ABSTRACT
Antibody-drug conjugates (ADCs) have emerged as a family of compounds with promise as efficient immunotherapies. First-generation ADCs were generated mostly via reactions on either lysine side-chain amines or cysteine thiol groups after reduction of the interchain disulfide bonds, resulting in heterogeneous populations with a variable number of drug loads per antibody. To control the position and the number of drug loads, new conjugation strategies aiming at the generation of more homogeneous site-specific conjugates have been developed. We report here the first multi-level characterization of a site-specific ADC by state-of-the-art mass spectrometry (MS) methods, including native MS and its hyphenation to ion mobility (IM-MS). We demonstrate the versatility of native MS methodologies for site-specific ADC analysis, with the unique ability to provide several critical quality attributes within one single run, along with a direct snapshot of ADC homogeneity/heterogeneity without extensive data interpretation. The capabilities of native IM-MS to directly access site-specific ADC conformational information are also highlighted. Finally, the potential of these techniques for assessing an ADC's heterogeneity/homogeneity is illustrated by comparing the analytical characterization of a site-specific DAR4 ADC to that of first-generation ADCs. Altogether, our results highlight the compatibility, versatility, and benefits of native MS approaches for the analytical characterization of all types of ADCs, including site-specific conjugates. Thus, we envision integrating native MS and IM-MS approaches, even in their latest state-of-the-art forms, into workflows that benchmark bioconjugation strategies.
KEYWORDS: Antibody-drug conjugate (ADC), ion mobility-mass spectrometry (IM-MS), middle level, native mass spectrometry, site-specific bioconjugation, top level
Abbreviations
- mAb
monoclonal antibody
- ADC
antibody-drug conjugate
- IM-MS
ion mobility-mass spectrometry
- MS
mass spectrometry
- DAR
drug-to-antibody ratio
- fGly
formylglycine
- FGE
formylglycine-generating enzyme
- TCEP
tris(2-carboxyethy1)phosphine
- LC
light chain
- HC
heavy chain
- HPLC
high performance liquid chromatography
- IdeS
immunoglobulin degrading enzyme from Streptococcus pyogenes
- Fc
fragment crystallizable
- SEC
size exclusion chromatography
- Q-TOF
quadrupole-time-of-flight
- ATD
arrival time distribution
- CIU
collision induced unfolding
- CCS
collision cross-section
- IM
ion mobility
- TFA
trifluoroacetic acid
Introduction
With advances in protein engineering technologies, monoclonal antibodies (mAbs) and their derivatives have emerged as the largest drug class in human therapeutics.1,2 For oncology therapy, however, first-generation mAbs are often inefficient or encounter disease resistance. To overcome these limitations, several families of armed antibodies have been developed, among which antibody-drug conjugates (ADCs)3 have so far proved the most successful, with 2 drugs currently on the market (Adcetris® and Kadcyla®). Typically, ADCs are ∼154 kDa tripartite molecules comprising a recombinant mAb, which specifically targets the cancer cell, attached to a highly cytotoxic drug (payload) that inhibits cancer cell growth. The payload and antibody are connected via a synthetic linker (cleavable or non-cleavable) that covalently links the drug to the mAb. ADCs in development target a wide range of cancers.4 Many of these new compounds have emerged from a better understanding of structure-function relationships, which have been achieved in large part thanks to state-of-the art mass spectrometry (MS) methods,5 as well as lessons learned from pharmaceutical and clinical developments.6
ADC development directly benefits from the intense exploration of new conjugation techniques.7 For first-generation ADCs, drug conjugation is most frequently achieved via reactions on either lysine side-chain amines or cysteine thiol groups after reduction of the interchain disulfide bonds.8 These approaches result in heterogeneous mixtures containing average drug-to-antibody ratios (DARs) ranging from zero to 8, as illustrated by the 2 marketed ADCs (Adcetris® and Kadcyla®, cysteine- and lysine-conjugates, respectively). One main concern with these first-generation ADCs stems from the complexity of the randomly-conjugated product, because each DAR species has the potential to exhibit different toxicities and properties relating to the absorption, distribution, metabolism, and excretion of the molecules.9 Thus, second-generation technologies have moved toward producing more homogeneous and monodisperse ADCs by using site-specific conjugation. A number of site-specific bioconjugation strategies have been reported.10,11 These include the addition of engineered cysteine residues at specific sites without disruption of the interchain disulfide bonds,12 the addition of engineered peptide tags recognized by microbial transglutaminases to specifically transamidate amine-containing drug linkers attached to glutamine residues,13,14 the insertion of unnatural amino acids into the primary sequence of mAbs to provide a chemical handle for bioconjugation,15,16 and the development of new heterobifunctional reagents that facilitate the production of more stable ADCs.17,18 To our knowledge, 8 of these next-generation ADCs have reached the stage of clinical trials (phase 1 to 3)19 and have shown an improved therapeutic index in preclinical toxicology studies.20 Such site-specific conjugation technologies are also used for antibody antibiotic conjugates,21 bispecific ADCs22 and antibody dual drug conjugates.23
Here, we report the analytical characterization, using native mass spectrometry approaches, of a site-specific DAR4 ADC generated through aldehyde-specific bioconjugation (Fig. 1).24-26 Briefly, the formylglycine (fGly) amino acid residue is produced through the highly selective oxidation of a cysteine residue found within a specific pentapeptide consensus sequence by formylglycine-generating enzyme (FGE).27,28 The fGly-containing protein is then further modified using aldehyde-specific chemistries.25,29 ADCs generated using these methods possess increased therapeutic indices and increased therapeutic activities.11,30,39,40
Figure 1.
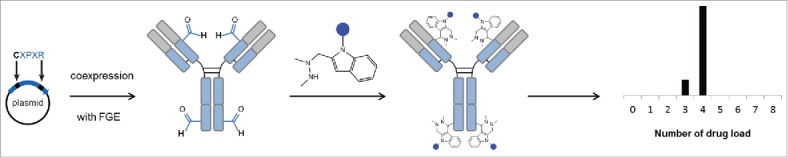
Schematic overview of the bioconjugation strategy. A formylglycine (fGly) amino acid residue is produced through the highly selective oxidation of a cysteine residue found within a specific pentapeptide consensus sequence by formylglycine-generating enzyme (FGE). The fGly-containing protein is then further modified using aldehyde specific chemistries.
