C, Liu C, et al. Genetic risk factors for the development of osteonecrosis in children under age 10 treated for acute lymphoblastic leukemia. Blood. 2016;127(5):558–64. doi: 10.1182/blood-2015-10-673848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Te Winkel ML, Pieters R, Wind EJ, Bessems JH, van den Heuvel-Eibrink MM. Management and treatment of osteonecrosis in children and adolescents with acute lymphoblastic leukemia. Haematologica. 2014;99(3):430–6. doi: 10.3324/haematol.2013.095562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman-Uribe P, Vargas-Ruiz AG. Thrombosis in leukemia: incidence, causes, and practical management. Curr Oncol Rep. 2015;17(5):444. doi: 10.1007/s11912-015-0444-2. [DOI] [PubMed] [Google Scholar]
- 29.Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360(26):2730–41. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawedia JD, Liu C, Pei D, Cheng C, Fernandez CA, Howard SC, et al. Dexamethasone exposure and asparaginase antibodies affect relapse risk in acute lymphoblastic leukemia. Blood. 2012;119(7):1658–64. doi: 10.1182/blood-2011-09-381731. Epub 2011/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arpe ML, Rorvig S, Kok K, Molgaard C, Frandsen TL. The association between glucocorticoid therapy and BMI z-score changes in children with acute lymphoblastic leukemia. Support Care Cancer. 2015;23(12):3573–80. doi: 10.1007/s00520-015-2718-5. [DOI] [PubMed] [Google Scholar]
- 32.Withycombe JS, Post-White JE, Meza JL, Hawks RG, Smith LM, Sacks N, et al. Weight patterns in children with higher risk ALL: A report from the Children’s Oncology Group (COG) for CCG 1961. Pediatr Blood Cancer. 2009;53(7):1249–54. doi: 10.1002/pbc.22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Touyz LM, Cohen J, Neville KA, Wakefield CE, Garnett SP, Mallitt KA, et al. Changes in body mass index in long-term survivors of childhood acute lymphoblastic leukemia treated without cranial radiation and with reduced glucocorticoid therapy. Pediatr Blood Cancer. 2017;64(4) doi: 10.1002/pbc.26344. [DOI] [PubMed] [Google Scholar]
- 34.Howard SC, Pui CH. Endocrine complications in pediatric patients with acute lymphoblastic leukemia. Blood Rev. 2002;16(4):225–43. doi: 10.1016/s0268-960x(02)00042-5. [DOI] [PubMed] [Google Scholar]
- 35.Trevino LR, Shimasaki N, Yang W, Panetta JC, Cheng C, Pei D, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27(35):5972–8. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng C, Pounds SB, Boyett JM, Pei D, Kuo ML, Roussel MF. Statistical significance threshold criteria for analysis of microarray gene expression data. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1064. Article36. Epub 2006/05/02. [DOI] [PubMed] [Google Scholar]
- 37.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, Lam KC, et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS genetics. 2011;7(1):e1001279. doi: 10.1371/journal.pgen.1001279. Epub 2011/02/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iatan I, Dastani Z, Do R, Weissglas-Volkov D, Ruel I, Lee JC, et al. Genetic variation at the proprotein convertase subtilisin/kexin type 5 gene modulates high-density lipoprotein cholesterol levels. Circ Cardiovasc Genet. 2009;2(5):467–75. doi: 10.1161/CIRCGENETICS.109.877811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer ND, Goodarzi MO, Langefeld CD, Wang N, Guo X, Taylor KD, et al. Genetic Variants Associated With Quantitative Glucose Homeostasis Traits Translate to Type 2 Diabetes in Mexican Americans: The GUARDIAN (Genetics Underlying Diabetes in Hispanics) Consortium. Diabetes. 2015;64(5):1853–66. doi: 10.2337/db14-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45(5):501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quach JM, Walker EC, Allan E, Solano M, Yokoyama A, Kato S, et al. Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. J Biol Chem. 2011;286(6):4186–98. doi: 10.1074/jbc.M110.178251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vera A, Recabal A, Saldivia N, Stanic K, Torrejon M, Montecinos H, et al. Interaction between SCO-spondin and low density lipoproteins from embryonic cerebrospinal fluid modulates their roles in early neurogenesis. Front Neuroanat. 2015;9:72. doi: 10.3389/fnana.2015.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim BJ, Scott DA. Mouse model reveals the role of RERE in cerebellar foliation and the migration and maturation of Purkinje cells. PLoS One. 2014;9(1):e87518. doi: 10.1371/journal.pone.0087518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89(5):607–18. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Sha Q, Zhang S. Joint Analysis of Multiple Traits Using “Optimal” Maximum Heritability Test. PLoS One. 2016;11(3):e0150975. doi: 10.1371/journal.pone.0150975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11(4):407–9. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Sluis S, Posthuma D, Dolan CV. TATES: efficient multivariate genotype-phenotype analysis for genome-wide association studies. PLoS genetics. 2013;9(1):e1003235. doi: 10.1371/journal.pgen.1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, Bai Y, Pan W. An Adaptive Association Test for Multiple Phenotypes with GWAS Summary Statistics. Genet Epidemiol. 2015;39(8):651–63. doi: 10.1002/gepi.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korte A, Vilhjalmsson BJ, Segura V, Platt A, Long Q, Nordborg M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet. 2012;44(9):1066–71. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glueck CJ, Freiberg RA, Fontaine RN, Tracy T, Wang P. Hypofibrinolysis, thrombophilia, osteonecrosis. Clin Orthop. 2001;(386):19–33. doi: 10.1097/00003086-200105000-00004. [DOI] [PubMed] [Google Scholar]
- 54.Miyanishi K, Yamamoto T, Irisa T, Yamashita A, Jingushi S, Noguchi Y, et al. A high low-density lipoprotein cholesterol to high-density lipoprotein cholesterol ratio as a potential risk factor for corticosteroid-induced osteonecrosis in rabbits. Rheumatology(Oxford) 2001;40(2):196–201. doi: 10.1093/rheumatology/40.2.196. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto T, Irisa T, Sugioka Y, Sueishi K. Effects of pulse methylprednisolone on bone and marrow tissues: corticosteroid-induced osteonecrosis in rabbits. Arthritis and Rheumatism. 1997;40(11):2055–64. doi: 10.1002/art.1780401119. [DOI] [PubMed] [Google Scholar]
- 56.Janke LJ, Liu C, Vogel P, Kawedia J, Boyd KL, Funk AJ, et al. Primary epiphyseal arteriopathy in a mouse model of steroid-induced osteonecrosis. Am J Pathol. 2013;183(1):19–25. doi: 10.1016/j.ajpath.2013.03.004. Epub 2013/05/16. doi: S0002–9440(13)00259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerachian MA, Seguin C, Harvey EJ. Glucocorticoids in osteonecrosis of the femoral head: a new understanding of the mechanisms of action. J Steroid Biochem Mol Biol. 2009;114(3–5):121–8. doi: 10.1016/j.jsbmb.2009.02.007. Epub 2009/05/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith RW, Margulis RR, Brennan MJ, Monto RW. The influence of ACTH and cortisone on certain factors of blood coagulation. Science. 1950;112(2907):295–7. doi: 10.1126/science.112.2907.295. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto T, Hirano K, Tsutsui H, Sugioka Y, Sueishi K. Corticosteroid enhances the experimental induction of osteonecrosis in rabbits with Shwartzman reaction. Clin Orthop Relat Res. 1995;316:235–43. [PubMed] [Google Scholar]
- 60.Huang J, Hsu YH, Mo C, Abreu E, Kiel DP, Bonewald LF, et al. METTL21C is a potential pleiotropic gene for osteoporosis and sarcopenia acting through the modulation of the NF-kappaB signaling pathway. J Bone Miner Res. 2014;29(7):1531–40. doi: 10.1002/jbmr.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helgadottir A, Gretarsdottir S, Thorleifsson G, Hjartarson E, Sigurdsson A, Magnusdottir A, et al. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat Genet. 2016;48(6):634–9. doi: 10.1038/ng.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knight J, Barnes MR, Breen G, Weale ME. Using functional annotation for the empirical determination of Bayes Factors for genome-wide association study analysis. PLoS One. 2011;6(4):e14808. doi: 10.1371/journal.pone.0014808. Epub 2011/05/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kindt AS, Navarro P, Semple CA, Haley CS. The genomic signature of trait-associated variants. BMC genomics. 2013;14:108. doi: 10.1186/1471-2164-14-108. Epub 2013/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS genetics. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. Epub 2010/04/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Georgy SR, Pagel CN, Ghasem-Zadeh A, Zebaze RM, Pike RN, Sims NA, et al. Proteinase-activated receptor-2 is required for normal osteoblast and osteoclast differentiation during skeletal growth and repair. Bone. 2012;50(3):704–12. doi: 10.1016/j.bone.2011.11.023. Epub 2011/12/17. [DOI] [PubMed] [Google Scholar]
- 66.Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480(7376):201–8. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