The development and optimization of ADCs rely on improving their analytical and bioanalytical characterization by assessing several critical quality attributes, namely the distribution and position of the drug, the amount of naked antibody, the average DAR, and the residual drug-linker and related product proportions. A large number of orthogonal analytical and bioanalytical methods, often based on state-of-the-art chromatographic, electrophoretic, and mass spectrometric techniques, are needed for the characterization of ADCs at multiple levels (intact, middle and top).5
Although theoretically more homogeneous and thus amenable to standard techniques for their analytical characterization, we perform here a thorough characterization of a site-specific ADC and highlight the benefits of these still emergent approaches for the direct assessment of their increased homogeneity. This represents the first report describing a straightforward middle-up analysis of a DAR4 site-specific ADC (CBW-03–106) using tris(2-carboxyethy1)phosphine (TCEP) reduction. Next, we applied native MS and ion mobility MS for full characterization of the ADC at the intact level under non-denaturing conditions. Finally, the potential to highlight an ADC's heterogeneity/homogeneity was illustrated by comparing the analytical characterization of a site-specific DAR4 ADC to that of first-generation ADCs. Results obtained for the site-specific ADC revealed a far more homogeneous sample as compared with Adcetris® and Kadcyla®. Altogether, our results highlight the compatibility, versatility, and benefits of native MS approaches for the analytical characterization of all types of ADCs, including site-specific conjugates.
Results
Middle-up LC-MS analysis of the DAR4 site-specific ADC, CBW-03–106
As middle level analysis consisting of the analysis of large ADC fragments (25–50 kDa) obtained by either reduction or enzymatic cleavage are amenable to routine HPLC-MS analysis, this approach is often used as first line for ADC characterization in our laboratory. We thus performed a middle level characterization of intact CBW-03–106 using a classical tris(2-carboxyethy1)phosphine (TCEP) reduction procedure (Fig. 2). Three peaks were observed corresponding to light chain (LC) and heavy chain (HC) fragments. Peak A (23403.9 ± 0.3 Da) corresponds to the theoretical mass of the LC, while peak C could be unambiguously attributed to the HC + 2 RED-106 linker/payload molecules (54092.7 ± 1.2 Da). A minor peak (Peak B) corresponding to HC + one RED-106 (52869.2 ± 4.0 Da) was also detected. No signals corresponding to HC without any ligated RED-106 could be detected from extracted ion chromatograms. Middle-up analysis thus confirmed that the bioconjugation was located on the HC and that no unconjugated HC was detected. Middle-up analysis of CBW-03–106 after TCEP reduction allowed the estimation of an average DAR of 3.9 ± 0.1. Of note, middle-up analysis of CBW-03–106 after immunoglobulin degrading enzyme from Streptococcus pyogenes (IdeS) enzymatic digestion followed by TCEP reduction was also performed in parallel (Fig. S1) and enabled the precise localization of each RED-106 molecule, one being linked to the Fc/2 fragment and the other bound to the Fd fragment, as expected according to the placement of the aldehyde tags within the heavy chain antibody sequence. However, coelution of Fc/2 and LC precluded facile data interpretation from IdeS digestion for the studied ADC.
Figure 2.

Middle-up analysis of CBW-03–106. (a) UV chromatogram of TCEP-reduced CBW-03–106 (a). LC without drug load and HC fragments with one or 2 RED-106 bound molecules were observed. Theoretical and experimental masses of the G0F glycoform obtained by middle-up analyses of CBW-03–106 (b).
Altogether, middle-up analysis of the CBW-03–106 site-specific ADC revealed a highly homogeneous sample, with mostly 2 RED-106 molecules bound per HC. No signal corresponding to unbound HC was detected, leading to an average DAR of 3.9 ± 0.1 in good agreement with HIC data (Fig. S2).
Intact top level MS characterization
As middle-up analysis always leads to indirect average DAR determination that can be biased by peak integration, we next aimed at performing a top-level characterization of the CBW-03–106 site-specific ADC using denaturing and native MS.
Denaturing MS analysis of CBW-03–106
As covalent modifications are involved in CBW-03–106 formation, we first performed a classical HPLC-MS analysis in denaturing conditions.
In classical denaturing conditions (without previous reduction step, see Experimental Section), the most intense charge envelope between m/z 1500 and 3500 (Fig. S3) corresponds to intact CBW-03–106 ADC bearing 4 RED-106 molecules (152 767 ± 1 Da). No additional minor ion series corresponding to D0-D3 species could be detected. However, as already reported for ADCs, MS data interpretation is not straightforward and is hampered by the superimposition of the 2 broad charge state distributions of DAR4 and DAR3 species. An average DAR of 4.0 could be deduced from the intact HPLC-MS analysis in denaturing conditions.
Native MS analysis of CBW-03–106
We evaluated the versatility of native MS analysis for site-specific ADC characterization, drug load profile determination, and average DAR assessment. First, we analyzed intact deglycosylated CBW-03–106 by online size exclusion chromatography (SEC) hyphenated to native MS on a quadrupole-time-of-flight (Q-TOF) instrument (Fig. 3a). A main ion series with charge states ranging from 25+ to 32+ with a measured mass of 152 773 ± 1 Da was observed, in agreement with the expected mass of the mAb with 4 conjugated drugs (expected mass 152 771 Da). A minor ion series (less than 10% of total ion signals) corresponding to the binding of 3 drugs was also detected (151 546 ± 3 Da, Fig. 3b). No signals corresponding to either unconjugated mAb or mAb bearing 1 or 2 drugs were observed. Next, semi-quantitation based on peak intensities was performed, leading to an average measured DAR of 3.9 ± 0.1 (Fig. 3c), which is in good agreement with values obtained previously. Then, a second native MS analysis on a high resolution orbitrap instrument was performed (Fig. 3d–f). As already described, better mass accuracies (−7 ppm instead of +13 ppm) were obtained on the high resolution orbitrap instrument as compared with the Q-TOF. No additional ion series except D3 and D4 could be detected with high resolution native MS. In agreement with the data obtained on the Q-TOF instrument, the average DAR value calculated on the high resolution orbitrap instrument for deglycosylated CBW-03–106 was 3.9 ± 0.1. Native MS enables easy detection of drug binding stoichiometry and drug load homogeneity, providing an instantaneous snapshot of the drug-load distribution. In addition, CBW-03–106 was also analyzed without deglycosylation. When high resolution native MS was performed on this sample (Fig. S4), each drug load, and all glycoforms, were baseline resolved. More accurate average DAR values were thus obtained from high resolution native mass spectra.
Figure 3.

Native mass spectrometry analysis of deglycosylated CBW-03–106. Full scan ESI mass spectra on the m/z range [4 500 – 7 500] of deglycosylated CBW-03–106 in native conditions obtained either on a Q-TOF (a-c) or an orbitrap (d-f) instrument. Zoom on the most intense charge states showing drug load profiles (b,e). Native MS derived drug load profile and subsequent average DAR (c,f): relative intensities of each drug load as a function of the number of drugs loaded onto the mAb. •: non-identified impurity; *: loss of one fucose (−146 Da) and **: glycation (+162 Da).
Altogether, our results illustrate that native MS affords better resolution of superimposed glycosylation and conjugation heterogeneities of site-specific ADCs within a single run. The improved mass accuracy capabilities of high resolution native MS are of utmost importance for the characterization of all types of conjugated ADCs, including site-specific ADCs (Fig. 4).
Figure 4.

Native MS and IM-MS for benchmarking bioconjugation strategies. (a-c) Deconvoluted native mass spectra from the Orbitrap showing drug load profiles (left panels) and native IM-MS plots of m/z vs. drift time from the Q-TOF instrument for brentuximab vedotin (a), trastuzumab emtansine (b) and CBW-03–106 DAR4 site-specific ADC (c).
Intact top level native IM-MS characterization
We described previously the use of emergent IM-MS for characterization of cysteine- and lysine-ADC conformational heterogeneity.31,32 Here, we present a global native IM-MS conformational characterization of a deglycosylated DAR4 site-specific ADC and its unconjugated mAb counterpart.
Global IM-MS conformational characterization of CBW-03–106 and the parental mAb
A comparison of the IM-MS plots obtained for deglycosylated DAR4 site-specific CBW-03–106 (Fig. 5a) and unconjugated mAb (Fig. 5b) shows that IM-MS provided a direct picture of DAR4 site-specific ADC homogeneity. Unlike for cysteine- or lysine- reference conjugates (Fig. 4), one unique population, corresponding to the binding of 4 drug loads (DAR4) was clearly observed for the most intense charge states (24+ to 28+; Fig. 5a).
Figure 5.
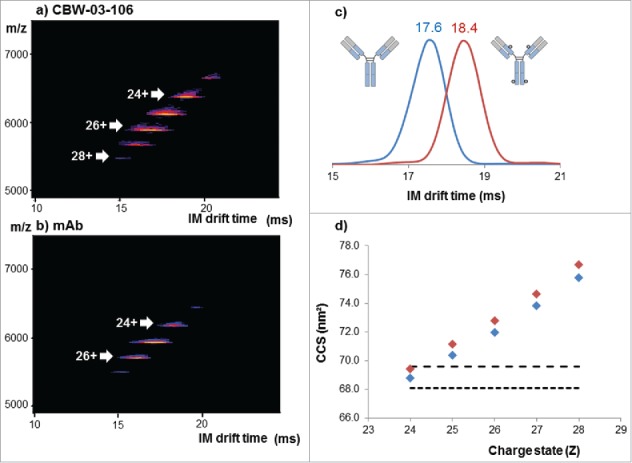
Native IM-MS analysis of deglycosylated CBW-03-106 and its parental mAb form. Plots of m/z vs. drift time for CBW-03-106 (a) and mAb (b). Extracted ATDs corresponding to the 24+ charge state of the parental mAb (blue) and CBW-03–106 (red) at a trap collision voltage of 4 V (c). Measured TWCCSN2 (nm2) of the parental mAb (blue diamonds) and CBW-03–106 (red diamonds) as a function of ESI charge state (d).
Overlaid extracted arrival time distributions (ATDs) of the 24+ charge state of the deglycosylated DAR4 site-specific ADC and its corresponding unconjugated mAb are depicted in Fig. 5c. When comparing the deglycosylated DAR4 site-specific ADC and the unconjugated mAb, the ATDs are resolved at 58% of the valley for the 24+ charge state with the ion mobility (IM) cell resolution used here (∼18). All ATDs were symmetric and significantly different IM drift times were measured for CBW-03–106 and for its unconjugated mAb (Fig. 5c, Table 1). A reproducible and constant difference in IM drift times of 0.8 ± 0.1 ms was observed for the addition of 4 drugs for the 24+ charge state, corresponding to 0.2 ± 0.1 ms per drug load of 1217 Da.
Table 1.
IM drift times and TWCCSN2 values obtained for the 24+ charge states of deglycosylated site-specific CBW-03–106 ADC and its unconjugated mAb form.
| mAb | CBW-03-106 | |
|---|---|---|
| IM drift time (ms) | 17.6 ± 0.1a | 18.4 ± 0.1a |
| Δ(IM drift time)b (ms) | – | 0.8 |
| CCS from IM-MS (nm2) | 68.8 ± 0.1a | 69.4 ± 0.1a |
| ΔCCSb (nm2) | – | 0.6 |
| Predicted CCSc (nm2) | 68.1 | 69.6 |
| Predicted ΔCCSb (nm2) | – | 1.5 |
| Average DAR from IM-MS | 0 | 4.0a |
Uncertainty values represent the variability as a result of repeat injections (n = 3).
Δ(IM drift times) and ΔCCS values represent differences between D4 ADC and unconjugated parental mAb.
CCS were predicted considering ADCs as spherical protein through the equation 33 CCS = 2.435 × M2/3.
Corresponding TWCCSN2 values were calculated for all the charge states of the DAR4 site-specific ADC and for the unconjugated mAb (Fig. 5d and Table 1) and compared with already published ones. TWCCSN2 values ranging from 68.8 to 75.8 nm2 and from 69.4 to 76.7 nm2 were obtained for the unconjugated mAb and the DAR4 ADC, respectively. Measured TWCCSN2 values and corresponding ΔTWCCSN2 were in agreement with both predicted collision cross-section (CCS) from mAbs considered as spherical proteins33 (Fig. 5d) and with already reported data on other mAbs34 or ADCs.31,32 Of note, the TWCCSN2 reported for CBW-03–106 DAR4 species (69.4 ± 0.1 nm2 for the 24+ charge state) correlated well with the one reported for the reference cysteine ADC (69.5 ± 0.1 nm2 for the 24+ charge state). The agreement between mass-predicted and IM-MS-measured CCSs also strongly argues for a mass increase effect rather than slight conformational changes induced upon drug binding. Similar to IM drift time variations, the addition of 4 drugs induced a slight but constant and reproducible TWCCSN2 variation of 0.6 nm2 observed on all charge states (Table 1, Fig. 5d). Slightly larger ΔTWCCSN2s between DAR0 and DAR4 species were reported for brentuximab vedotin (1.5 nm2 for 4 drugs of 1317 Da each) and ado-trastuzumab emtansine (1.2 nm2 for 4 drugs of 958 Da each), which could be a first hint of increased conformational heterogeneity within the preparations of those molecules.
Average DAR determination by IM-MS
As already performed on lysine- and cysteine conjugates,31,32 we used IM-MS data for average DAR determination of CBW-03–106. The intensities of the drift peaks extracted for each charge state were plotted across the series for DAR0–DAR4 drug binding stoichiometries, and a Gaussian curve was fitted to the resulting distribution (Fig. S5). The relative intensity of these ATDs indicates the relative abundance of each species. As only ATDs for DAR4 were detected (Fig. S5), an average DAR of 4.0 could be deduced from IM-MS data (Table 1), in good agreement with the expected value obtained from other MS approaches.
CIU experiments of CBW-03–106 and the parental mAb
We next performed collision induced unfolding (CIU) experiments to compare the gas-phase conformational stability of CBW-03–106 and parental mAb. CIU experiments consist of gas-phase collisional activation of the ions in the trap T-Wave of the instrument before IM separation. CIU experiments have already been reported for the rapid analysis of antibody structure and chemical modifications.35 Here, a comparison of the CIU patterns of CBW-03–106 ADC with those of the parental mAb allowed for an investigation into the effect of conjugation on mAb gas-phase stability.
First, we analyzed CIU data acquired for CBW-03–106 and parental mAb to highlight differences in the CIU fingerprints. As shown in Fig. 6a–b, the 24+ intact antibody ions for parental mAb and CBW-03–106 exhibited different low energy IM migration times (17.6 and 18.6 ms, respectively), and possessed significantly different CIU fingerprints, as highlighted by the high root-mean-square deviation value shown in the CIU difference plot (Fig. 6c).
Figure 6.

CIU experiments performed on deglycosylated CBW-03–106 site-specific ADC and its unconjugated mAb form. CIU fingerprints of the 24+ charge state for the unconjugated mAb (a) and the site-specific ADC, CBW-03–106 (b). CIU difference plot between CBW-03–106 and unconjugated mAb (c). Extracted arrival time distributions (ATD) of the unconjugated mAb (blue) and CBW-03–106 (red) at different trap collision voltages (d).
Three main CIU features were observed for the unconjugated mAb (Fig. 6a), while CBW-03–106 only displayed 2 states (Fig. 6b) over the collision voltage range 0–200 V.
For unconjugated mAb, the first transition from the most compact initial state 0 (Fig. 6a and 6d) to the first unfolded state I happens between 40 and 75 V, the unfolded state I being then stable over the 75–100 V Trap CE range. When collision energies increased from trap CE 100 V to 140 V, ATDs became broader. At 150 V, the ATD of the 24+ charge state of parental mAb presented a double distribution corresponding to the coexistence of at least 2 conformational families (Fig. 6d) that could be observed throughout the trap CE 150–190 V range, highlighting a second transition between trap CE 140 and 150 V from unfolded state I to the final unfolded state II. The IM drift time related to the most unfolded species was 20.7 ms (Fig. 6d), which corresponded to a total CCS increase of 3.3% as compared with the initial state 0.
For CBW-03–106, the initial most compact state had an IM migration time of 18.6 ms (Fig. 6b), which is ∼ 1 ms on average longer than parental mAb IM drift time (Fig. 6c). These differences could be attributed to the mass increase due to drug conjugation, as already concluded from CCS calculations. This conformational family started progressively and continuously unfolding from 40 V to 100 V, leading to the unfolded state I at 21.0 ms (Fig. 6c). By contrast to the parental mAb, the ADC unfolded state I was then stable over the 100–190 V range (Fig. 6b). The drift time variation observed during this transition implies a relative CCS variation of 2.6%.
Altogether, CIU results highlight that conjugation seems to stabilize the mAb structure for the following reasons: i) even if the DAR4 ADC begins unfolding from state 0 to state I at similar trap CE voltage (40 V), the unfolding process is distributed over a broader trap CE range (40–100V) compared with the parent mAb (40–75 V), which is favor of a global stabilization effect of the mAb conformation upon conjugation; and ii) the site-specific ADC is less prone to unfolding events (fewer unfolding intermediates) than its unconjugated counterpart mAb. Taken together and noting the high structural and sequence identity (same number of interchain disulfide bonds and same amino acid sequence) between the CBW-03–106 and parental mAb, the data presented here strongly support the hypothesis that drug conjugation stabilizes mAb conformation.
Discussion
We report here the first multi-level characterization of a new generation of site-specific antibodies by state-of-the-art MS methods. A combination of HPLC-MS methods and non-denaturing MS approaches were used to provide a comprehensive characterization of a site-specific ADC at both intact and middle levels. Middle level analysis, which is now the first line strategy used in most laboratories for mAb and ADC analytical characterization, revealed a highly homogeneous sample in terms of drug load, with mainly DAR4 detected and low amounts of DAR3. Of note, for this type of ADC, the classical reduction strategy that results in LC and HC fragments was more appropriate than the IdeS strategy, which led to the coelution of LC and Fc fragments. Native MS was next applied to site-specific ADC characterization leading to a direct observation of the drug load profile compared with the middle-up indirect approach. Native MS is now easily amenable to high throughput through its hyphenation to SEC chromatography for rapid and efficient buffer exchange. Thus, we believe that SEC-native MS will more and more challenge classical HPLC-MS techniques. Finally, extensive conformational characterization by IM-MS of our site-specific ADC enabled detection of different drift times for unconjugated and conjugated ADC. Again, average DAR values along with drug load distributions could be achieved.
We demonstrate here the capabilities of native IM-MS to directly access site-specific ADC conformational information through the measurement of IM drift times, CCS calculations and CIU fingerprints. However, as the site-specific ADC and its unconjugated mAb version exhibited similar intact CCS, it was hard to distinguish between a mass effect and a real conformational change induced upon drug conjugation. We demonstrate here for the first time the capabilities of CIU in the context of intact ADC analysis. CIU allows one to circumvent the lack of IM resolution through the establishment of unfolding patterns. We demonstrate here the ability of CIU to differentiate a site-specific ADC from its parental unconjugated mAb along with increase in ADC stability toward unfolding as compared with the unconjugated mAb. This could be concluded from the CIU landscapes of the ADC presenting fewer numbers of unfolding states than the unconjugated mAb, along with broader unfolding transitions for the ADC than for its mAb counterpart. Thus, we envision the use of CIU protocols to optimize conjugation strategies for next-generation ADC development, accompanied by a fast assessment of ADC gas phase stability.
Our group has already reported the in-depth characterization of first-generation heterogeneous lysine- or cysteine-conjugated ADCs.5,31,32,36 Here, we demonstrate the versatility of native MS methodologies (native MS and IM-MS) for site-specific ADC analysis. These methodologies present the advantages of providing a direct snapshot of ADC homogeneity/heterogeneity without extensive data interpretation. As an illustration, Fig. 4 shows native MS and IM-MS data obtained for brentuximab vedotin (Adcetris®, the reference cysteine conjugate), ado-trastuzumab emtansine (Kadcyla®, the reference lysine-ADC), and the CBW-03–106 site-specific ADC. At a glance, the increased homogeneity of the site-specific ADC is obvious from either native mass spectra or IM-MS plots. Some of the most important quality attributes, including the presence/amount of unconjugated mAb, the drug load profile, the average DAR, and even payload degradation/modification can be assessed within a few minutes from one single SEC-native MS run. Thus, we believe that native MS and its hyphenation to ion mobility are ready to serve for the benchmarking of conjugation strategies, for the evaluation of drug binding heterogeneity, and for the fast conformational characterization of ADCs. We envision integrating native MS and IM-MS approaches even in their latest state-of-the-art forms (like CIU, or SEC-native IM-MS) to evaluate bioconjugation strategies.
Materials and methods
Chemicals
All chemicals were purchased from Sigma-Aldrich: ammonium acetate (A1542), cesium iodide (21004), 2-propanol (I9516), sodium phosphate dibasic (S7907), sodium chloride (S7653), guanidine hydrochloride (G4505), TCEP hydrochloride (C4706), trifluoroacetic acid (TFA; 74564) and HPLC-grade acetonitrile (34851). IgGZERO (A0-IZ1–010) and IdeS (A0-FR1–008) enzymes were obtained from Genovis. All the aqueous solutions were prepared using an ultra-pure water system (Sartorius, Göttingen, Germany).
Middle level UHPLC-MS analysis
Middle level analysis
CBW-03–106 ADC reduction
Thirty µg of ADC in 50 mM Na2HPO4 and 150 mM NaCl at pH 6.6 were denatured by the addition of 5 M guanidine hydrochloride. The reduction was then performed using 56 mM of TCEP-HCl for 60 minutes at 57 °C. Finally, the reaction was quenched by adding 1% TFA.
Liquid chromatography coupled to mass spectrometry
UHPLC-MS analysis was performed using an Acquity H-class (Waters, Manchester, UK) coupled to the Synapt G2 high definition (HD) MS (Waters, Manchester, UK). A volume equivalent to 9 µg of mAb in sample preparation was injected on a PLRP-S column (2.1 × 150 mm, 8 µm, 1000 Å from Agilent, Waldbronn, Germany) set at 80 °C. The gradient was generated at a flow rate of 250 µL/min using water containing 0.1% TFA for mobile phase A and acetonitrile containing 0.08% TFA for mobile phase B. The latter was raised from 10 to 30% over 8 minutes, 30 to 34% over 12 minutes, and 34 to 50% over 20 minutes, followed by a 3 minute washing step at 90% B and a 12 minute re-equilibration period. The Synapt G2 HDMS was operated in positive mode with a capillary voltage of 3.2 kV. Acquisitions were performed in the mass range of 500–5000 m/z with a 1-s scan time. Signal acquisition was realized by UV absorbance measurement at 214 nm. Calibration was performed using the singly charged ions produced by a tuning mix. MS data interpretations were performed using Mass Lynx V4.1 (Waters, Manchester, UK).
Intact mass analysis
Deglycosylation
The deglycosylation was performed by incubating for 30 min at 37 °C one unit of IgGZERO per microgram of mAb and ADC.
Buffer exchange
Prior to native MS and IM-MS experiments, mAb and ADC were desalted against a 150 mM ammonium acetate solution at pH 6.9 using 6 cycles of concentration/dilution using a microconcentrator (Vivaspin, 10-kD cutoff, Sartorius, Göttingen, Germany). Protein concentration was determined by UV absorbance using a NanoDrop spectrophotometer (Thermo Fisher Scientific, France).
Native MS analysis
High resolution native MS (native-HRMS) of intact mAb and ADC were performed on an Orbitrap Exactive Plus EMR (Thermo Fisher Scientific, Bremen, Germany) mass spectrometer coupled to an automated chip-based nanoelectrospray device (Triversa Nanomate, Advion Bioscience, Ithaca, USA) operating in the positive ion mode. The capillary voltage was set at 1.86 kV and nitrogen nanoflow at 0.15 psi for the Orbitrap. The in-source collision-induced dissociation and the higher-energy collisional dissociation cells were set to 200 eV and 50 eV, respectively. The trapping gas pressure was set to 3 a.u. (which corresponds to an Ultra High Vacuum of ∼4.10−10 mbar). To improve the transmission of the high mass species, the voltages on the injection-, inter-, and bent- flatapoles were fixed to 8, 7, and 6 V, respectively. Samples were diluted in 150 mM NH4OAc at pH 6.9 and infused at 10 µM. External calibration was performed using singly charged ions produced by a 2 g/L solution of cesium iodide in 2-propanol/water (50/50 v/v). Orbitrap MS data interpretation was performed using Protein Deconvolution 4.0 available on BioPharmaFinder 1.0 SP1 (Thermo Fisher Scientific, Bremen, Germany). The parameters of the software were optimized for each spectrum.
Size exclusion chromatography coupled to native mass spectrometry
An ACQUITY UPLC H-class system (Waters, Manchester, UK) comprising a quaternary solvent manager, a sample manager operating at 10 °C, a column oven maintained at 50 °C and a TUV detector operating at 280 nm and 214 nm hyphenated to a Synapt G2 HDMS mass spectrometer (Waters, Manchester, UK) was used for the online buffer exchange and intact mass measurement. 51 µg of glycosylated CBW-03–106 and 45 µg of its deglycosylated version were loaded on the ACQUITY UPLC Protein BEH SEC column (2.1 × 150 mm, 1.7 µm particle size, 200 Å pore size from Waters, Manchester, UK) using an isocratic elution of 100 mM NH4OAc (pH 6.8) at a flow rate of 0.25 mL/min over 4.1 min. Then, the flow rate was decreased to 0.10 mL/min over 5.5 min and finally increased by 0.25 mL/min over 2.4 min. The Synapt G2 HDMS was operated in positive mode with a capillary voltage of 3.0 kV. Acquisitions were performed in the m/z range of 1000–8000 with a 1.5 s scan time. External calibration was performed using singly charged ions produced by a 2 g/L solution of cesium iodide in 2-propanol/water (50/50 v/v). MS data interpretations were performed using Mass Lynx V4.1 (Waters, Manchester, UK).
Average DAR calculation
Average DAR values were calculated by using Equation 1. For native MS, these results were derived from the relative peak intensities measured from deconvoluted mass spectra. For middle level analysis, these results were based on the area of each peak corresponding to the drug loaded fragments obtained on the UV chromatogram.
| (1) |
Ion mobility mass spectrometry
Ion mobility experiments were performed on a TWIMS-MS Synapt G2 instrument (Waters, Manchester, UK). The instrument was carefully tuned to achieve a good trade-off between ion separation and TWIMS resolution. Low accelerating voltages were used, especially before IM separation, to guide the ions through the mobility cell to the TOF analyzer without ion activation. The backing pressure of the Z-spray source was 6.0 mbar, and the sampling cone was operated at 80 V. The argon flow rate in the traveling-wave-based ion trap was 5 mL/min, and the trap collision energy was set at 4 V. Prior to IM separation, ions were thermalized in the helium cell (130 mL/min). Ion separation was performed in the pressurized ion mobility cell using a constant N2 flow rate of 45 mL/min. The IM wave height and velocity were 40 V and 923 m/s, respectively. Transfer collision energy was fixed to 2 V to extract the ions from the IM cell to the TOF analyzer. IM data were calibrated to perform CCS calculations using the most intense charge states of 3 external calibrants (concanavaline A, pyruvate kinase, and alcohol dehydrogenase) in non-denaturing conditions as described elsewhere.31,32,37 IM-MS experiments were performed in triplicate under identical conditions.
Drug-to-antibody ratio calculations were also performed from ion mobility data. ATDs corresponding to each charge state were fitted on a Gaussian profile. The delimited areas were quantified and used to calculate average DAR from Equation 1.
Collision induced unfolding experiments
ATDs were recorded by progressively increasing the trap collision energy in the traveling-wave-based ion trap before the IM cell. All the ions were accelerated into the ion trap without any previous selection; however, only ion mobility data corresponding to the 24+ charge state were compiled to create the CIU fingerprint. The trap collision energy was ramped from 0 to 200 V using an energy interval of 5 V. This voltage interval corresponds to the best trade-off between CIU fingerprint resolution and acquisition time.
CIU fingerprint data
2D CIU fingerprint plots were analyzed using the open source CIUSuite software, especially the CIUSuite_stats and CIUSuite_compare modules.38 Briefly, these modules allow for drift time data extraction at each trap CE collision voltage. Ion intensities were normalized to a maximum value of 1 and the ATDs were smoothed using a Savitzky-Golay filter with a window length of 3 and polynomial order of 2. The individual IM data were gathered to create the CIU data shown. Drift time (ms) is shown in the size axis versus trap collision energy (V) in the x-axis. The relative intensities of the ATD distributions are denoted by a color-coded axis. CIU fingerprint plots and standard deviations were generated for multiple replicates using CIUSuite_stats function to quantify the reproducibility of the experiment. Comparison of 2 different CIU fingerprint plots was achieved using the CIUSuite_compare module.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the several sponsors (Région Alsace, Communauté Urbaine de Strasbourg, CNRS, and the Université de Strasbourg), by the Agence Nationale de la Recherche (ANR) and the French Proteomic Infrastructure (ProFI; ANR-10-INBS-08–03). We thank GIS IBiSA and Région Alsace for financial support in purchasing a Synapt G2 HDMS instrument and an Exactive Plus EMR Orbitrap spectrometer, respectively. T. Botzanowski and A. Ehkirch acknowledge Institut de Recherche Servier and Syndivia for funding of their PhD fellowships, respectively.
References
- 1.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol 2010; 10:345-52; PMID:20414207; https://doi.org/ 10.1038/nri2747 [DOI] [PubMed] [Google Scholar]
- 2.Reichert JM. Antibodies to watch in 2017. MAbs 2017; 9:167-81; PMID:27960628; https://doi.org/ 10.1080/19420862.2016.1269580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck A, Reichert JM. Antibody-drug conjugates: Present and future. MAbs 2014; 6:15-7; PMID:24423577; https://doi.org/ 10.4161/mabs.27436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Junutula JR, Gerber HP. Next-Generation antibody-drug conjugates (ADCs) for cancer therapy. ACS Med Chem Lett 2016; 7:972-3; PMID:27882192; https://doi.org/ 10.1021/acsmedchemlett.6b00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck A, Terral G, Debaene F, Wagner-Rousset E, Marcoux J, Janin-Bussat MC, Colas O, Van Dorsselaer A, Cianférani S. Cutting-edge mass spectrometry methods for the multi-level structural characterization of antibody-drug conjugates. Expert Rev Proteomics 2016; 13:157-83; PMID:26653789; https://doi.org/ 10.1586/14789450.2016.1132167 [DOI] [PubMed] [Google Scholar]
- 6.Loganzo F, Sung M, Gerber HP. Mechanisms of resistance to antibody-drug conjugates. Mol Cancer Ther 2016; 15:2825-34; PMID:27780876; https://doi.org/ 10.1158/1535-7163.MCT-16-0408 [DOI] [PubMed] [Google Scholar]
- 7.Dennler P, Fischer E, Schibli R. Antibody conjugates: From heterogeneous populations to defined reagents. Antibodies 2015; 4:197-224; https://doi.org/ 10.3390/antib4030197 [DOI] [Google Scholar]
- 8.Beck A, Lambert J, Sun M, Lin K. Fourth world antibody-drug conjugate summit: February 29-March 1, 2012, Frankfurt, Germany. MAbs 2012; 4:637-47; PMID:22909934; https://doi.org/ 10.4161/mabs.21697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts SA, Andrews PA, Blanset D, Flagella KM, Gorovits B, Lynch CM, Martin PL, Kramer-Stickland K, Thibault S, Warner G. Considerations for the nonclinical safety evaluation of antibody drug conjugates for oncology. Regul Toxicol Pharmacol 2013; 67:382-91; PMID:24012707; https://doi.org/ 10.1016/j.yrtph.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 10.Gordon MR, Canakci M, Li L, Zhuang J, Osborne B, Thayumanavan S. Field guide to challenges and opportunities in antibody-drug conjugates for chemists. Bioconjug Chem 2015; 26:2198-215; PMID:26308881; https://doi.org/ 10.1021/acs.bioconjchem.5b00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, et al.. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol 2008; 26:925-32; PMID:18641636; https://doi.org/ 10.1038/nbt.1480 [DOI] [PubMed] [Google Scholar]
- 12.Panowski S, Bhakta S, Raab H, Polakis P, Junutula JR. Site-specific antibody drug conjugates for cancer therapy. MAbs 2014; 6:34-45; PMID:24423619; https://doi.org/ 10.4161/mabs.27022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennler P, Chiotellis A, Fischer E, Bregeon D, Belmant C, Gauthier L, Lhospice F, Romagne F, Schibli R. Transglutaminase-based chemo-enzymatic conjugation approach yields homogeneous antibody-drug conjugates. Bioconjug Chem 2014; 25:569-78; PMID:24483299; https://doi.org/ 10.1021/bc400574z [DOI] [PubMed] [Google Scholar]
- 14.Farias SE, Strop P, Delaria K, Galindo Casas M, Dorywalska M, Shelton DL, Pons J, Rajpal A. Mass spectrometric characterization of transglutaminase based site-specific antibody-drug conjugates. Bioconjug Chem 2014; 25:240-50; PMID:24359082; https://doi.org/ 10.1021/bc4003794 [DOI] [PubMed] [Google Scholar]
- 15.Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, et al.. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. P Natl Acad Sci USA 2012; 109:16101-6; PMID:22988081; https://doi.org/ 10.1073/pnas.1211023109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith EL, Giddens JP, Iavarone AT, Godula K, Wang LX, Bertozzi CR. Chemoenzymatic Fc glycosylation via engineered aldehyde tags. Bioconjug Chem 2014; 25:788-95; PMID:24702330; https://doi.org/ 10.1021/bc500061s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodych S, Koniev O, Baatarkhuu Z, Bonnefoy JY, Debaene F, Cianferani S, Van Dorsselaer A, Wagner A. CBTF: New amine-to-thiol coupling reagent for preparation of antibody conjugates with increased plasma stability. Bioconjug Chem 2015; 26:197-200; PMID:25614935; https://doi.org/ 10.1021/bc500610g [DOI] [PubMed] [Google Scholar]
- 18.Koniev O, Kolodych S, Baatarkhuu Z, Stojko J, Eberova J, Bonnefoy JY, Cianférani S, Van Dorsselaer A, Wagner A. MAPN: First-in-Class reagent for kinetically resolved thiol-to-thiol conjugation. Bioconjug Chem 2015; 26:1863-7; PMID:26335849; https://doi.org/ 10.1021/acs.bioconjchem.5b00440 [DOI] [PubMed] [Google Scholar]
- 19.Beck A, Goetsch L, Dumontet C, Corvaïa N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat Rev Drug Discov 2017; in press; PMID:28303026; https://doi.org/ 10.1038/nrd.2016.268 [DOI] [PubMed] [Google Scholar]
- 20.Strop P, Tran TT, Dorywalska M, Delaria K, Dushin R, Wong OK, Ho WH, Zhou D, Wu A, Kraynov E, et al.. RN927C, a site-specific trop-2 antibody-drug conjugate (ADC) with enhanced stability, is highly efficacious in preclinical solid tumor models. Mol Cancer Ther 2016; 15:2698-708; PMID:27582525; https://doi.org/ 10.1158/1535-7163.MCT-16-0431 [DOI] [PubMed] [Google Scholar]
- 21.Zhou C, Lehar S, Gutierrez J, Rosenberger CM, Ljumanovic N, Dinoso J, Koppada N, Hong K, Baruch A, Carrasco-Triguero M, et al.. Pharmacokinetics and pharmacodynamics of DSTA4637A: A novel THIOMAB antibody antibiotic conjugate against Staphylococcus aureus in mice. MAbs 2016; 8:1612-9; PMID:27653831; https://doi.org/ 10.1080/19420862.2016.1229722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Goeij BE, Vink T, Ten Napel H, Breij EC, Satijn D, Wubbolts R, Miao D, Parren PW. Efficient payload delivery by a bispecific antibody-drug conjugate targeting HER2 and CD63. Mol Cancer Ther 2016; 15:2688-97; PMID:27559142; https://doi.org/ 10.1158/1535-7163.MCT-16-0364 [DOI] [PubMed] [Google Scholar]
- 23.Levengood MR, Zhang X, Hunter JH, Emmerton KK, Miyamoto JB, Lewis TS, Senter PD. Orthogonal cysteine protection enables homogeneous multi-drug antibody-drug conjugates. Angew Chem Int Ed Engl 2017; 56:733-7; PMID:27966822; https://doi.org/ 10.1002/anie.201608292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabuka D, Rush JS, deHart GW, Wu P, Bertozzi CR. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat Protoc 2012; 7:1052-67; PMID:22576105; https://doi.org/ 10.1038/nprot.2012.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal P, Kudirka R, Albers AE, Barfield RM, de Hart GW, Drake PM, Jones LC, Rabuka D. Hydrazino-pictet-spengler ligation as a biocompatible method for the generation of stable protein conjugates. Bioconjug Chem 2013; 24:846-51; PMID:23731037; https://doi.org/ 10.1021/bc400042a [DOI] [PubMed] [Google Scholar]
- 26.Kudirka R, Barfield RM, McFarland J, Albers AE, de Hart GW, Drake PM, Holder PG, Banas S, Jones LC, Garofalo AW, et al.. Generating site-specifically modified proteins via a versatile and stable nucleophilic carbon ligation. Chem Biol 2015; 22:293-8; PMID:25619935; https://doi.org/ 10.1016/j.chembiol.2014.11.019 [DOI] [PubMed] [Google Scholar]
- 27.York D, Baker J, Holder PG, Jones LC, Drake PM, Barfield RM, Bleck GT, Rabuka D. Generating aldehyde-tagged antibodies with high titers and high formylglycine yields by supplementing culture media with copper(II). BMC Biotechnol 2016; 16:23; PMID:26911368; https://doi.org/ 10.1186/s12896-016-0254-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holder PG, Jones LC, Drake PM, Barfield RM, Banas S, de Hart GW, Baker J, Rabuka D. Reconstitution of formylglycine-generating enzyme with copper(II) for aldehyde tag conversion. J Biol Chem 2015; 290:15730-45; PMID:25931126; https://doi.org/ 10.1074/jbc.M115.652669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudirka RA, Barfield RM, McFarland JM, Drake PM, Carlson A, Banas S, Zmolek W, Garofalo AW, Rabuka D. Site-specific tandem knoevenagel condensation-michael addition to generate antibody-drug conjugates. ACS Med Chem Lett 2016; 7:994-8; PMID:27882197; https://doi.org/ 10.1021/acsmedchemlett.6b00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillow TH, Tien J, Parsons-Reponte KL, Bhakta S, Li H, Staben LR, Li G, Chuh J, Fourie-O'Donohue A, Darwish M, et al.. Site-specific trastuzumab maytansinoid antibody-drug conjugates with improved therapeutic activity through linker and antibody engineering. J Med Chem 2014; 57:7890-9; PMID:25191794; https://doi.org/ 10.1021/jm500552c [DOI] [PubMed] [Google Scholar]
- 31.Marcoux J, Champion T, Colas O, Wagner-Rousset E, Corvaia N, Van Dorsselaer A, Beck A, Cianférani S. Native mass spectrometry and ion mobility characterization of trastuzumab emtansine, a lysine-linked antibody drug conjugate. Protein Sci 2015; 24:1210-23; PMID:25694334; https://doi.org/ 10.1002/pro.2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debaene F, Boeuf A, Wagner-Rousset E, Colas O, Ayoub D, Corvaia N, Van Dorsselaer A, Beck A, Cianférani S. Innovative native MS methodologies for antibody drug conjugate characterization: High resolution native MS and IM-MS for average DAR and DAR distribution assessment. Anal Chem 2014; 86:10674-83; PMID:25270580; https://doi.org/ 10.1021/ac502593n [DOI] [PubMed] [Google Scholar]
- 33.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc 2008; 3:1139-52; PMID:18600219; https://doi.org/ 10.1038/nprot.2008.78 [DOI] [PubMed] [Google Scholar]
- 34.Pacholarz KJ, Porrini M, Garlish RA, Burnley RJ, Taylor RJ, Henry AJ, Barran PE. Dynamics of intact immunoglobulin G explored by drift-tube ion-mobility mass spectrometry and molecular modeling. Angew Chem Int Ed Engl 2014; 53:7765-9; PMID:24916519; https://doi.org/ 10.1002/anie.201402863 [DOI] [PubMed] [Google Scholar]
- 35.Tian Y, Han L, Buckner AC, Ruotolo BT. Collision induced unfolding of intact antibodies: Rapid characterization of disulfide bonding patterns, glycosylation, and structures. Anal Chem 2015; 87:11509-15; PMID:26471104; https://doi.org/ 10.1021/acs.analchem.5b03291 [DOI] [PubMed] [Google Scholar]
- 36.Terral G, Beck A, Cianferani S. Insights from native mass spectrometry and ion mobility-mass spectrometry for antibody and antibody-based product characterization. J Chromatogr B Analyt Technol Biomed Life Sci 2016; 1032:79-90; PMID:27108304; https://doi.org/ 10.1016/j.jchromb.2016.03.044 [DOI] [PubMed] [Google Scholar]
- 37.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal Chem 2010; 82:9557-65; PMID:20979392; https://doi.org/ 10.1021/ac1022953 [DOI] [PubMed] [Google Scholar]
- 38.Eschweiler JD, Rabuck-Gibbons JN, Tian Y, Ruotolo BT. CIUSuite: A quantitative analysis package for collision induced unfolding measurements of gas-phase protein ions. Anal Chem 2015; 87:11516-22; PMID:26489593; https://doi.org/ 10.1021/acs.analchem.5b03292 [DOI] [PubMed] [Google Scholar]
- 39.Albers AE, Garofalo AW, Drake PM, Kudirka R, de Hart GW, Barfield RM, Baker J, Banas S, Rabuka D. Exploring the effects of linker composition on site-specifically modified antibody-drug conjugates. Eur J Med Chem 2014; 88:3-9; PMID:25176286; https://doi.org/ 10.1016/j.ejmech.2014.08.062 [DOI] [PubMed] [Google Scholar]
- 40.Drake PM, Albers AE, Baker J, Banas S, Barfield RM, Bhat AS, de Hart GW, Garofalo AW, Holder P, Jones LC, et al.. Aldehyde tag coupled with HIPS chemistry enables the production of ADCs conjugated site-specifically to different antibody regions with distinct in vivo efficacy and PK outcomes. Bioconjug Chem 2014; 25:1331-41; PMID:24924618; https://doi.org/ 10.1021/bc500189z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


